Abstract
Ultrasound-Fluoroscopy fusion is a key step toward intra-operative dosimetry for prostate brachytherapy. We propose a method for intensity-based registration of fluoroscopy to ultrasound that obviates the need for seed segmentation required for seed-based registration. We employ image thresholding and morphological and Gaussian filtering to enhance the image intensity distribution of ultrasound volume. Finally, we find the registration parameters by maximizing a point-to-volume similarity metric. We conducted an experiment on a ground truth phantom and achieved registration error of 0.7±0.2 mm. Our clinical results on 5 patient data sets show excellent visual agreement between the registered seeds and the ultrasound volume with a seed-to-seed registration error of 1.8±0.9 mm. With low registration error, high computational speed and no need for manual seed segmentation, our method is promising for clinical application.
1 Introduction
Low dose rate prostate brachytherapy is a treatment for prostate cancer involving permanent implantation of radioactive seeds of 125I or 103Pd inside the prostate and periprostatic tissue. The seeds are implanted using needles that pass through a guiding template, according to a plan, to create an appropriate dose distribution inside and around the prostate. The procedure is performed under transrectal ultrasound (TRUS) visual guidance. C-arm fluoroscopy images are frequently used for gross visualization of the implant. The treatment quality depends on accurate seed placement which is a challenging task due to problems such as prostate motion and deformation during insertion, needle deflection and edema. Seed misplacement can cause excessive radiation to the healthy tissue that leads to consequent complications, or can result in insufficient radiation to the cancerous prostate (producing “cold spots”) that leads to treatment failure.
Intraoperative dosimetry and planning has the potential to overcome the seed misplacement problem and improve the treatment quality by intermittently calculating the delivered dose and optimizing the treatment plan in order to compensate for the emerging cold spots [1].
Although TRUS enables visualization of prostate boundary, seed segmentation in TRUS is not robust due to significant number of seed-like artifacts (false positives) created by calcifications and needle tracks, and also missing seeds [2]. However, C-arm images can be used to localize the seeds in 3D space (henceforth, called reconstructed seeds) [3, 4]. Spatial registration of reconstructed seeds to the prostate – delineated in the TRUS - can combine the benefits of these two imaging modalities and provide intraoperative dose evaluation.
Lead markers attached to the probe [5] or radio-opaque fiducials attached to the guiding template [6] have been proposed in the past for ultrasound-fluoroscopy registration. However, markers or fiducials need segmentation and their images may overlap with the seeds and complicate the seed reconstruction. Moreover, the radio-opaque fiducial cannot compensate for the prostate motion caused by probe retraction. As a solution, Su et al. [7] and Tutar et al. [8] used point-to-point registration and registered the reconstructed seeds to a set of manually segmented seeds from TRUS images. Manual seed selection in TRUS is tedious as TRUS images are rife with false positives. Fallavollita et al. [9] proposed intensity-based registration of CT or fluoroscopy to TRUS. Their method showed successful registration between CT and TRUS in a phantom study and qualitative agreement between the reconstructed seeds and TRUS for a single patient data set.
In this paper, we introduce a point-to-volume intensity-based rigid registration method with application to prostate brachytherapy. We use image thresholding combined with morphological and Gaussian filtering to enhance the quality of TRUS images – without removing the false positives. Except for manual selection of the region of interest, our algorithm is fully automatic and eliminates the need for seed segmentation.
Our registration results on phantom and patient data sets not only show excellent visual agreement between the reconstructed seeds and TRUS images, but also show quantitative registration errors below clinically acceptable levels. In contrast to the work of Fallavollita et al. [9], we use different preprocessing steps, similarity metric, and optimizer. In addition, our trials on patient data show smaller registration error and faster computational speed. Considering its low registration error, robustness, and high computational speed, our method is suitable for intraoperative dosimetry.
2 Methods
The following work-flow is envisioned for data acquisition for intraoperative dosimetry. The physician acquires several slices of TRUS images by retracting the probe from the prostate base to its apex (41–57 slices with spacing of 1 mm in this work). In a preprocessing step, these slices are processed and compounded into a volume. The probe is fully retracted and several C-arm images are taken from different angles. The seeds are reconstructed from 5–6 images in 3D using available methods such as [3, 4]. The reconstructed seeds comprise a set of 3D points that should be registered to the ultrasound volume.
2.1 Preprocessing
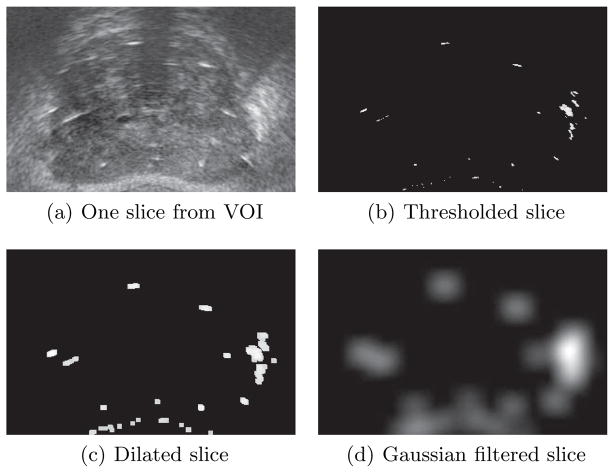
In the preprocessing phase, we follow several steps, as shown in Fig. 1, to enhance the quality of TRUS images. A region of interest is manually selected from a mid-gland slice of the TRUS volume to limit the search region during optimization and increase the likelihood of convergence. This is the only manual intervention needed in our registration method. Corresponding regions are cropped in all the slices and compounded together to create the volume of interest (VOI). Although calcifications and air bubbles trapped in the needle tracks have strong reflections, most of the bright areas in TRUS images belong to seeds. Based on this intuition, we apply a threshold (T) to the images to enhance seed visibility (see Fig. 1(b)). We define:
| (1) |
where, μ and σ are the mean and standard deviation of intensity in the VOI, respectively, and α is a parameter chosen based on the characteristics of the TRUS images (α = 3 in this work). Note that we do not try to remove false positives such as calcifications or air bubbles. The thresholded images are then dilated using a disk structural element (r = 3 pixels in this work) to increase the size of the bright areas (see Fig. 1(c)). Finally, the dilated images are filtered using a Gaussian filter (standard deviation = 10 pixels) in order to spread the bright areas (see Fig. 1(d)). The Gaussian filter is applied to provide a smooth change of intensity in the image in order to increase the capture range and enhance the convergence of the optimization algorithm (details in Sec. 2.2). The image dilation and Gaussian filtering are applied slice by slice. We sub-sample the filtered TRUS slices with a factor of 2 to gain computational speed.
Fig. 1.
Preprocessing steps.
2.2 Intensity-based Point-to-Volume Registration
Since transformation of a set of points is computationally faster than the transformation of a volume, we consider the ultrasound VOI as the fixed volume and register the reconstructed seeds to this volume.
The transformation from the C-arm homogeneous coordinate system to the TRUS homogeneous coordinate system is defined as T (θ, δ), where θ = [θR, θP, θY] represents the roll, pitch and yaw angles, respectively, and δ = [δx, δy, δz] represents the translation along x, y and z axes, respectively. We assume that the x axis is parallel to the horizontal axis of the template, the y axis is parallel to the vertical axis of the template from bottom to the top, and the z axis is parallel to the long axis of the probe from the base of the prostate to its apex. Now, consider a mapping Ψ from every point in the TRUS coordinate system to the indices of the voxel corresponding to that point in the VOI. Therefore, the indices of the voxel corresponding to each reconstructed seed can be calculated as:
| (2) |
where is the coordinates of the nth seed in the C-arm homogeneous coordinate system, vn represents the indices of the voxel corresponding to this point in the VOI (henceforth a seed voxel) and N is the number of implanted seeds. We assume rectangular cuboids with dimensions of (2qi + 1) × (2qj + 1) × (2qk + 1) voxels centered at each seed voxel. The integers qi, qj and qk are calculated so that each cuboid has dimensions of approximately 2×2×6mm3 (slightly thicker and longer than a seed). The similarity metric is evaluated as:
| (3) |
where, I(·) is the VOI intensity at given indices. In other words, the similarity metric is the summation of the intensities of the voxels inside all cuboids around all the seed voxels. This similarity metric quantifies the overlap between the cuboids and bright regions in the VOI and hence, guides the reconstructed seeds toward the center of the bright regions.
We employ the Covariance Matrix Adaptation Evolution Strategy (CMA-ES) [10] in order to maximize the similarity metric. CMA-ES is a stochastic optimization method suitable for nonlinear and non-convex problems and was previously used for registration purpose in [11]. This algorithm samples the search region using a normal distribution, the covariance matrix of which is adapted iteratively. If the bright regions in the VOI are not expanded and spread using image dilation and Gaussian filtering, the changes in the optimization parameters may result in insignificant or no change in the similarity metric since the bright regions are sparsely located in the VOI. Therefore, it is difficult for the optimization algorithm to select the path to improve the similarity metric. However, image dilation and Gaussian filtering provide a smooth and discernible change of similarity metric over the VOI and help the optimization algorithm to hone in on the optimal parameters.
3 Results
3.1 Phantom Study
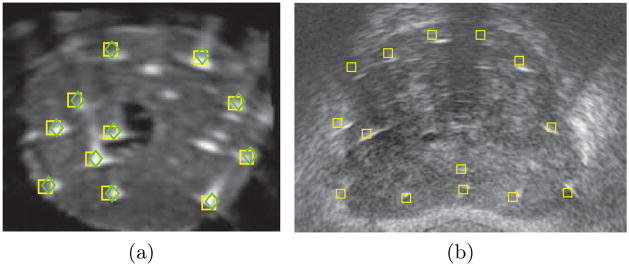
First, we tested our registration method on a phantom implanted with 48 dummy seeds. Both TRUS and CT images of the phantom were acquired. The registration ground truth between the TRUS and CT volumes was established using a tracked probe and fiducials that were attached to the phantom box [9]. The seeds in the CT volume were segmented using thresholding to yield a set of points similar to the outcome of seed reconstruction using C-arm imaging. These seed positions were assumed to be the ground truth. We applied independent perturbations of −10 to 10 mm, with steps of 1 mm, along each axis and rotations of −10° to 10°, with steps of 1°, around each axis to the ground truth seeds and tried the registration algorithm. The registration algorithm successfully converged close to the ground truth for all of the perturbations. The registration error defined as the distance between the registered seeds and the ground truth was on average 0.7 mm (STD = 0.2 mm). The successful convergence of the algorithm to the global optimum despite the applied perturbations shows its robust performance and wide capture range. Figure 2(a) shows the ground truth and registered seeds overlaid on a slice of ultrasound volume.
Fig. 2.
Registered seeds overlaid on TRUS images. (a) Phantom result. Green diamonds are ground truth seeds, yellow squares are registered seeds. (b) Patient result. Yellow squares are registered seeds.
3.2 Study on Clinical Data
We also applied our algorithm to clinical data sets. We collected data from 5 patients implanted with 64 to 105 103Pd seeds. The transverse images – acquired using a BK Pro Focus (BK Medical, Peabody, MA) ultrasound machine - were automatically captured at 1 mm intervals by reading the TRUS stepper position from the encoder while the surgeon continuously retracted the TRUS probe from the prostate base to apex. The transverse slices were processed and compounded into a volume. Several C-arm images were acquired from different angles within a 20° cone around the anterior-posterior axis (AP-axis) using a pre-calibrated GE OEC 9600 mobile C-arm. The C-arm poses were computed using a radio-opaque fiducial [12] that was attached to the template. The C-arm images were preprocessed to correct the image distortion, segment the 2D seed locations and estimate the image poses. The seeds were reconstructed in 3D from 5–6 images by solving a seed matching problem using a dimensionality reduced linear programming algorithm (called REDMAPS) [4].
The registration algorithm was initialized by coinciding the center of mass of the reconstructed seeds with the center of the VOI. The initial rotation angles were provided by the radio-opaque fiducial which was attached to the guiding template. The registration results showed excellent visual agreement between the reconstructed seeds and the TRUS images as it can be seen in Fig. 2(b). Since no ground truth was available at this stage, we manually identified several seeds from the TRUS images (henceforth called the selected seeds) and measured their distances from the closest registered seed. The average and standard deviation of these seed-to-seed distances are reported in Table 1. The registration error had an overall average of 1.8 mm (STD = 0.9 mm). We also reported the mean and standard deviation of the magnitude of the registration error vectors projected along each axis as shown in Table 1. The registration error along the long axis of the probe (the z axis) is the most significant error.
Table 1.
Clinical results, showing the mean and standard deviation of registration error, and mean and standard deviation of absolute error along each axis.
| Abs. Proj. Error (mm)
|
||||||
|---|---|---|---|---|---|---|
| Patient Num. | Total Seeds | Selected Seeds | Reg. Err. (mm) Mean±STD |
x Mean±STD |
y Mean±STD |
z Mean±STD |
| 1 | 81 | 12 | 1.3 ± 0.4 | 0.5 ± 0.3 | 0.7 ± 0.4 | 0.7 ± 0.7 |
| 2 | 76 | 33 | 1.7 ± 0.7 | 0.5 ± 0.5 | 0.6 ± 0.4 | 1.3 ± 0.8 |
| 3 | 90 | 37 | 2.1 ± 1.2 | 0.6 ± 0.5 | 1.3 ± 1.1 | 1.1 ± 0.9 |
| 4 | 64 | 20 | 1.9 ± 1.2 | 0.5 ± 0.3 | 0.4 ± 0.3 | 1.7 ± 1.3 |
| 5 | 105 | 23 | 1.5 ± 0.5 | 0.7 ± 0.4 | 0.7 ± 0.5 | 0.8 ± 0.5 |
|
| ||||||
| Total | 416 | 125 | 1.8 ± 0.9 | 0.6 ± 0.4 | 0.8 ± 0.7 | 1.2 ± 0.9 |
4 Discussion
Su et al. showed that the deviation in D90 (the minimum dose delivered to 90% of the prostate volume) is less than 5% for seed localization errors of less than 2 mm [13]. Our registration errors for 4 of the 5 patients studied are below this limit. Patient 3 had an average registration error slightly greater than 2 mm. This patient had a relatively large prostate that was significantly deformed by the probe pressure. Such patients require a deformable registration between the reconstructed seeds and the TRUS volume. We expect to describe the statistical pattern of deformation from a handful of patients, as the boundary conditions are fairly similar across cases. We also suspect that, in addition to statistical observation, a simple deformation model will explain the primary effect of the probe pressure. Note that we need a registration error smaller than 2 mm [13] for which, a 1D deformation model along the AP-axis can suffice. Alternatively, as recommended in [14], the physician can lower the probe posteriorly in order to decrease the pressure on the prostate. The prostate deformation for the other patients was negligible as the small registration errors along x and y axes confirm.
As it can be seen in Table 1, the error along the z axis is on average 1.2 mm, while the error along x and y axes are on average less than 1 mm. It should be noted that our TRUS volume has a slice spacing of 1 mm along the z axis. In addition, it is difficult to accurately select the center of a 5 mm long seed image which is usually elongated by needle tracks. Therefore our manual seed segmentation can have an error of the same order of magnitude along the z axis, that contributes to the measured registration error.
The algorithm was programmed in MATLAB on a computer with an Intel Core 2 CPU (2 GHz) with 2 GB of RAM. The registration – excluding the manual VOI selection - runs, on average, in approximately 40 s. Our seed-to-seed registration error is less than or equal to the results reported in [7, 8]. Note that [7] and [8] are based on manually selected seeds in TRUS images. Our registration error is also smaller than 2.8 mm reported by Fallavollita et al. [9]. The registration speed, accuracy and robustness are vital in a clinical setting. Our registration method with low registration errors, wide capture range and fast computational speed is promising for clinical application.
We reported our results based on 5 patient data sets. We expect to get similar results for a larger number of patients. However, we will test our algorithm on a statistically more significant number of patient data sets in the future as data collection is currently underway.
5 Conclusions and Future Work
We presented an intensity-based method for registration of ultrasound to fluoroscopy for intraoperative dosimetry in prostate brachytherapy. Our method obviates the need for tedious manual seed segmentation required for seed-based registration. We applied thresholding, and morphological and Gaussian filtering to the TRUS images to enhance the quality of the images and increase the capture range of the algorithm without removing the false positives or identifying the missing seeds. On a ground truth phantom, the algorithm converged to an average registration error of 0.7 mm despite perturbations of −10 to 10 mm along each axis and −10° to 10° around each axis. This demonstrates the wide capture range and robustness of our algorithm. In a clinical study on 5 patient data sets, we achieved average registration error of 1.8 mm in approximately 40 s that shows the feasibility of our method for clinical application.
Extensive tests on more clinical data sets, automatic selection of region of interest and accommodation of prostate deformation as well as sensitivity analysis to the chosen parameters are parts of the future work.
Footnotes
E. Dehghan was supported as an Ontario Ministry of Research and Innovation Fellow. G. Fichtinger was supported as Cancer Care Ontario Research Chair. This work was also supported by National Institutes of Health/National Cancer Institute (NIH/NCI) under Grants 2R44CA099374 and 1R01CA151395.
References
- 1.Polo A, et al. Review of intraoperative imaging and planning techniques in permanent seed prostate brachytherapy. Radiother Oncol. 2010;94(1):12–23. doi: 10.1016/j.radonc.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Han BH, et al. Prostate brachytherapy seed identification on post-implant TRUS images. Med Phys. 2003;30(5):898–900. doi: 10.1118/1.1568976. [DOI] [PubMed] [Google Scholar]
- 3.Dehghan E, et al. Prostate brachytherapy seed reconstruction using c-arm rotation measurement and motion compensation. In: Jiang T, et al., editors. Medical Image Computing and Computer-Assisted Intervention MICCAI 2010. Volume 6361 of LNCS. Springer; Berlin/Heidelberg: 2010. pp. 283–290. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, et al. REDMAPS: Reduced-dimensionality matching for prostate brachytherapy seed reconstruction. IEEE Trans Med Imaging. 2011;30(1):38–51. doi: 10.1109/TMI.2010.2059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todor DA, et al. Intraoperative dynamic dosimetry for prostate implants. Phys Med Biol. 2003;48(9):1153–1171. doi: 10.1088/0031-9155/48/9/306. [DOI] [PubMed] [Google Scholar]
- 6.Jain A, et al. Intra-operative 3D guidance and edema detection in prostate brachytherapy using a non-isocentric C-arm. Med Image Anal. 2010 doi: 10.1016/j.media.2010.07.011. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su Y, et al. Seed localization and TRUS-fluoroscopy fusion for intraoperative prostate brachytherapy dosimetry. Comput Aided Surg. 2007;12(1):25–34. doi: 10.3109/10929080601168239. [DOI] [PubMed] [Google Scholar]
- 8.Tutar IB, et al. Seed-based transrectal ultrasound-fluoroscopy registration method for intraoperative dosimetry analysis of prostate brachytherapy. Med Phys. 2008;35:840–848. doi: 10.1118/1.2836422. [DOI] [PubMed] [Google Scholar]
- 9.Fallavollita P, et al. Registration between ultrasound and fluoroscopy or CT in prostate brachytherapy. Med Phys. 2010;37:2749–2760. doi: 10.1118/1.3416937. [DOI] [PubMed] [Google Scholar]
- 10.Hansen N. The CMA evolution strategy: a comparing review. In: Lozano JA, et al., editors. Towards a new evolutionary computation Advances on estimation of distribution algorithms. Volume 192 of Studies in Fuzziness and Soft Computing. Springer; Berlin/Heidelberg: 2006. pp. 75–102. [Google Scholar]
- 11.Gong RH, Abolmaesumi P. 2D/3D registration with the CMA-ES method. Vol. 6918. SPIE; 2008. p. 69181M. [Google Scholar]
- 12.Jain AK, et al. FTRAC – A robust fluoroscope tracking fiducial. Med Phys. 2005;32(10):3185–3198. doi: 10.1118/1.2047782. [DOI] [PubMed] [Google Scholar]
- 13.Su Y, et al. Dosimetry accuracy as a function of seed localization uncertainty in permanent prostate brachytherapy: increased seed number correlates with less variability in prostate dosimetry. Phys Med Biol. 2007;52(11):3105–3119. doi: 10.1088/0031-9155/52/11/012. [DOI] [PubMed] [Google Scholar]
- 14.Wallner K, Blasko JC, Dattoli M. Prostate Brachytherapy Made Complicated. 2. Smartmedicine Press; 2001. [Google Scholar]