Abstract
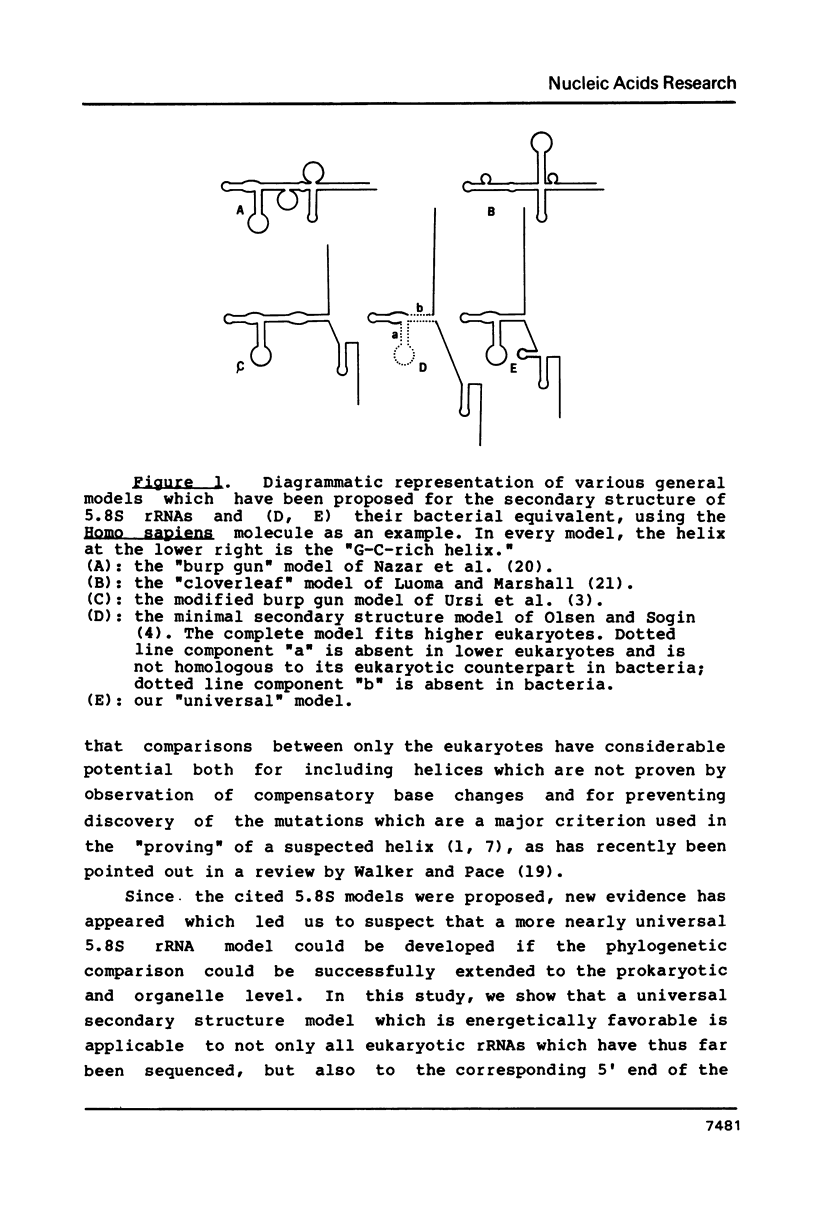
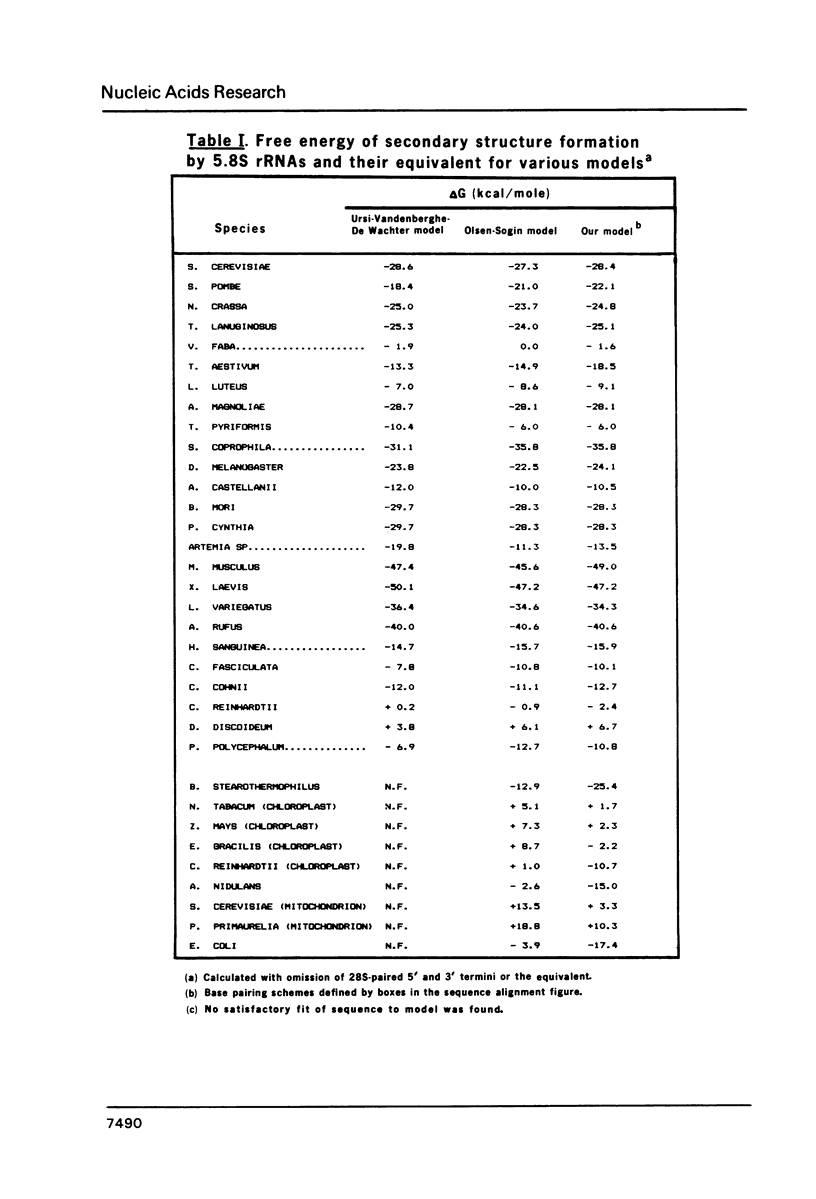
The phylogenetic approach (ref. 1) has been utilized in construction of a universal 5.8S rRNA secondary structure model, in which about 65% of the residues exist in paired structures. Conserved nucleotides primarily occupy unpaired regions. Multiple compensating base changes are demonstrated to be present in each of the five postulated helices, thereby forming a major basis for their proof. The results of chemical and enzymatic probing of 5.8S rRNAs (ref. 13, 32) are fully consistent with, and support, our model. This model differs in several ways from recently proposed 5.8S rRNA models (ref. 3, 4), which are discussed. Each of the helices in our model has been extended to the corresponding bacterial, chloroplast and mitochondrial sequences, which are demonstrated to be positionally conserved by alignment with their eukaryotic counterparts. This extension is also made for the base paired 5.8S/28S contact points, and their prokaryotic and organelle counterparts. The demonstrated identity of secondary structure in these diverse molecules strongly suggests that they perform equivalent functions in prokaryotic and eukaryotic ribosomes.
Full text
PDF





















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. L., Olvera J., Wool I. G. The structure of rat 28S ribosomal ribonucleic acid inferred from the sequence of nucleotides in a gene. Nucleic Acids Res. 1983 Nov 25;11(22):7819–7831. doi: 10.1093/nar/11.22.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981 Jun 25;9(12):2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r105–r133. [PMC free article] [PubMed] [Google Scholar]
- Feng Y. X., Krupp G., Gross H. J. The nucleotide sequence of 5.8S rRNA from the posterior silk gland of the silkworm Philosamia cynthia ricini. Nucleic Acids Res. 1982 Oct 25;10(20):6383–6387. doi: 10.1093/nar/10.20.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Ishikawa H. Primary and secondary structures of Tetrahymena and aphid 5.8S rRNAs: structural features of 5.8S rRNA which interacts with the 28S rRNA containing the hidden break. Nucleic Acids Res. 1982 Sep 11;10(17):5173–5182. doi: 10.1093/nar/10.17.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Kawata Y., Ishikawa H. Primary and secondary structure of 5.8S rRNA from the silkgland of Bombyx mori. Nucleic Acids Res. 1982 Apr 10;10(7):2415–2418. doi: 10.1093/nar/10.7.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev O. I., Nikolaev N., Hadjiolov A. A., Skryabin K. G., Zakharyev V. M., Bayev A. A. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981 Dec 21;9(24):6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf L., Kössel H., Stutz E. Sequencing of 16S--23S spacer in a ribosomal RNA operon of Euglena gracilis chloroplast DNA reveals two tRNA genes. Nature. 1980 Aug 28;286(5776):908–910. doi: 10.1038/286908a0. [DOI] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindenach B. R., Stafford D. W. Nucleotide sequence of the 18S-26S rRNA intergene region of the sea urchin. Nucleic Acids Res. 1984 Feb 10;12(3):1737–1747. doi: 10.1093/nar/12.3.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq B. Sequence homologies between eukaryotic 5.8S rRNA and the 5' end of prokaryotic 23S rRNa: evidences for a common evolutionary origin. Nucleic Acids Res. 1981 Jun 25;9(12):2913–2932. doi: 10.1093/nar/9.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Coding and spacer sequences in the 5.8S-2S region of Sciara coprophila ribosomal DNA. Nucleic Acids Res. 1980 Aug 25;8(16):3565–3573. doi: 10.1093/nar/8.16.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano M., Tomioka N., Sugiura M. The complete nucleotide sequence of a 23S rRNA gene from a blue-green alga, Anacystis nidulans. Gene. 1983 Oct;24(2-3):219–225. doi: 10.1016/0378-1119(83)90082-3. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly P., Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983 Nov 11;11(21):7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F., Renalier M. H. Homology of the 5'-terminal sequence of 28 S rRNA of mouse with yeast and Xenopus. Implication for the secondary structure of the 5.8 S--28 S RNA complex. FEBS Lett. 1982 Apr 19;140(2):193–197. doi: 10.1016/0014-5793(82)80892-2. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'terminal domain. Organisation of the 5.8S-28S rRNA complex. Nucleic Acids Res. 1982 Sep 11;10(17):5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Hassouna N., Bachellerie J. P. Secondary structure of mouse 28S rRNA and general model for the folding of the large rRNA in eukaryotes. Nucleic Acids Res. 1984 May 25;12(10):4259–4279. doi: 10.1093/nar/12.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N. A 5.8 S rRNA-like sequence in prokaryotic 23 S rRNA. FEBS Lett. 1980 Oct 6;119(2):212–214. doi: 10.1016/0014-5793(80)80254-7. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., McCarroll R., Sogin M. L. Secondary structure of the Dictyostelium discoideum small subunit ribosomal RNA. Nucleic Acids Res. 1983 Nov 25;11(22):8037–8049. doi: 10.1093/nar/11.22.8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Sogin M. L. Nucleotide sequence of Dictyostelium discoideum 5.8S ribosomal ribonucleic acid: evolutionary and secondary structural implications. Biochemistry. 1982 May 11;21(10):2335–2343. doi: 10.1021/bi00539a010. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Rushlow K. E., Dodd J. R., Hallick R. B. Euglena gracilis chloroplast ribosomal RNA transcription units. II. Nucleotide sequence homology between the 16 S--23 S ribosomal RNA spacer and the 16 S ribosomal RNA leader regions. J Biol Chem. 1980 Nov 25;255(22):10997–11003. [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Sakaki Y., Takagi Y. Nucleotide sequence of Physarum polycephalum 5.8S rRNA gene and its flanking regions. Nucleic Acids Res. 1982 Apr 10;10(7):2379–2385. doi: 10.1093/nar/10.7.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. A., Walker T. A., Pace N. R. Independent binding sites in mouse 5.8S ribosomal ribonucleic acid for 28S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2329–2335. doi: 10.1021/bi00539a009. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D., Darlix J. L. Composite structure of the chloroplast 23 S ribosomal RNA genes of Chlamydomonas reinhardii. Evolutionary and functional implications. J Mol Biol. 1982 Aug 15;159(3):383–395. doi: 10.1016/0022-2836(82)90290-x. [DOI] [PubMed] [Google Scholar]
- Seilhamer J. J., Cummings D. J. Structure and sequence of the mitochondrial 20S rRNA and tRNA tyr gene of Paramecium primaurelia. Nucleic Acids Res. 1981 Dec 11;9(23):6391–6406. doi: 10.1093/nar/9.23.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Complete DNA sequence coding for the large ribosomal RNA of yeast mitochondria. Nucleic Acids Res. 1983 Jan 25;11(2):339–348. doi: 10.1093/nar/11.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam C. S., Cassidy B., Busch H., Rothblum L. I. Nucleotide sequence of the region between the 18S rRNA sequence and the 28S rRNA sequence of rat ribosomal DNA. Nucleic Acids Res. 1982 Jun 25;10(12):3667–3680. doi: 10.1093/nar/10.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F., Sugiura M. The complete nucleotide sequence of a 23-S rRNA gene from tobacco chloroplasts. Eur J Biochem. 1982 May;124(1):13–19. doi: 10.1111/j.1432-1033.1982.tb05901.x. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Ursi D., Vandenberghe A., De Wachter R. Nucleotide sequences of the 5.8S rRNAs of a mollusc and a porifer, and considerations regarding the secondary structure of 5.8S rRNA and its interaction with 28S rRNA. Nucleic Acids Res. 1983 Nov 25;11(22):8111–8120. doi: 10.1093/nar/11.22.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursi D., Vandenberghe A., De Wachter R. The sequence of the 5.8 S ribosomal RNA of the crustacean Artemia salina. With a proposal for a general secondary structure model for 5.8 S ribosomal RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3517–3530. doi: 10.1093/nar/10.11.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. A., Johnson K. D., Olsen G. J., Peters M. A., Pace N. R. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2320–2329. doi: 10.1021/bi00539a008. [DOI] [PubMed] [Google Scholar]
- Walker T. A., Pace N. R. 5.8S ribosomal RNA. Cell. 1983 Jun;33(2):320–322. doi: 10.1016/0092-8674(83)90413-0. [DOI] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Studies on the secondary structure of 5.8 S rRNA from a thermophile, Thermomyces lanuginosus. J Biol Chem. 1981 Jun 10;256(11):5675–5682. [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Studies on the secondary structure of wheat 5.8 S rRNA. Conformational changes in the A + U-rich stem during ribosome assembly. Eur J Biochem. 1982 Jan;121(2):357–363. doi: 10.1111/j.1432-1033.1982.tb05794.x. [DOI] [PubMed] [Google Scholar]



