Abstract
Defining the sites of action of ethanol on brain proteins is a major prerequisite to understanding the molecular pharmacology of this drug. The main barrier to reaching an atomic-level understanding of alcohol action is the low potency of alcohols, ethanol in particular, which is a reflection of transient, low-affinity interactions with their targets. These mechanisms are difficult or impossible to study with traditional techniques such as radioligand binding or spectroscopy. However, there has been considerable recent progress in combining X-ray crystallography, structural modeling, and site-directed mutagenesis to define the sites and mechanisms of action of ethanol and related alcohols on key brain proteins. We review such insights for several diverse classes of proteins including inwardly rectifying potassium, transient receptor potential, and neurotransmit-ter-gated ion channels, as well as protein kinase C epsilon. Some common themes are beginning to emerge from these proteins, including hydrogen bonding of the hydroxyl group and van der Waals interactions of the methylene groups of ethanol with specific amino acid residues. The resulting binding energy is proposed to facilitate or stabilize low-energy state transitions in the bound proteins, allowing ethanol to act as a “molecular lubricant” for protein function. We discuss evidence for characteristic, discrete alcohol-binding sites on protein targets, as well as evidence that binding to some proteins is better characterized by an interaction region that can accommodate multiple molecules of ethanol.
Keywords: Protein Binding, Protein Structure, Glycine Receptor, TRP Channel, Protein Kinase C, GIRK, IRK, Ligand-Gated Ion Channel
Alcohol (Ethanol) has a wide range of pharmacological effects on the body, with the brain as a primary target. Initial ethanol exposure increases central nervous system (CNS) activity by a variety of measures; conversely, prolonged exposure results in CNS depression, most likely due to increased signaling through GABAergic and other inhibitory pathways (Deitrich et al., 1989). Although behavioral responses to ethanol are well characterized, the molecular mechanisms by which ethanol alters neuronal activity in the brain are poorly understood. One purpose of this review is to provide a brief account of recent findings concerning the interactions of ethanol with prototype brain proteins thought to underlie alcohol actions in the brain, which were presented at the “Alcohol binding sites: The quest for atomic level resolution” symposium at the Research Society on Alcoholism 2010 Annual Meeting.
It is remarkable that ethanol exerts such robust pharmacology with so little volume or distinguishing stereochemistry. Indeed, ethanol is notable for its low potency: the common legal threshold for intoxication is 0.08% volume/volume or about 17 mM in the blood (Harris et al., 2008), and the anesthetic concentration is about 190 mM (Alifimoff et al., 1989). The low potency and simple molecular structure of ethanol led to early suggestions that it might interact nonspecifically with membrane lipids, rather than acting, as do all other psychoactive drugs, at specific protein sites. However, there is now abundant evidence that ethanol can bind to proteins and change their function (Harris et al., 2008).
Along with its familiar effects on human intoxication, low-millimolar alcohol has significant effects on behavioral and electrophysiological measures of CNS function. For example, approximating from the effective dose for immobility implies a binding affinity of about 0.1 M (Eckenhoff and Johansson, 1997), suggesting that relevant protein targets are modulated by equivalent concentrations. This weak presumed binding affinity gives rise to 2 important implications. First, it implies a binding energy of only about 1 kcal/mol (Eckenhoff and Johansson, 1997), such that the effect of ethanol binding on a protein will be only slightly more than that of thermal energy (0.6 kcal/mol at room temperature). Of course, a single binding site may accommodate multiple ethanol molecules, providing somewhat more binding energy. Still, ethanol will likely alter the function of a protein only if it binds to a region critical for a low-energy transition between states— “lubricating” a common functional transition via local structural change. Second, low-affinity binding must impart rapid rates of association or dissociation (or both). To make it easier to “catch” an alcohol molecule in a protein structure, for example by X-ray crystallography, a longer-chain alcohol is often used rather than ethanol. Still, until recently, detailed structures of alcohol-binding sites on proteins were limited to alcohol dehydrogenase (Plapp, 2010) and the Drosophila odorant-binding protein, LUSH (Kruse et al., 2003), neither of which has been implicated in direct actions of ethanol on brain function.
This review focuses on brain proteins that are targets of alcohol action, and for which emerging structural studies are elucidating binding sites for alcohol. Recent dramatic advances in structural characterization of membrane-associated proteins are proving critical to defining mechanisms of low-affinity binding interactions such as those of anesthetics (Vemparala et al., 2010) and, more recently, alcohols. The proteins we have selected for review include potassium (K+) channels (inwardly rectifying K+ channels, IRK and GIRK), an enzyme (protein kinase C epsilon, PKCε), a nonselective cation channel (transient receptor potential vanilloid receptor type 1, TRPV1), and ligandgated ion channels (LGICs) (glycine receptors, GlyRs).
GIRK CHANNELS: STRUCTURAL EVIDENCE FOR ALCOHOL MODULATION VIA A DISCRETE CYTOPLASMIC HYDROPHOBIC POCKET
The critical role of K+ channels in regulating neuronal excitability (Misonou, 2010) has led to substantial interest in their modulation by drugs such as ethanol. In 1999, 2 groups reported for the first time that ethanol activates G protein-gated inwardly rectifying K+ (GIRK or Kir3) channels (Kobayashi et al., 1999; Lewohl et al., 1999). Subsequent work implicated GIRK channels in ethanol-related behaviors (Blednov et al., 2001, 2003; Kozell et al., 2009). GIRK channels belong to a large family of inwardly rectifying K+ channels (Kir1 to Kir7) that conduct K+ ions more efficiently into the cell than out (“inward rectification”). At resting membrane potentials, however, they conduct small outward K+ currents that decrease the excitability of neurons. GIRK channels are typically activated directly by the Gβγ subunit dimers following stimulation of neurotransmitters receptors that couple to Gαi/o G proteins (Logothetis et al., 1987; Reuveny et al., 1994). There are 3 principal GIRK subunits in the brain (GIRK1 to GIRK3); these assemble as heterotetramers of GIRK1/2, 1/3, 2/3, or homotetramers of GIRK2 (Lüscher and Slesinger, 2010).
Inwardly rectifying K+ channels have recently proven fruitful targets for investigating the structural properties of alcohol binding. Although a full-length GIRK structure has yet to be determined at high resolution, various groups have reported crystal structures of the tetrameric cytoplasmic domains of GIRK1, GIRK2, and a related G protein-insensitive inwardly rectifying K+ channel (IRK1 or Kir2.1) (Inanobe et al., 2007; Nishida and MacKinnon, 2002; Pegan et al., 2005). Pegan and colleagues (2006) recently solved the structure (2.0 Å) of an IRK1 variant bound to 4 molecules of an alcohol, 2-methyl-2,4-pentanediol (MPD), via nearly symmetrical hydrophobic pockets. The water-accessible pockets are located on the surface of the cytoplasmic domain, each formed by the N-terminus, the βD-βE domain, and the βL-βM domain of 2 adjacent subunits. Seven amino acids in each pocket interact with MPD. Of these, the hydrophobic side chains of 5 residues point toward MPD and form a hydrophobic pocket. In addition, the hydroxyl groups of MPD can form hydrogen bonds with nearby tyrosine residues and backbone carbonyls, in some cases mediated by resolved water molecules (Pegan et al., 2006).
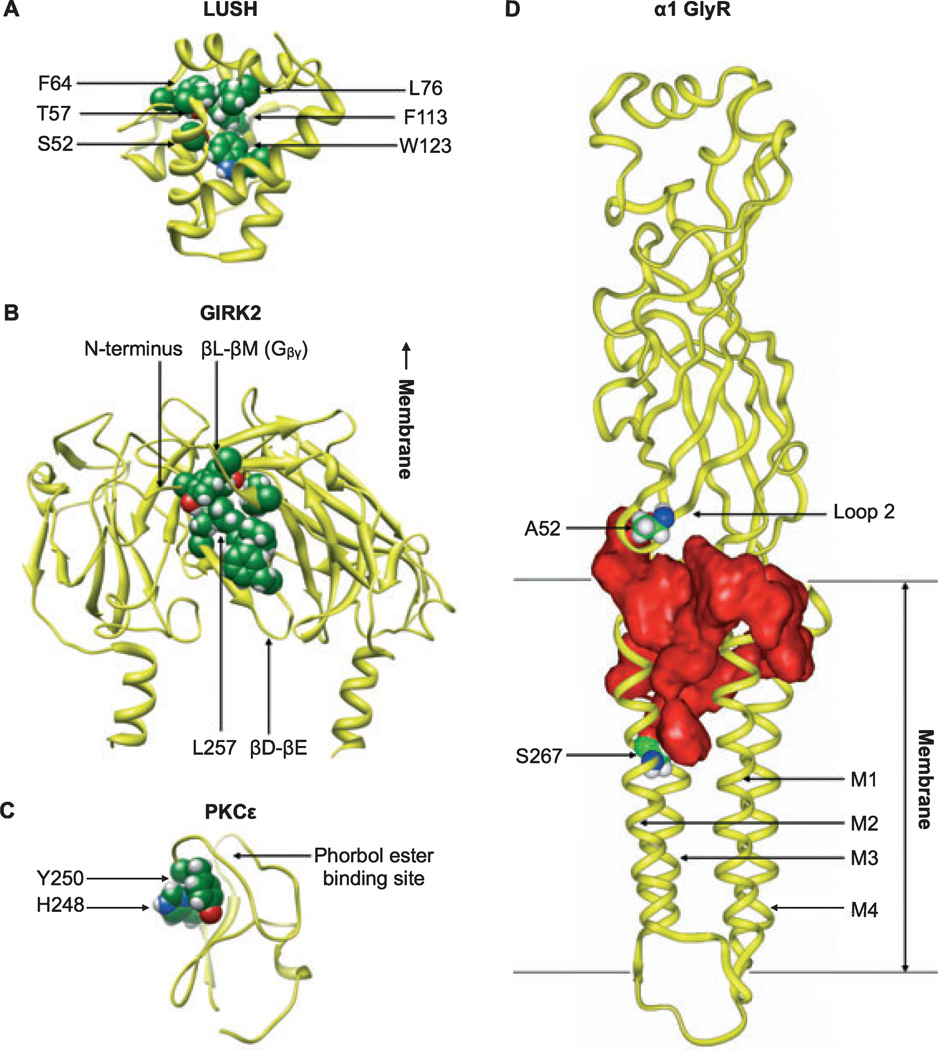
The MPD-bound IRK1 structure is a particularly valuable model, as there are currently no structures of LGICs or voltage-gated ion channels bound to alcohol. Structural models of alcohol binding were previously derived from soluble proteins such as LUSH (Kruse et al., 2003). Intriguingly, the MPD-bound pocket in IRK1 shares structural features with the ethanol-bound pocket of LUSH, which is formed by hydrogen-bonding polar groups surrounded by a cluster of hydrophobic amino acids (Fig. 1A).
Fig. 1.
Sites of alcohol binding to various protein targets. (A) Coordination of n-alcohols (ethanol, n-propanol, or n-butanol) in the odorant-binding protein LUSH (PDB ID 1OOH) includes hydrogen bonding of S52 and T57 with the alcohol hydroxyl group and hydrophobic interactions of F64, L76, F113, and W123 with the alkyl chain. Residues interacting with n-butanol in the crystal structure are shown as spheres. (B) Binding of alcohol to the GIRK2 intracellular domain (PDB ID 2E4F) may involve rearrangement of residues in the N-terminal domain, the βD-βE loop (including the critical alcohol-binding residue L257), and the βL- βM loop (including sites involved in Gβγ binding). Residues interacting with MPD in the IRK1 crystal structure are mapped onto GIRK2 and shown as spheres. For clarity, only the 2 proximal subunits of the tetramer are shown. (C) Amino acid residues in the C1B domain of PKCε (molecular model based on PKCδ, PDB ID 1PTQ) shown by photolabeling and mass spectrometry to interact with alcohol include H248 and Y250, shown as spheres. Residues in the same groove interact with cyclopropanemethanol in a PKCδ co-crystal structure. (D) A large water-filled cavity (red) contained within a single subunit of the homopentameric α1 GlyR (modeled after the nicotinic acetylcholine ˚ receptor, PDB ID 2BG9) is implicated in ethanol binding. Alcohol-binding residues A52 (Loop 2) and S267 (M2), shown as spheres, are separated by ~28 Å but border a contiguous pocket.
Structural similarities between the IRK1 and GIRK2 cytoplasmic domains (Inanobe et al., 2007) led investigators to test whether an alcohol-binding site similar to that in IRK1 is the site for alcohol-mediated activation in GIRK2. The presence of a homologous interaction site was initially supported by the ability of MPD to activate GIRK2 channels, similar to other small n-alcohols: bath application of MPD (100 mM) increased basal GIRK2 current amplitudes, with a dose–response curve between those of ethanol and 1-propanol (Aryal et al., 2009). Structural analysis of the GIRK2 cytoplasmic domain structure (Inanobe et al., 2007) further revealed a hydrophobic pocket, homologous to the MPD-binding site in IRK1, at each of the 4 subunit interfaces in the GIRK2 cytoplasmic domain structure (Fig. 1B). To determine whether the hydrophobic pocket in GIRK2 mediates alcohol activation, investigators examined the effects of changing side-chain volume using small (ala-nine) or large (tryptophan) side chain substitutions at various sites. Of the 4 resulting mutants that produced functional channels activated by ethanol, tryptophan substitution at residue L257 significantly reduced activation by ethanol, MPD, and 1-propanol. In fact, 100 mM MPD no longer activated but rather inhibited basal currents for GIRK2 mutant L257W. This finding indicates that L257, located in the bD-bE ribbon, is a key residue for alcohol-dependent activation of GIRK2 channels. Systematic substitution of GIRK2 residue L257 with amino acids of increasing molecular volume showed that smaller-volume substitutions such as alanine, cysteine, and methionine yielded channels indistinguishable from wild-type, whereas substitution with tyrosine reduced MPD and 1-propanol activation (though ethanol activation was unchanged). For both GIRK2 mutants L257W and L257Y, activation by MPD or 1-propanol decreased for a full range of concentrations (25, 125, or 250 mM), indicating a significant impairment in alcohol sensitivity. Thus, it was suggested that substitutions of bulky residues at position L257 block a functionally important binding site for short-chain n-alco-hols (Aryal et al., 2009).
Because MPD is bound to a hydrophobic pocket in the crystal structure of IRK1, it was further suggested that MPD might modulate IRK1 channels. Indeed, bath application of 100 mM MPD inhibited the basal inwardly rectifying K+ current through IRK1 channels by nearly 50% in a concentration-dependent fashion with an IC50 of 104 ± 23 mM and a Hill coefficient of 0.93±0.02 (n=8). However, tryp-tophan substitutions at various sites in the hydrophobic MPD-binding pocket of IRK1 failed to alter MPD-dependent inhibition (Aryal et al., 2009). Thus, alcohol inhibition in IRK1 channels may operate by an alternative mechanism not involving the hydrophobic pocket.
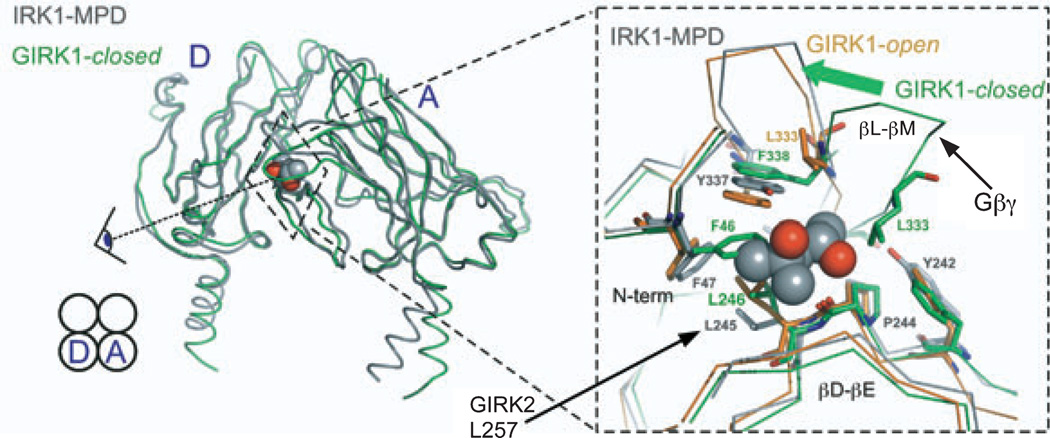
Comparing the high-resolution IRK1-MPD structure (Pegan et al., 2006) with that of a full-length Kir channel chimera, comprising the cytoplasmic domain of GIRK1 and the transmembrane (TM) domain of the bacterial inwardly rectifying K+ channel homolog KirBac1.3 (Nishida et al., 2007), provides some clues into possible mechanisms for etha-nol-dependent channel gating. Two different conformational states of GIRK were described in the chimera: one, considered “closed,” features a constricted cytoplasmic gate too narrow to allow the passage of even a dehydrated K+ ion; in the other, considered “open,” the gate is sufficiently dilated to allow ions to pass (Nishida et al., 2007; Pegan et al., 2005). As shown in Fig. 2, the IRK1-MPD structure aligns well with the putative open-state GIRK1 structure in the region of the hydrophobic alcohol-binding pocket. By contrast, the alignment with the putative closed structure shows striking differences in the hydrophobic alcohol pocket. In particular, amino acid side chains from the N-terminal domain (F46), βD-βE ribbon (L246), and βL-βM ribbon (L333) fill the hydrophobic pocket of the closed state of GIRK1. In the open state, structural rearrangements of these residues create a cavity that could accommodate MPD.
Fig. 2.
Comparison of chimeric KirBac-GIRK1 structures in “closed” (green) and “open” (orange) states with MPD-bound IRK1 (gray). MPD is shown as a CPK model with hydroxyl groups in red. IRK1 is more similar to the putative open- than closed-state GIRK1 structures in the region of the hydrophobic MPD-binding pocket. Note movement of the βL-βM domain (top), implicated previously in Gβγ activation, between the putative open and closed configurations. In the closed state, several amino acid side chains occupy the pocket in which, in IRK1, MPD is bound. Figure modified from Aryal and colleagues (2009).
These mutagenesis data and structural analyses support a working model for ethanol-dependent activation of GIRK channels. At rest, GIRK2 channels undergo infrequent structural rearrangements in the region of the hydrophobic pocket that correlate with open and closed positions of the cytoplas-mic gate (Nishida et al., 2007; Pegan et al., 2005) and pore-lining TM domains (Jin et al., 2002; Sadja et al., 2001; Yi et al., 2001). In a simple 2-state model, ethanol entering the pocket could act as a “lubricant” and facilitate transitions into the open state, either by lowering the activation barrier for channel opening or by stabilizing the open state, and generate ethanol-activated currents. Thus, ethanol may overcome its low-binding energy (1 to 1.5 kcal/mol) by targeting a transition site for GIRK channels that is important for channel activation. Bulky substitutions at positions such as GIRK2 L257, located at the base of the alcohol pocket, would hinder ethanol from filling the pocket. High-resolution structures of GIRK channels in different conformational states in complex with ethanol will be needed to further explore the mechanism of activation. It will be exciting to see in future how the mechanism of ethanol-dependent GIRK modulation compares to other ethanol-sensitive ion channels and proteins.
PROTEIN KINASE C EPSILON: EVIDENCE FOR DIRECT ALCOHOL BINDING TO A REGULATORY PROTEIN
PKCε is a member of the serine/threonine kinase family of signaling proteins that has been implicated in regulating the behavioral effects of alcohol (Newton and Ron, 2007). Hodge and colleagues (1999) observed that PKCε null mice consumed 75% less alcohol than wild type; furthermore, positive allosteric modulators of γ-aminobutyric acid type A receptors (GABAARs) increased alcohol sensitivity in the knockout mice compared to wild type, suggesting that modulation of PKCε may underlie GABAA-mediated behavioral effects of alcohol. Recent behavioral data based on local knockdown (Lesscher et al., 2009) and conditional rescue (Choi et al., 2002) experiments have further established a clear role of PKCε in the regulation of alcohol effects. Although the mechanism of modulation is unknown, at the cellular level ethanol affects translocation of PKCε to the cytosolic compartment (Gordon et al., 1997; Yao et al., 2008), possibly by direct interaction with the enzyme. Slater and colleagues (1993, 1997, 2004) used biochemical experiments to reveal a hydrophobic-binding site for n-alcohols in the C1 domain of the related enzyme PKC alpha, raising the possibility that a similar interaction site may mediate alcohol modulation in PKCε.
Recent studies have pursued identification of the PKCε alcohol-binding site in an in vitro system using photolabeling and X-ray crystallography. Labeling with the photoactive alcohols 3-azibutanol and 3-azioctanol, followed by trypsin digestion and mass spectrometric peptide sequencing, enabled the identification of alcohol-binding sites at positions H248 and Y250 in the PKCε C1B domain, which is also the activator-binding domain (Fig. 1C). Mutation of these positions to alanine reduced alcohol binding (Das et al., 2009). The importance of this region was further validated by a high-resolution (1.3 Å) crystal structure of cyclopropanemethanol complexed with the C1B domain of the closely-related PKC delta (PKCδ) (Das et al., 2007). The alcohol-bound PKCδ crystal structure revealed van der Waals interactions between the methylene groups of the alcohol and those of residue M239. In addition, the alcohol forms hydrogen bonds with the hydroxyl group of Y236 and a nearby water molecule. The Y236 residue in PKCδ is homologous to H248 in PKCε, and is similarly labeled by photoactive alcohols (Das et al., 2004). When Y236 in the PKCδ C1B domain was mutated to phenylalanine, no bound alcohol was observed in the resulting co-crystal structure, highlighting the importance of hydrogen bonding in alcohol– protein interactions (Das et al., 2007).
These biochemical and crystallographic results support the existence of an alcohol-binding site in PKCε involving specific histidine and tyrosine residues. Overall, the binding motif is reminiscent of the IRK1 cytoplasmic domain described above (Fig. 1B), in which the alcohol forms hydrogen bonds directly with 1 tyrosine residue and via a water molecule with another tyrosine residue, and is otherwise stabilized by van der Waals contacts with nearby hydrophobic residues (Pegan et al., 2006). Further structural information will establish the consistency of alcohol-binding sites across related enzymes and other protein targets of ethanol.
TRPV1: A NEWLY PROPOSED ION CHANNEL TARGET FOR ALCOHOL
Interactions between alcohol and cannabinoids in the brain have long been proposed (Vengeliene et al., 2008), but are poorly understood at a molecular level. Ion channels in the TRP superfamily comprise relatively new targets for both cannabinoid (Akopian et al., 2009) and alcohol research (Benedikt et al., 2007). One family member in particular, TRPV1, displays dramatic modulation by ethanol with apparent behavioral consequences in genetically engineered mice (Blednov and Harris, 2009; Ellingson et al., 2008). Recent preliminary investigations challenge several possible mechanisms of action for alcohol on TRPV1, and suggest that this channel comprises a novel paradigm for direct alcohol modulation.
Combined effects of cannabinoids and alcohol on the brain are partly of interest due to their frequently combined recreational use (Poulin and Elliott, 1997) and similar range of physiological effects (Heishman et al., 1988). They also involve some of the same signaling pathways: for example, mice develop cross-tolerance between ethanol and the marijuana component Δ9-tetrahydrocannabinol (THC) (Hungund and Basavarajappa, 2000), and chronic alcohol increases the production of the endogenous cannabinoid anandamide (AEA) (Basavarajappa and Hungund, 1999). Alcohol and cannabinoids also modulate function and/or expression of some of the same receptors (Basavarajappa and Hungund, 2002; Weil et al., 2005). Substantial work remains to be done to characterize these interactions at a molecular level.
Although the most well-known molecular targets for cannabinoids are the G protein-coupled receptors CB1 (Matsuda et al., 1990) and CB2 (Munro et al., 1993), TRP channels have attracted recent attention as additional targets (Akopian et al., 2009). TRP channels are nonselective ion channels that directly gate the influx of cations into the cell in response to diverse stimuli (Bandell et al., 2007). Notably, several TRP channels have been cloned as molecular sensors for temperature and temperature-mimicking chemicals: for example, TRPM8 opens in response to cold or menthol (McKemy et al., 2002), while TRPV1 opens in response to heat or capsaicin (Caterina et al., 1997). TRPA1 has been proposed as a sensor for painfully cold temperatures (Kwan and Corey, 2009) along with pungent chemicals such as those found in mustard (Jordt et al., 2004) and garlic (Macpherson et al., 2005). These channels are also gated or modulated by cannabinoids such as THC (Bisogno et al., 2001; De Petrocellis et al., 2008) and AEA (Zygmunt et al., 1999). Of course, extreme temperatures and phytochemicals are primarily encountered in peripheral tissues; however, increasing evidence supports the expression of TRP channels, especially TRPV1, in brain (Kauer and Gibson, 2009), where they may play roles in synaptic transmission. In the brain, endogenous cannabinoids such as AEA are likely to be the predominant agonists for TRP channel activation (van der Stelt and Di Marzo, 2004).
Several of the same TRP channels that are gated by cannabinoids are also modulated by alcohols. TRPM8 is inhibited by high concentrations of ethanol (Weil et al., 2005), a phenomenon that involves the PIP2-binding domain in the channel’s C-terminus (Benedikt et al., 2007). Conversely, n-alcohols and volatile anesthetics activate TRPA1 (Matta et al., 2008) and potentiate TRPV1 (Cornett et al., 2008; Trevisani et al., 2002). Indeed, potentiation of TRPV1 by ethanol may underlie the burning sensation that can accompany strong alcoholic drinks (Trevisani et al., 2002).
Recent behavioral evidence further substantiates a role for TRPV1 in alcohol effects. TRPV1 null mutant mice were protected from aversive alcohol taste (Ellingson et al., 2008); moreover, they were less sensitive than wild-type C57BL/6J mice to behavioral measures of alcohol-induced intoxication (Blednov and Harris, 2009). This response is specific to TRPV1, as injecting wild-type mice with a TRPV1 antagonist reproduced the knockouts’ resistance to intoxication, while injecting capsaicin made them more sensitive to alcohol (Blednov and Harris, 2009). Thus TRPV1 appears to mediate intoxicating effects of alcohol in the brain, most likely by potentiation of AEA signaling (Blednov and Harris, 2009).
Elucidation of TRPV1 modulation has been aided by recent structural data. Binding sites for ligands such as capsaicin, AEA, and protons (H+), initially defined by biochemical and physiological experiments (Jordt and Julius, 2002; Wang et al., 2010; Welch et al., 2000), can now be placed in an increasingly well-defined structural context. Cryo-electron microscopy (EM) of TRPV1 at 19 Å resolution (Moiseenkova-Bell et al., 2008) shows it comprises a compact TM domain with large intracellular N- and C-termini. The TM domain is homologous to that of the Kv1.2 channel, the structure of which is known (Long et al., 2005) and fits the EM model. Much of the N-terminal domain consists of an ankyrin repeat motif, whose crystal structure reveals sites for interaction with adenosine triphosphate and calmodulin (CaM) (Lishko et al., 2007). Although the structure of the TRPV1 C-terminus is unknown, models based on the tumor suppressor protein FHIT (Vlachová et al., 2003) and on the cyclic nucleotide-gated channel C-terminus (Garcıa-Sanz et al., 2004) have been proposed. Full-length TRPV1 was recently expressed and purified in insect cells (Korepanova et al., 2009), suggesting that high-resolution structural data may soon emerge. Still, a detailed picture of the diverse factors involved in TRPV1 modulation remains elusive. In particular, sites of action for alcohol are poorly understood, and depend on further structural and functional investigation.
There are several possible mechanisms of alcohol modulation of TRPV1, including indirect effects on kinases, phosphoinositides, or CaM, or direct binding to the channel. Preliminary investigations of some of these mechanisms have been performed by 2-electrode voltage clamp electrophysiology in Xenopus laevis oocytes (Wagner et al., 2000), a system previously shown to efficiently produce functional TRPV1 currents (Caterina et al., 1997). Calcium-free buffers containing AEA or low pH induced robust, reversible currents in TRPV1-injected oocytes with sigmoidal concentration dependence. Consistent with previous results in neurons and mammalian cells (Trevisani et al., 2002), high (50 to 200 mM) concentrations of ethanol potentiated activation of TRPV1 in oocytes by both AEA and H+, with 200 mM ethanol potentiating about 2-fold (RJ Howard and RA Harris, unpublished data). The consistency of this result indicates that ethanol potentiation of AEA activation is not an artifact of ligand solubilization (Glaser et al., 2005) or interaction with intracellu-lar factors (Premkumar and Ahern, 2000), but occurs independent of the mechanism of activation.
One possible mechanism for alcohol modulation of TRPV1 involves phosphorylation by PKC, which sensitizes the channel (Bhave et al., 2003). Indeed, ablation of PKC phosphorylation by treatment with the nonspecific kinase inhibitor staurosporine, or by mutation of serine residues S502 and S800 to alanine, was recently shown to completely inhibit PKC enhancement of TRPV1 activity (Studer and McNaugh-ton, 2010). Given the well-documented association between PKC and alcohol abuse (Newton and Ron, 2007, and elsewhere in this review), it is possible that alcohol modulates TRPV1 indirectly via kinase modulation. However, either treatment with staurosporine or mutation of phosphorylation sites (S502A/S800A) failed to remove ethanol potentiation of TRPV1 (RJ Howard and RA Harris, unpublished data). Thus, alcohol modulation is unlikely to depend on PKC phosphorylation.
There is controversial evidence that PIP2, a component of membrane lipids, inhibits TRPV1 via an intracellular motif (Lukacs et al., 2007; Prescott and Julius, 2003). This mechanism seemed a particularly promising target for alcohol modulation, given the parallel modulation of the related channel TRPM8. As described above, ethanol disrupts PIP2-mediated enhancement of TRPM8 currents, thereby inhibiting TRPM8 function. Accordingly, depleting oocytes of PIP2 (e.g., with the kinase inhibitor wortmannin) decreases ethanol inhibition of TRPM8 (Benedikt et al., 2007). Vetter and colleagues (2008) recently suggested that wortmannin acts similarly on ethanol potentiation of TRPV1, though the demonstrated effect was small and in a different experimental system. In oo-cytes, treatment with wortmannin inhibited rather than sensitized TRPV1 currents; moreover, wortmannin did not decrease subsequent potentiation by ethanol. Deletion of a 16-residue region implicated in PIP2 inhibition (ΔL777 to ΔL792) (Prescott and Julius, 2003), or indeed deletion of 16 residues on either side of this region (ΔE761 to ΔS776, ΔV793 to ΔQ808), also failed to significantly reduce ethanol potentiation (RJ Howard and RA Harris, unpublished data). These findings are more consistent with an alternative, well-supported model in which PIP2 primarily serves to potentiate, rather than inhibit, TRPV1 activity, probably via a distinct structural motif close to the plasma membrane (Lukacs et al., 2007; Stein et al., 2006), and in which ethanol does not disrupt modulation of the channel.
Another important modulator of TRPV1 is the calcium-binding protein CaM, which interacts with 1 or more intracellular sites (Lishko et al., 2007; Numazaki et al., 2003). Because CaM binding mediates calcium-dependent desensitization of TRPV1 (Rosenbaum et al., 2004), disrupting this interaction could enhance channel function. An arginine residue in the putative C-terminal CaM-binding motif was recently shown to be essential for CaM binding (Grycova et al., 2008); however, mutating this residue to alanine did not reduce ethanol potentiation (RJ Howard and RA Harris, unpublished data). In fact, the CaM-binding domain overlaps with the putative PIP2-inhibition domain shown above to have no effect on ethanol modulation. The lack of involvement of putative CaM-binding sites, along with the robust effect of ethanol in calcium-free oocyte electrophysiology experiments, renders a model in which alcohol potentiates TRPV1 by disrupting CaM binding unlikely.
Lack of evidence for an indirect mechanism of alcohol modulation of TRPV1 leaves open the important possibility that TRPV1, like other protein targets described in this review, may bind alcohol directly to modulate function. As described above, serial deletion experiments have ruled out a considerable region of the C-terminus, and may help identify a direct site of action for alcohol on TRPV1. Recent advances in identifying sites of alcohol action in other protein targets, such as kinases, GIRK channels, and LGICs, further suggest that identification of similar sites in TRPV1 will benefit greatly from structural information. Such insight will be valuable to alcohol as well as TRP channel research, and may present a novel paradigm for alcohol modulation of an ion channel involved in intoxication.
GLYCINE RECEPTORS: DEFINING ALCOHOL BINDING THROUGH MUTAGENESIS AND MODELING
Building evidence supports a role for GlyRs in mediating the effects of ethanol. Indeed, behaviorally relevant concentrations of ethanol positively modulate GlyR function in a variety of brain and spinal cord preparations (Aguayo and Pancetti, 1994; Eggers et al., 2000; Engblom and Akerman, 1991; McCool et al., 2003; Molander et al., 2007). The molecular sites and mechanisms that initiate ethanol action in GlyRs represent an active area of investigation extensively covered in a recent review (Perkins et al., 2010).
Early molecular investigations focused on ethanol-sensitive regions within the TM domains of GlyRs, comprising helices M1 to M4 in each subunit. Chimeric and mutagenic strategies demonstrated that amino acid residues S267 in M2 and A288 in M3 of the α1 GlyR were critical for allosteric modulation by ethanol (Mihic et al., 1997). Moreover, the molecular volume of the residue at position 267 in GlyRs determined alcohol cutoff (Wick et al., 1998) as well as the sensitivity and qualitative response (potentiating vs. inhibitory) to ethanol. Systematic substitution of serine at position 267 with each of the other 19 amino acids revealed an inverse correlation between molecular volume at this position and ethanol effects (Ye et al., 1998). Subsequently, use of the substituted cysteine accessibility method (SCAM) (Karlin and Akabas, 1998) in combination with the anesthetic-like reagent propyl methan-ethiosulfonate (PMTS) demonstrated that labeling of α1 GlyR mutant S267C produced irreversible alcohol-like potentiation and reduced alcohol cutoff (Mascia et al., 2000). This result provided further evidence that position 267 in the GlyR TM domain contributes to an ethanol “binding/action” pocket. Potentiation by alcohols was also altered by mutations at position A288 (Yamakura et al., 1999) and proximal sites in M1 (I229) (Lobo et al., 2004a) and M4 (W407, Y410) (Lobo et al., 2006). These and other studies provided structural context for these findings by characterizing the orientation of TM domain residues and their importance in channel function (Lobo and Harris, 2005; Lobo et al., 2004a,b, 2006, 2008; McCracken et al., 2010).
An additional area of recent focus in GlyR alcohol research concerns the extracellular (EC) domain. Mascia and colleagues (1996) found that mutation of position 52 in the EC domain of the α1 GlyR from alanine to serine (A52S) significantly changed the sensitivity of the receptor to low ethanol concentrations. Further studies of the involvement of the GlyR EC domain in ethanol action have taken advantage of the pharmacological phenomenon that increased atmospheric pressure (pressure) acts as a direct mechanistic antagonist to ethanol. Pressures up to 12 times normal antagonized the behavioral and biochemical actions of ethanol (Alkana and Malcolm, 1981; Alkana et al., 1992; Bejanian et al., 1993; Davies and Alkana, 1998, 2001) without altering ethanol pharmacokinetics, behavioral or electrophysiological baselines, or CNS excitation (Davies et al., 1994, 1999; Syapin et al., 1996). Thus, if a GlyR site is involved in ethanol modulation, mutating it should alter the effects of pressure as well as ethanol. In agreement with this prediction, pressure antagonized ethanol effects in the wild-type α1 GlyR (Davies et al., 2003), but not in the A52S mutant (Davies et al., 2004). Interestingly, pressure did not antagonize ethanol effects in the α2 GlyR, which has a threonine (T59) instead of alanine at the position homologous to α1 A52 (Davies et al., 2004; Perkins et al., 2008). Given the otherwise high homology between α1 and α2 GlyRs, these findings support the involvement of α1 A52 as a target for ethanol action, and suggest that limited amino acid differences between α1 and α2 GlyRs in the EC domain contribute to differences in sensitivity, both to etha-nol and to pressure antagonism of ]ethanol.
The use of SCAM techniques with PMTS labeling facilitated incorporation of TM and EC domain sites into a structural model of alcohol modulation of the GlyR. Exposure to PMTS caused significant irreversible alcohol-like potentiation and decreased alcohol cutoff in the A52C α1 GlyR, implicating alcohol binding at this site in GlyR modulation (Crawford et al., 2007). Furthermore, PMTS treatment of the double mutant A52C/S267C α1 GlyR decreased alcohol cutoff even more than either substitution alone, consistent with the notion that EC domain position 52 and TM domain position 267 participate in forming a single interaction pocket for alcohol action (Crawford et al., 2007) (Fig. 1D). Notably, the key determinant of sensitivity to both ethanol and pressure antagonism of ethanol was the polarity of the EC domain site (α1 52 or α2 59) (Perkins etal., 2008). This finding contrasts with the TM domain site (α1 267) where, as described above, molecular volume rather than polarity determined ethanol sensitivity (Ye et al., 1998). Thus, different physicochemical properties influence ethanol action at EC and TM domain sites, despite their participation in a common interaction pocket.
The demonstrated role of α1 GlyR position 52 in ethanol sensitivity brought focus to the role of Loop 2, the EC domain region containing the ethanol sensitivity site, in function and modulation of the GlyR and other receptors. As an alternative template for alcohol effects, investigators took advantage of recent evidence for high ethanol sensitivity in δ subunit-containing GABAARs. Although not without controversy (Baur et al., 2009; Borghese et al., 2006) (for review, see Borghese and Harris, 2007), δ subunit-containing GABAARs (e.g., α4β3δ) were reported to be sensitive to ethanol concentrations as low as 1 to 3 mM (Hanchar et al., 2005, 2006; Sundstrom-Poromaa et al., 2002; Wei et al., 2004). Perkins and colleagues (2009) predicted that, if the Loop 2 sequence plays a key role in determining ethanol sensitivity, then mutating Loop 2 of the α1 GlyR to the homologous sequence in the high-sensitivity δ GABAAR would increase ethanol effects. As predicted, substituting the δGABAAR Loop 2 into the α1 GlyR reduced the threshold for ethanol response from 30 to 1 mM or less. In addition, the mutation significantly increased the degree of ethanol potentiation at the low concentrations tested (1 to 30 mM) (Perkins et al., 2009). Taken together, these findings indicate that the structure of Loop 2 in the EC domain can have a marked influence on ethanol sensitivity. This finding led to new models that suggest physi-cochemical mechanisms by which ethanol modulates receptor responses to agonists, as briefly described below (see also Perkins et al., 2009).
Recent studies have begun to investigate the role of charge at position 52 in Loop 2 of the α1 GlyR in sensitivity to and pressure antagonism of ethanol. Substitution of A52 with charged residues, as with polar uncharged residues (Perkins et al., 2008), abolished pressure antagonism of ethanol. Ethanol sensitivity, on the other hand, did not vary directly with charge: effects of A52 substitutions did not depend on the presence or absence of charge per se, and were not influenced by the nature (positive or negative) of the charge. Instead (DI Perkins, JR Trudell, DL Davies, and RL Alkana, in preparation), the effects of A52 substitutions appear to be influenced by subtle structural differences in amino acid side chains. Taken together, these findings suggest that the structural bases for ethanol action and pressure antagonism at position 52 in Loop 2 of GlyRs may be different (DI Perkins, JR Trudell, DL Davies, and RL Alkana, in preparation).
The growing field of LGIC research has firmly established the importance of multiple sites of action in ethanol modulation of GlyRs, involving regions of both the EC and TM domains. Recent findings described above indicate that the structure of Loop 2 in the EC domain, including position 52 in the α1 GlyR, influences ethanol sensitivity. Continuing investigations will establish the physicochemical properties and mechanistic outcomes of alcohol interaction with various regions of these receptors.
FUTURE DIRECTIONS: MODELING STRUCTURAL AND FUNCTIONAL OUTCOMES OF ALCOHOL BINDING
The preceding sections describe recent progress in identifying alcohol-binding sites in brain ion channels and signaling proteins. The next key question to be addressed is: what happens to the protein when ethanol binds to the alcohol site? The answer is likely to be complicated, as a large number of high-resolution structures have revealed surprisingly small changes in tertiary structure following ligand binding. For example, high-resolution structures are available for the acetylcholine-binding protein (AChBP), a prototype structure for the ligand-binding domain of the LGIC superfamily (Brejc et al., 2001), bound to ligands as small as nicotine (Celie et al., 2004) and as large as cobratoxin (Bertaccini et al., 2008). While binding of cobratoxin to the acetylcholine-binding site in AChBP caused little change in the static structure, it did change the axially symmetric receptor motions associated with channel gating (Bertaccini et al., 2008). A similar mechanism may account for the effects of alcohol binding in receptors discussed in this review.
The LGICs are among the best-characterized model systems for alcohol binding, and share several critical features. As described above, binding energy derived from hydrogen bonding and van der Waals interactions between a protein and alcohol is proposed to “lubricate” a functional transition, either by lowering an energy barrier or stabilizing the facilitated state. However, alcohol imparts weak binding energy and is likely to be promiscuous (Harris et al., 2008). Binding of alcohol to LGICs may be facilitated by displacement of water molecules from partially hydrated internal cavities (Trudell and Harris, 2004). Still, even with the entropic contribution of liberating water molecules, the free energy available for alcohol to exert its effects may be small. Notably, thermal energy accounts for all major functional transitions in LGICs, including binding of agonist, gating of the ion channel, unbinding of agonist, and binding and unbinding of alcohol. In contrast, external driving forces such as light absorption and TM electrostatic gradients aid the activation of proteins such as rhodopsin (Pepe, 2001) and voltage-gated K+ channels (Bezanilla, 2008).
Because of these considerations, researchers must search for pathways along which the amino acid residues of the receptors can move without encountering high free energy barriers. One way to achieve a low free energy pathway is to describe a collective motion where all residues in a receptor move essentially simultaneously. For example, as residues in a particular ion channel subunit rearrange from a closed to an open state, residues throughout that subunit may rearrange to accommodate the new local conformation. By doing so, they can avoid van der Waals collisions with neighboring residues. Such van der Waals collisions are unforgiving: while a hydrogen bond could contribute 3 to 5 kcal/mol of binding energy, a collision could subtract hundreds of kcal/mol. The concept of a collective motion could include the conformational “wave” mode of activation postulated by Grosman and colleagues (2000) as well as the initial deformation of a single subunit described by Nury and colleagues (2010).
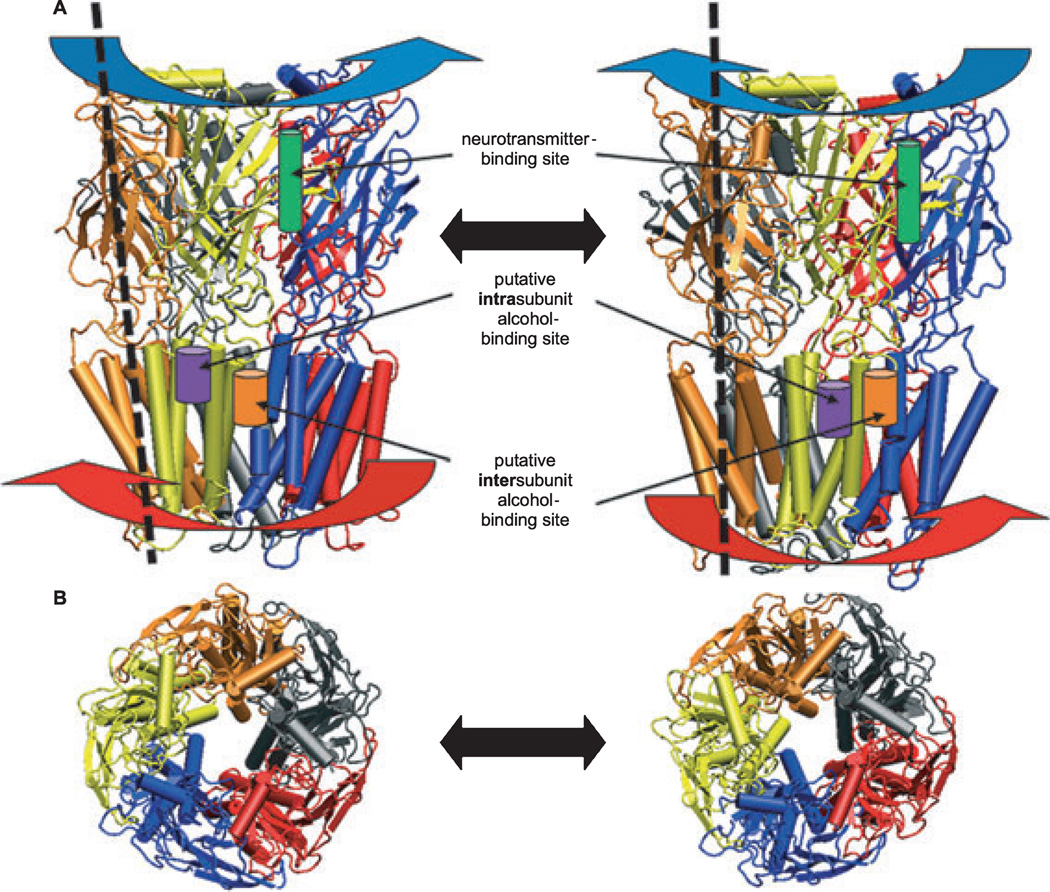
One way to search for a low free energy pathway is to perform elastic network calculations (Lindahl et al., 2006). Figure 3 shows 1 high-amplitude/low-frequency motion in a model of the α1 GlyR suspended in a fully hydrated lipid bilayer (Bertaccini et al., 2010a). Normal mode analysis of this model revealed an iris-like gating motion, in which the EC and TM domains twist in opposite directions. These motions create a harmonic spiral motion that results in channel gating (Bertaccini et al., 2007, 2010a). Similar twisting motions were demonstrated in models of the α7 nicotinic acetylcholine receptor (Taly et al., 2005) and the prokaryotic ion channel GLIC (Zhu and Hummer, 2009).
Fig. 3.
Models of 2 states of a ligand-gated ion channel with pentamer subunits colored individually (orange, gray, red, blue, yellow) illustrating “wringing” or iris-like gating motions. (A) Lateral view from the plane of the membrane. The ligand-binding extracellular (EC) domain (blue arrows) and transmembrane (TM) domain (red arrows) twist in opposite directions. The dotted lines illustrate tilted axes of spiral motion of 1 subunit in each state. These motions create a harmonic spiral motion that results in channel gating. One of 5 native agonist ligand-binding sites in the EC domain is illustrated as a cylinder (green) between 2 subunits. Putative binding sites for alcohols and anesthetics are illustrated as cylinders at intrasubunit (purple) and intersubunit (orange) helix interfaces within the TM domain. Note that, although only 1 example of each binding site is shown, the 5-fold symmetry or pseudosymmetry of the channel may create as many as 5 such sites either within or between subunits. (B) Top view of the ion pore from the EC side. Figure modified from Bertaccini and colleagues (2010a).
A second way to seek low free energy pathways for the gating transition is to use molecular dynamics simulations. There have been several successful applications of molecular dynamics to the study of alcohol (Cheng et al., 2008) and anesthetic (Liu et al., 2009, 2010; Mowrey et al., 2010) effects on LGICs. Tools such as the molecular simulation package GROMACS (Hess et al., 2008; van der Spoel et al., 2005) allow long time scale simulations of alcohol binding to receptors, even in fully-hydrated phospholipid bilayers (Murail et al., 2011). However, even a 1-microsecond simulation using 128 cores on a large Linux cluster presently takes about 6 weeks, whereas a simulation of 1 millisecond is required to observe the effects of alcohol on the gating transition (Chakrapani and Auer-bach, 2005). In future, new methods may dramatically improve simulation times. For example, coarse-grained methods, such as elastic networks, may be used to generate reasonable pathways and then divide them into segments amenable to detailed molecular dynamics (Arora and Brooks, 2007). Alternately, a biasing force may be used to move elements of a model in specific directions to distinct “open” and “closed” states (Nury et al., 2010). The Climber algorithm developed by Weiss and Levitt (2009) may also prove useful in generating sub states along a low-energy pathway.
Functional experiments that uncouple the effects of agonist versus alcohol binding on channel opening may further simplify computational modeling. For example, the finding that the D97R α1 GlyR mutant is spontaneously active provided a means to study alcohol effects independent of glycine binding (Beckstead et al., 2002). Molecular models of the α1 GlyR helped to explain how the mutation D97R causes spontaneous channel gating and allowed prediction of residues in proximity to position 97 that could interact with it electrostatically (Todorovic et al., 2010). Mutagenesis and electrophysiologi-cal analysis of the proposed interacting sites revealed that reversing the charge pair in the mutant D97R/R119E stabilized the channels in their closed states. Models of these mutations will aid future molecular dynamics simulations of alcohol’s effects.
The selective effects of alcohols and anesthetics on various LGICs (Belelli et al., 1999; Flood et al., 1997; Krasowski and Harrison, 1999; Peoples and Weight, 1999) imply the existence of binding sites that differ in specificity by virtue of their molecular properties. Common features of alcohol-binding sites are evident in Fig. 1A–C, where the alcohol-binding regions of LUSH, GIRK2, and PKCe are similarly characterized by shallow hydrophobic cavities on the cytoplasmic protein surface, surrounding a limited number of hydrogen-bonding groups. However, although we have referred to “alcohol-binding sites” as though they comprise discrete, well-defined structures, at least 3 observations suggest that their properties can be more variable and complex. First, alcohols can have receptor-specific effects, for example modulating closely related GlyRs and GABAARs receptors in different ways (reviewed in Perkins et al., 2010). Second, similar alcohols can have different effects on the same receptor: for example, ethanol and octanol have opposite modulatory effects on the nicotinic acetylcholine receptor (Borghese et al., 2003). Recent evidence also shows opposite effects of ethanol and hexanol on ATP-gated purinergic P2×4 receptors (P2×4Rs) (Asatryan et al., 2010); this work enabled generation of the first molecular model of an ethanol-binding site in P2XRs, and demonstrated that diverse effects of closely related alcohols are a consistent theme among LGICs. Third, molecular dynamics simulations of LGIC alcohol binding show multiple alcohol molecules fluctuating between various preferred orientations in a single protein cavity (Cheng et al., 2008). The large water-accessible cavity in Fig. 1D associated with alcohol modulation of the GlyR illustrates one such alternative model of alcohol binding. Allowing models to accommodate multiple alcohol molecules in a range of orientations may help deal with the complexity of LGICs and rationalize the spectrum of in vivo responses to alcohol.
Recent computational research has made substantial progress in modeling effects of alcohol on GlyRs and GABAARs (Bertaccini et al., 2010b; Harris et al., 2008; Perkins et al., 2010). Emerging structural templates such as GIRK2 (Aryal et al., 2009), PKCε (Das et al., 2009), and P2XRs (Asatryan et al., 2010) continue to enrich studies of alcohol binding and modulation, and in future may inform new targets such as TRP channels. Further integration of structural, functional, and computational approaches promises to elucidate in detail the molecular mechanisms underlying alcohol effects on the brain.
ACKNOWLEDGMENTS
This work was supported by NIH/NIAAA grants F31 AA017042 (P Aryal), R21 AA016140-02 (JD), R01 AA013890 and R01 AA013922 (DLD), T32 AA007471 (R Gonzales, RJH), R01 AA06399 (RAH, RJH), R01 AA013378 (RAH, JRT), and R01 AA018734 (PAS); NIH grant R01 GM056653 (S Choe); and the University of Southern California School of Pharmacy (DLD). Molecular representations were made using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH grant P41 RR01081); the PyMOL Molecular Graphics System, Schröd inger, LLC, New York, NY; and Discovery Studio 2.5, Accelrys Inc., San Diego, CA. We thank Dr. Edward J. Bertaccini for preparing Fig. 3 and for helpful discussions.
Footnotes
This review summarizes the proceedings of a symposium (“Alcohol binding sites: The quest for atomic level resolution”) organized and chaired by R. Adron Harris and James R. Trudell and presented at the 2010 annual meeting of the Research Society on Alcoholism in San Antonio, Texas.
Contributor Information
Rebecca J. Howard, The Waggoner Center for Alcohol and Addiction Research, The University of Texas at Austin, Austin, Texas
Paul A. Slesinger, The Salk Institute, La Jolla, California
Daryl L. Davies, Titus Family Department of Clinical Pharmacy and Pharmaceutical Economics and Policy, University of Southern California, Los Angeles, California
Joydip Das, Department of Pharmacological and Pharmaceutical Sciences, University of Houston, Houston, Texas.
James R. Trudell, Department of Anesthesia, Stanford University, Stanford, California
R. Adron Harris, The Waggoner Center for Alcohol and Addiction Research, The University of Texas at Austin, Austin, Texas.
REFERENCES
- Aguayo LG, Pancetti FC. Ethanol modulation of the gamma-aminobutyric acidA- and glycine-activated Cl- current in cultured mouse neurons. J Pharmacol Exp Ther. 1994;270(1):61–69. [PubMed] [Google Scholar]
- Akopian AN, Ruparel NB, Jeske NA, Patwardhan A, Hargreaves KM. Role of ionotropic cannabinoid receptors in peripheral antinociception and antihyperalgesia. Trends Pharmacol Sci. 2009;30(2):79–84. doi: 10.1016/j.tips.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifimoff JK, Firestone LL, Miller KW. Anaesthetic potencies of primary alkanols: implications for the molecular dimensions of the anaesthetic site. Br J Pharmacol. 1989;96(1):9–16. doi: 10.1111/j.1476-5381.1989.tb11777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkana RL, Jones BL, Palomares ML, Crabbe JC, Bejanian M. Mechanism of low level hyperbaric ethanol antagonism: specificity versus other drugs. Alcohol Alcohol. 1992;(Suppl 1):63. [Google Scholar]
- Alkana RL, Malcolm RD. Low-level hyperbaric ethanol antagonism in mice. Dose and pressure response. Pharmacology. 1981;22(3):199–208. doi: 10.1159/000137491. [DOI] [PubMed] [Google Scholar]
- Arora K, Brooks CL. Large-scale allosteric conformational transitions of adenylate kinase appear to involve a population-shift mechanism. Proc Natl Acad Sci USA. 2007;104(47):18496–18501. doi: 10.1073/pnas.0706443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S, Slesinger PA. A discrete alcohol pocket involved in GIRK channel activation. Nat Neurosci. 2009;12(8):988–995. doi: 10.1038/nn.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asatryan L, Popova M, Perkins D, Trudell JR, Alkana RL, Davies DL. Ivermectin antagonizes ethanol inhibition in purinergic P2×4 receptors. J Pharmacol Exp Ther. 2010;334(3):720–728. doi: 10.1124/jpet.110.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandell M, Macpherson LJ, Patapoutian A. From chills to chilis: mechanisms for thermosensation and chemesthesis via thermoTRPs. Curr Opin Neurobiol. 2007;17(4):490–497. doi: 10.1016/j.conb.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa BS, Hungund BL. Chronic ethanol increases the canna-binoid receptor agonist anandamide and its precursor N-arachidonoylphos-phatidylethanolamine in SK-N-SH cells. J Neurochem. 1999;72(2):522–528. doi: 10.1046/j.1471-4159.1999.0720522.x. [DOI] [PubMed] [Google Scholar]
- Basavarajappa BS, Hungund BL. Neuromodulatory role of the endo-cannabinoid signaling system in alcoholism: an overview. Prostaglandins Leukot Essent Fatty Acids. 2002;66(2–3):287–299. doi: 10.1054/plef.2001.0352. [DOI] [PubMed] [Google Scholar]
- Baur R, Kaur KH, Sigel E. Structure of alpha6 beta3 delta GABA(A) receptors and their lack of ethanol sensitivity. J Neurochem. 2009;111(5):1172–1181. doi: 10.1111/j.1471-4159.2009.06387.x. [DOI] [PubMed] [Google Scholar]
- Beckstead MJ, Phelan R, Trudell JR, Bianchini MJ, Mihic SJ. Anesthetic and ethanol effects on spontaneously opening glycine receptor channels. J Neurochem. 2002;82(6):1343–1351. doi: 10.1046/j.1471-4159.2002.01086.x. [DOI] [PubMed] [Google Scholar]
- Bejanian M, Jones BL, Alkana RL. Low-level hyperbaric antagonism of ethanol-induced locomotor depression in C57BL/6J mice: dose response. Alcohol Clin Exp Res. 1993;17(5):935–939. doi: 10.1111/j.1530-0277.1993.tb05644.x. [DOI] [PubMed] [Google Scholar]
- Belelli D, Pistis M, Peters JA, Lambert JJ. General anaesthetic action at transmitter-gated inhibitory amino acid receptors. Trends Pharmacol Sci. 1999;20(12):496–502. doi: 10.1016/s0165-6147(99)01405-4. [DOI] [PubMed] [Google Scholar]
- Benedikt J, Teisinger J, Vyklicky L, Vlachova V. Ethanol inhibits cold-menthol receptor TRPM8 by modulating its interaction with membrane phosphatidylinositol 4,5-bisphosphate. J Neurochem. 2007;100(1):211–224. doi: 10.1111/j.1471-4159.2006.04192.x. [DOI] [PubMed] [Google Scholar]
- Bertaccini EJ, Lindahl E, Sixma T, Trudell JR. Effect of cobratoxin binding on the normal mode vibration within acetylcholine binding protein. J Chem Inf Model. 2008;48(4):855–860. doi: 10.1021/ci700456s. [DOI] [PubMed] [Google Scholar]
- Bertaccini EJ, Trudell JR, Lindahl E. Normal-mode analysis of the gly-cine alpha1 receptor by three separate methods. J Chem Inf Model. 2007;47(4):1572–1579. doi: 10.1021/ci600566j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaccini EJ, Trudell JR, Lindahl E. Normal mode gating motions of a ligand-gated ion channel persist in a fully hydrated lipid bilayer model. ACS Chem Neurosci. 2010a;1(8):552–558. doi: 10.1021/cn100026t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaccini EJ, Wallner B, Trudell JR, Lindahl E. Modeling anesthetic binding sites within the glycine alpha one receptor based on prokaryotic ion channel templates: the problem with TM4. J Chem Inf Model. 2010b;50(12):2248–2255. doi: 10.1021/ci100266c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. How membrane proteins sense voltage. Nat Rev Mol Cell Biol. 2008;9(4):323–332. doi: 10.1038/nrm2376. [DOI] [PubMed] [Google Scholar]
- Bhave G, Hu H-J, Glauner KS, Zhu W, Wang H, Brasier DJ, Oxford GS, Gereau RW. Protein kinase C phosphorylation sensitizes but does not activate the capsaicin receptor transient receptor potential vanilloid 1 (TRPV1) Proc Natl Acad Sci USA. 2003;100(21):12480–12485. doi: 10.1073/pnas.2032100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisogno T, Hanus L, De Petrocellis L, Tchilibon S, Ponde DE, Brandi I, Moriello AS, Davis JB, Mechoulam R, Di Marzo V. Molecular targets for cannabidiol and its synthetic analogues: effect on vanilloid VR1 receptors and on the cellular uptake and enzymatic hydrolysis of ananda-mide. Br J Pharmacol. 2001;134(4):845–852. doi: 10.1038/sj.bjp.0704327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Harris RA. Deletion of vanilloid receptor (TRPV1) in mice alters behavioral effects of ethanol. Neuropharmacology. 2009;56(4):814–820. doi: 10.1016/j.neuropharm.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Alva H, Harris RA. A pervasive mechanism for analgesia: activation of GIRK2 channels. Proc Natl Acad Sci USA. 2003;100(1):277–282. doi: 10.1073/pnas.012682399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Chang SR, Harris RA. Potassium channels as targets for ethanol: studies of G-protein-coupled inwardly rectifying potassium channel 2 (GIRK2) null mutant mice. J Pharmacol Exp Ther. 2001;298(2):521–530. [PubMed] [Google Scholar]
- Borghese CM, Harris RA. Studies of ethanol actions on recombinant delta-containing gamma-aminobutyric acid type A receptors yield contradictory results. Alcohol. 2007;41(3):155–162. doi: 10.1016/j.alcohol.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese CM, Henderson LA, Bleck V, Trudell JR, Harris RA. Sites of excitatory and inhibitory actions of alcohols on neuronal alpha2beta4 nico-tinic acetylcholine receptors. J Pharmacol Exp Ther. 2003;307(1):42–52. doi: 10.1124/jpet.102.053710. [DOI] [PubMed] [Google Scholar]
- Borghese CM, Storustovu Sı, Ebert B, Herd MB, Belelli D, Lambert JJ, Marshall G, Wafford KA, Harris RA. The delta subunit of gamma-aminobutyric acid type A receptors does not confer sensitivity to low concentrations of ethanol. J Pharmacol Exp Ther. 2006;316(3):1360–1368. doi: 10.1124/jpet.105.092452. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411(6835):269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389(6653):816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41(6):907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Chakrapani S, Auerbach A. A speed limit for conformational change of an allosteric membrane protein. Proc Natl Acad Sci USA. 2005;102(1):87–92. doi: 10.1073/pnas.0406777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MH, Coalson RD, Cascio M. Molecular dynamics simulations of ethanol binding to the transmembrane domain of the glycine receptor: implications for the channel potentiation mechanism. Proteins. 2008;71(2):972–981. doi: 10.1002/prot.21784. [DOI] [PubMed] [Google Scholar]
- Choi D-S, Wang D, Dadgar J, Chang WS, Messing RO. Conditional rescue of protein kinase C epsilon regulates ethanol preference and hypnotic sensitivity in adult mice. J Neurosci. 2002;22(22):9905–9911. doi: 10.1523/JNEUROSCI.22-22-09905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett PM, Matta JA, Ahern GP. General anesthetics sensitize the capsaicin receptor transient receptor potential V1. Mol Pharmacol. 2008a;74:1261–1268. doi: 10.1124/mol.108.049684. [DOI] [PubMed] [Google Scholar]
- Crawford DK, Trudell JR, Bertaccini EJ, Li K, Davies DL, Alkana RL. Evidence that ethanol acts on a target in Loop 2 of the extracellular domain of alpha1 glycine receptors. J Neurochem. 2007;102(6):2097–2109. doi: 10.1111/j.1471-4159.2007.04680.x. [DOI] [PubMed] [Google Scholar]
- Das J, Addona GH, Sandberg WS, Husain SS, Stehle T, Miller KW. Identification of a general anesthetic binding site in the diacylglycerol-binding domain of protein kinase Cδ. J Biol Chem. 2004;279(36):37964–37972. doi: 10.1074/jbc.M405137200. [DOI] [PubMed] [Google Scholar]
- Das J, Pany S, Rahman GM, Slater SJ. PKC epsilon has an alcohol-binding site in its second cysteine-rich regulatory domain. Biochem J. 2009;421(3):405–413. doi: 10.1042/BJ20082271. [DOI] [PubMed] [Google Scholar]
- Das J, Shunmugasundararaj S, Stehle T, Miller KW. Crystal structure of an alcohol complexed with the second cysteine-rich domain of protein kinase C delta. Alcohol Clin Exp Res. 2007;31(6 Suppl):202A. abstract. [Google Scholar]
- Davies DL, Alkana RL. Direct antagonism of ethanol’s effects on GABA(A) receptors by increased atmospheric pressure. Alcohol Clin Exp Res. 1998;22(8):1689–1697. [PubMed] [Google Scholar]
- Davies DL, Alkana RL. Ethanol enhances GABAA receptor function in short sleep and long sleep mouse brain membranes. Alcohol Clin Exp Res. 2001;25(3):478–483. [PubMed] [Google Scholar]
- Davies DL, Bolger MB, Brinton RD, Finn DA, Alkana RL. In vivo and in vitro hyperbaric studies in mice suggest novel sites of action for etha-nol. Psychopharmacology (Berl) 1999;141(4):339–350. doi: 10.1007/s002130050843. [DOI] [PubMed] [Google Scholar]
- Davies DL, Crawford DK, Trudell JR, Mihic SJ, Alkana RL. Multiple sites of ethanol action in alpha1 and alpha2 glycine receptors suggested by sensitivity to pressure antagonism. J Neurochem. 2004;89(5):1175–1185. doi: 10.1111/j.1471-4159.2004.02390.x. [DOI] [PubMed] [Google Scholar]
- Davies DL, Morland J, Jones BL, Palomares ML, Alkana RL. Low-level hyperbaric antagonism of ethanol’s anticonvulsant property in C57BL /6J mice. Alcohol Clin Exp Res. 1994;18(2):446. doi: 10.1111/j.1530-0277.1994.tb00103.x. abstract. [DOI] [PubMed] [Google Scholar]
- Davies DL, Trudell JR, Mihic SJ, Crawford DK, Alkana RL. Ethanol potentiation of glycine receptors expressed in Xenopus oocytes antagonized by increased atmospheric pressure. Alcohol Clin Exp Res. 2003;27(5):743–755. doi: 10.1097/01.ALC.0000065722.31109.A1. [DOI] [PubMed] [Google Scholar]
- Deitrich RA, Dunwiddie TV, Harris RA, Erwin VG. Mechanism of action of ethanol: initial central nervous system actions. Pharmacol Rev. 1989;41(4):489–537. [PubMed] [Google Scholar]
- De Petrocellis L, Vellani V, Schiano-Moriello A, Marini P, Magherini PC, Orlando P, Di Marzo V. Plant-derived cannabinoids modulate the activity of transient receptor potential channels of ankyrin type-1 and melastatin type-8. J Pharmacol Exp Ther. 2008;325(3):1007–1015. doi: 10.1124/jpet.107.134809. [DOI] [PubMed] [Google Scholar]
- Eckenhoff RG, Johansson JS. Molecular interactions between inhaled anesthetics and proteins. Pharmacol Rev. 1997;49(4):343–367. [PubMed] [Google Scholar]
- Eggers ED, O’Brien JA, Berger AJ. Developmental changes in the modulation of synaptic glycine receptors by ethanol. J Neurophysiol. 2000;84(5):2409–2416. doi: 10.1152/jn.2000.84.5.2409. [DOI] [PubMed] [Google Scholar]
- Ellingson JM, Silbaugh BC, Brasser SM. Reduced oral ethanol avoidance in mice lacking transient receptor potential channel vanilloid receptor 1. Behav Genet. 2008;39(1):62–72. doi: 10.1007/s10519-008-9232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engblom AC, Akerman KE. Effect of ethanol on gamma-aminobutyric acid and glycine receptor-coupled Cl- fluxes in rat brain synaptoneuro-somes. J Neurochem. 1991;57(2):384–390. doi: 10.1111/j.1471-4159.1991.tb03764.x. [DOI] [PubMed] [Google Scholar]
- Flood P, Ramirez-Latorre J, Role L. Alpha 4 beta 2 neuronal nicotinic acetylcholine receptors in the central nervous system are inhibited by isoflu-rane and propofol, but alpha 7-type nicotinic acetylcholine receptors are unaffected. Anesthesiology. 1997;86(4):859–865. doi: 10.1097/00000542-199704000-00016. [DOI] [PubMed] [Google Scholar]
- Garcıa-Sanz N, Fernández-Carvajal A, Morenilla-Palao C, Planells-Cases R, Fajardo-Sanchez E, Fernández-Ballester G, Ferrer-Montiel A. Identification of a tetramerization domain in the C terminus of the vanilloid receptor. J Neurosci. 2004;24(23):5307–5314. doi: 10.1523/JNEUROSCI.0202-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser ST, Kaczocha M, Deutsch DG. Anandamide transport: a critical review. Life Sci. 2005;77(14):1584–1604. doi: 10.1016/j.lfs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gordon AS, Yao L, Wu ZL, Coe IR, Diamond I. Ethanol alters the subcellular localization of delta- and epsilon protein kinase C in NG108-15 cells. Mol Pharmacol. 1997;52(4):554–559. doi: 10.1124/mol.52.4.554. [DOI] [PubMed] [Google Scholar]
- Grosman C, Zhou M, Auerbach A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature. 2000;403(6771):773–776. doi: 10.1038/35001586. [DOI] [PubMed] [Google Scholar]
- Grycova L, Lansky Z, Friedlova E, Obsilova V, Janouskova H, Obsil T, Teisinger J. Ionic interactions are essential for TRPV1 C-terminus binding to calmodulin. Biochem Biophys Res Commun. 2008;375(4):680–683. doi: 10.1016/j.bbrc.2008.08.094. [DOI] [PubMed] [Google Scholar]
- Hanchar HJ, Chutsrinopkun P, Meera P, Supavilai P, Sieghart W, Wallner M, Olsen RW. Ethanol potently and competitively inhibits binding of the alcohol antagonist Ro15-4513 to alpha4 / 6beta3delta GABAA receptors. Proc Natl Acad Sci USA. 2006;103(22):8546–8551. doi: 10.1073/pnas.0509903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchar HJ, Dodson PD, Olsen RW, Otis TS, Wallner M. Alcohol-induced motor impairment caused by increased extrasynaptic GABA(A) receptor activity. Nat Neurosci. 2005;8(3):339–345. doi: 10.1038/nn1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Trudell JR, Mihic SJ. Ethanol’s molecular targets. Sci Signal. 2008;1(28):re7. doi: 10.1126/scisignal.128re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heishman SJ, Stitzer ML, Bigelow GE. Alcohol and marijuana: comparative dose effect profiles in humans. Pharmacol Biochem Behav. 1988;31(3):649–655. doi: 10.1016/0091-3057(88)90244-4. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hodge CW, Mehmert KK, Kelley SP, McMahon T, Haywood A, Olive MF, Wang D, Sanchez-Perez AM, Messing RO. Supersensitivity to alloste-ric GABA(A) receptor modulators and alcohol in mice lacking PKCepsilon. Nat Neurosci. 1999;2(11):997–1002. doi: 10.1038/14795. [DOI] [PubMed] [Google Scholar]
- Hungund BL, Basavarajappa BS. Are anandamide and cannabinoid receptors involved in ethanol tolerance? A review of the evidence. Alcohol Alcohol. 2000;35(2):126–133. doi: 10.1093/alcalc/35.2.126. [DOI] [PubMed] [Google Scholar]
- Inanobe A, Matsuura T, Nakagawa A, Kurachi Y. Structural diversity in the cytoplasmic region of G protein-gated inward rectifier K+ channels. Channels (Austin) 2007;1(1):39–45. [PubMed] [Google Scholar]
- Jin T, Peng L, Mirshahi T, Rohacs T, Chan KW, Sanchez R, Logothetis DE. The (beta)gamma subunits of G proteins gate a K(+) channel by pivoted bending of a transmembrane segment. Mol Cell. 2002;10(3):469–481. doi: 10.1016/s1097-2765(02)00659-7. [DOI] [PubMed] [Google Scholar]
- Jordt S-E, Bautista DM, Chuang H-H, McKemy DD, Zygmunt PM, Högestätt ED, Meng ID, Julius D. Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature. 2004;427(6971):260–265. doi: 10.1038/nature02282. [DOI] [PubMed] [Google Scholar]
- Jordt S-E, Julius D. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell. 2002;108(3):421–430. doi: 10.1016/s0092-8674(02)00637-2. [DOI] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Substituted-cysteine accessibility method. Meth Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- Kauer JA, Gibson HE. Hot flash: TRPV channels in the brain. Trends Neurosci. 2009;32(4):215–224. doi: 10.1016/j.tins.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ikeda K, Kojima H, Niki H, Yano R, Yoshioka T, Kumanishi T. Ethanol opens G-protein-activated inwardly rectifying K+ channels. Nat Neurosci. 1999;2(12):1091–1097. doi: 10.1038/16019. [DOI] [PubMed] [Google Scholar]
- Korepanova A, Pereda-Lopez A, Solomon LR, Walter KA, Lake MR, Bianchi BR, McDonald HA, Neelands TR, Shen J, Matayoshi ED, More-land RB, Chiu ML. Expression and purification of human TRPV1 in baculovirus-infected insect cells for structural studies. Protein Expr Purif. 2009;65(1):38–50. doi: 10.1016/j.pep.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Kozell LB, Walter NAR, Milner LC, Wickman K, Buck KJ. Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J Neurosci. 2009;29(37):11662–11673. doi: 10.1523/JNEUROSCI.1413-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Harrison NL. General anaesthetic actions on ligand-gated ion channels. Cell Mol Life Sci. 1999;55(10):1278–1303. doi: 10.1007/s000180050371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse SW, Zhao R, Smith DP, Jones DNM. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat Struct Biol. 2003;10(9):694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Corey DP. Burning cold: involvement of TRPA1 in noxious cold sensation. J Gen Physiol. 2009;133(3):251–256. doi: 10.1085/jgp.200810146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesscher HMB, Wallace MJ, Zeng L, Wang V, Deitchman JK, McMahon T, Messing RO, Newton PM. Amygdala protein kinase C epsilon controls alcohol consumption. Genes Brain Behav. 2009;8(5):493–499. doi: 10.1111/j.1601-183X.2009.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wilson WR, Mayfield RD, Brozowski SJ, Morrisett RA, Harris RA. G-protein-coupled inwardly rectifying potassium channels are targets of alcohol action. Nat Neurosci. 1999;2(12):1084–1090. doi: 10.1038/16012. [DOI] [PubMed] [Google Scholar]
- Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34:W52–W56. doi: 10.1093/nar/gkl082. Web Server Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54(6):905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Liu LT, Haddadian EJ, Willenbring D, Xu Y, Tang P. Higher susceptibility to halothane modulation in open- than in closed-channel alpha4beta2 nAChR revealed by molecular dynamics simulations. J Phys Chem B. 2010;114(1):626–632. doi: 10.1021/jp908944e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LT, Willenbring D, Xu Y, Tang P. General anesthetic binding to neuronal alpha4beta2 nicotinic acetylcholine receptor and its effects on global dynamics. J Phys Chem B. 2009;113(37):12581–12589. doi: 10.1021/jp9039513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo IA, Harris RA. Sites of alcohol and volatile anesthetic action on glycine receptors. Int Rev Neurobiol. 2005;65:53–87. doi: 10.1016/S0074-7742(04)65003-3. [DOI] [PubMed] [Google Scholar]
- Lobo IA, Harris RA, Trudell JR. Cross-linking of sites involved with alcohol action between transmembrane segments 1 and 3 of the glycine receptor following activation. J Neurochem. 2008;104(6):1649–1662. doi: 10.1111/j.1471-4159.2007.05090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo IA, Mascia MP, Trudell JR, Harris RA. Channel gating of the glycine receptor changes accessibility to residues implicated in receptor potentiation by alcohols and anesthetics. J Biol Chem. 2004a;279(32):33919–33927. doi: 10.1074/jbc.M313941200. [DOI] [PubMed] [Google Scholar]
- Lobo IA, Trudell JR, Harris RA. Cross-linking of glycine receptor transmembrane segments two and three alters coupling of ligand binding with channel opening. J Neurochem. 2004b;90(4):962–969. doi: 10.1111/j.1471-4159.2004.02561.x. [DOI] [PubMed] [Google Scholar]
- Lobo IA, Trudell JR, Harris RA. Accessibility to residues in transmem-brane segment four of the glycine receptor. Neuropharmacology. 2006;50(2):174–181. doi: 10.1016/j.neuropharm.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Logothetis DE, Kurachi Y, Galper J, Neer EJ, Clapham DE. The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 1987;325(6102):321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Lukacs V, Thyagarajan B, Varnai P, Balla A, Balla T, Rohacs T. Dual regulation of TRPV1 by phosphoinositides. J Neurosci. 2007;27(26):7070–7080. doi: 10.1523/JNEUROSCI.1866-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, Slesinger PA. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neu-rosci. 2010;11(5):301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Geierstanger BH, Viswanath V, Bandell M, Eid SR, Hwang S, Patapoutian A. The pungency of garlic: activation of TRPA1 and TRPV1 in response to allicin. Curr Biol. 2005;15(10):929–934. doi: 10.1016/j.cub.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Mascia MP, Mihic SJ, Valenzuela CF, Schofield PR, Harris RA. A single amino acid determines differences in ethanol actions on strychnine-sensitive glycine receptors. Mol Pharmacol. 1996;50(2):402–406. [PubMed] [Google Scholar]
- Mascia MP, Trudell JR, Harris RA. Specific binding sites for alcohols and anesthetics on ligand-gated ion channels. Proc Natl Acad Sci USA. 2000;97(16):9305–9310. doi: 10.1073/pnas.160128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346(6284):561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- Matta JA, Cornett PM, Miyares RL, Abe K, Sahibzada N, Ahern GP. General anesthetics activate a nociceptive ion channel to enhance pain and inflammation. Proc Natl Acad Sci USA. 2008;105(25):8784–8789. doi: 10.1073/pnas.0711038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool BA, Frye GD, Pulido MD, Botting SK. Effects of chronic ethanol consumption on rat GABA(A) and strychnine-sensitive glycine receptors expressed by lateral / basolateral amygdala neurons. Brain Res. 2003;963(1–2):165–177. doi: 10.1016/s0006-8993(02)03966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken LM, McCracken ML, Gong DH, Trudell JR, Harris RA. Linking of glycine receptor transmembrane segments three and four allows assignment of intrasubunit-facing residues. ACS Chem Neurosci. 2010;1(7):482–494. doi: 10.1021/cn100019g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKemy DD, Neuhausser WM, Julius D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 2002;416(6876):52–58. doi: 10.1038/nature719. [DOI] [PubMed] [Google Scholar]
- Mihic SJ, Ye Q, Wick MJ, Koltchine VV, Krasowski MD, Finn SE, Mascia MP, Valenzuela CF, Hanson KK, Greenblatt EP, Harris RA, Harrison NL. Sites of alcohol and volatile anaesthetic action on GABA(A) and gly-cine receptors. Nature. 1997;389(6649):385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- Misonou H. Homeostatic regulation of neuronal excitability by K(+) channels in normal and diseased brains. Neuroscientist. 2010;16(1):51–64. doi: 10.1177/1073858409341085. [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc Natl Acad Sci USA. 2008;105(21):7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molander A, Lidö HH, Löf E, Ericson M, Söderpalm B. The glycine reuptake inhibitor Org 25935 decreases ethanol intake and preference in male wistar rats. Alcohol Alcohol. 2007;42(1):11–18. doi: 10.1093/alcalc/agl085. [DOI] [PubMed] [Google Scholar]
- Mowrey D, Haddadian EJ, Liu LT, Willenbring D, Xu Y, Tang P. Unresponsive correlated motion in alpha7 nAChR to halothane binding explains its functional insensitivity to volatile anesthetics. J Phys Chem B. 2010;114(22):7649–7655. doi: 10.1021/jp1009675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365(6441):61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Murail S, Wallner B, Trudell JR, Bertaccini E, Lindahl E. Microsecond simulations indicate that ethanol binds between subunits and could stabilize an open-state model of a glycine receptor. Biophys J. 2011;100:1642–1650. doi: 10.1016/j.bpj.2011.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton PM, Ron D. Protein kinase C and alcohol addiction. Pharmacol Res. 2007;55(6):570–577. doi: 10.1016/j.phrs.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Nishida M, Cadene M, Chait BT, Mackinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26(17):4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M, MacKinnon R. Structural basis of inward rectification: cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 A resolution. Cell. 2002;111(7):957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- Numazaki M, Tominaga T, Takeuchi K, Murayama N, Toyooka H, Tomina-ga M. Structural determinant of TRPV1 desensitization interacts with calmodulin. Proc Natl Acad Sci USA. 2003;100(13):8002–8006. doi: 10.1073/pnas.1337252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nury H, Poitevin F, Van Renterghem C, Changeux J-P, Corringer P-J, Delarue M, Baaden M. One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc Natl Acad Sci USA. 2010;107(14):6275–6280. doi: 10.1073/pnas.1001832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegan S, Arrabit C, Slesinger PA, Choe S. Andersen’s syndrome mutation effects on the structure and assembly of the cytoplasmic domains of Kir2.1. Biochemistry. 2006;45(28):8599–8606. doi: 10.1021/bi060653d. [DOI] [PubMed] [Google Scholar]
- Pegan S, Arrabit C, Zhou W, Kwiatkowski W, Collins A, Slesinger PA, Choe S. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8(3):279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- Peoples RW, Weight FF. Differential alcohol modulation of GABA(A) and NMDA receptors. Neuroreport. 1999;10(1):97–101. doi: 10.1097/00001756-199901180-00019. [DOI] [PubMed] [Google Scholar]
- Pepe IM. Recent advances in our understanding of rhodopsin and pho-totransduction. Prog Retin Eye Res. 2001;20(6):733–759. doi: 10.1016/s1350-9462(01)00013-1. [DOI] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Alkana RL, Davies DL. Targets for ethanol action and antagonism in loop 2 of the extracellular domain of glycine receptors. J Neurochem. 2008;106(3):1337–1349. doi: 10.1111/j.1471-4159.2008.05476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Alkana RL, Davies DL. Molecular targets and mechanisms for ethanol action in glycine receptors. Pharmacol Ther. 2010;127(1):53–65. doi: 10.1016/j.pharmthera.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Asatryan L, Alkana RL, Davies DL. Loop 2 structure in glycine and GABA(A) receptors plays a key role in determining ethanol sensitivity. J Biol Chem. 2009;284(40):27304–27314. doi: 10.1074/jbc.M109.023598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plapp BV. Conformational changes and catalysis by alcohol dehydroge-nase. Arch Biochem Biophys. 2010;493(1):3–12. doi: 10.1016/j.abb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin C, Elliott D. Alcohol, tobacco and cannabis use among Nova Scotia adolescents: implications for prevention and harm reduction. CMAJ. 1997;156(10):1387–1393. [PMC free article] [PubMed] [Google Scholar]
- Premkumar LS, Ahern GP. Induction of vanilloid receptor channel activity by protein kinase C. Nature. 2000;408(6815):985–990. doi: 10.1038/35050121. [DOI] [PubMed] [Google Scholar]
- Prescott ED, Julius D. A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science. 2003;300(5623):1284–1288. doi: 10.1126/science.1083646. [DOI] [PubMed] [Google Scholar]
- Reuveny E, Slesinger PA, Inglese J, Morales JM, Iniguez-Lluhi JA, Lefkowitz RJ, Bourne HR, Jan YN, Jan LY. Activation of the cloned musca-rinic potassium channel by G protein beta gamma subunits. Nature. 1994;370(6485):143–146. doi: 10.1038/370143a0. [DOI] [PubMed] [Google Scholar]
- Rosenbaum T, Gordon-Shaag A, Munari M, Gordon SE. Ca2+ / calmodulin modulates TRPV1 activation by capsaicin. J Gen Physiol. 2004;123(1):53–62. doi: 10.1085/jgp.200308906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadja R, Smadja K, Alagem N, Reuveny E. Coupling Gbetagamma-dependent activation to channel opening via pore elements in inwardly rectifying potassium channels. Neuron. 2001;29(3):669–680. doi: 10.1016/s0896-6273(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Slater SJ, Cox KJ, Lombardi JV, Ho C, Kelly MB, Rubin E, Stubbs CD. Inhibition of protein kinase C by alcohols and anaesthetics. Nature. 1993;364(6432):82–84. doi: 10.1038/364082a0. [DOI] [PubMed] [Google Scholar]
- Slater SJ, Kelly MB, Larkin JD, Ho C, Mazurek A, Taddeo FJ, Yeager MD, Stubbs CD. Interaction of alcohols and anesthetics with protein kinase Calpha. J Biol Chem. 1997;272(10):6167–6173. doi: 10.1074/jbc.272.10.6167. [DOI] [PubMed] [Google Scholar]
- Slater SJ, Malinowski SA, Stubbs CD. The nature of the hydrophobic n-alkanol binding site within the C1 domains of protein kinase Calpha. Biochemistry. 2004;43(23):7601–7609. doi: 10.1021/bi049755z. [DOI] [PubMed] [Google Scholar]
- Stein AT, Ufret-Vincenty CA, Hua L, Santana LF, Gordon SE. Phosphoinositide 3-kinase binds to TRPV1 and mediates NGF-stimulated TRPV1 trafficking to the plasma membrane. J Gen Physiol. 2006;128(5):509–522. doi: 10.1085/jgp.200609576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M, McNaughton PA. Modulation of single-channel properties of TRPV1 by phosphorylation. J Physiol. 2010;588(19):3743–3756. doi: 10.1113/jphysiol.2010.190611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom-Poromaa I, Smith DH, Gong QH, Sabado TN, Li X, Light A, Wiedmann M, Williams K, Smith SS. Hormonally regulated alpha(4)beta(2)delta GABA(A) receptors are a target for alcohol. Nat Neu-rosci. 2002;5(8):721–722. doi: 10.1038/nn888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syapin PJ, Jones BL, Finn DA, Davies DL, Alkana RL. Effect of 12 atmospheres helium-oxygen on the response of mice to convulsant drugs. Undersea Hyperb Med. 1996;23(1):35–41. [PubMed] [Google Scholar]
- Taly A, Delarue M, Grutter T, Nilges M, Le Novere N, Corringer PJ, Chan-geux JP. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys J. 2005;88:3954–3965. doi: 10.1529/biophysj.104.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic J, Welsh BT, Bertaccini EJ, Trudell JR, Mihic SJ. Disruption of an intersubunit electrostatic bond is a critical step in glycine receptor activation. Proc Natl Acad Sci USA. 2010;107(17):7987–7992. doi: 10.1073/pnas.1001845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisani M, Smart D, Gunthorpe MJ, Tognetto M, Barbieri M, Campi B, Amadesi S, Gray J, Jerman JC, Brough SJ, Owen D, Smith GD, Randall AD, Harrison S, Bianchi A, Davis JB, Geppetti P. Ethanol elicits and potentiates nociceptor responses via the vanilloid receptor-1. Nat Neurosci. 2002;5(6):546–551. doi: 10.1038/nn0602-852. [DOI] [PubMed] [Google Scholar]
- Trudell JR, Harris RA. Are sobriety and consciousness determined by water in protein cavities? Alcohol Clin Exp Res. 2004;28(1):1–3. doi: 10.1097/01.ALC.0000108648.32241.BD. [DOI] [PubMed] [Google Scholar]
- van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- van der Stelt M, Di Marzo V. Endovanilloids putative endogenous ligands of transient receptor potential vanilloid 1 channels. Eur J Biochem. 2004;271:1827–1834. doi: 10.1111/j.1432-1033.2004.04081.x. [DOI] [PubMed] [Google Scholar]
- Vemparala S, Domene C, Klein ML. Computational studies on the interactions of inhalational anesthetics with proteins. Acc Chem Res. 2010;43(1):103–110. doi: 10.1021/ar900149j. [DOI] [PubMed] [Google Scholar]
- Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br J Pharmacol. 2008;154(2):299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I, Wyse BD, Roberts-Thomson SJ, Monteith GR, Cabot PJ. Mechanisms involved in potentiation of transient receptor potential vanil-loid 1 responses by ethanol. Eur J Pain. 2008;12(4):441–454. doi: 10.1016/j.ejpain.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Vlachová V, Teisinger J, Susánková K, Lyfenko A, Ettrich R, Vyklicky L. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J Neurosci. 2003;23:1340–1350. doi: 10.1523/JNEUROSCI.23-04-01340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CA, Friedrich B, Setiawan I, Lang F, Bröer S. The use of Xenopus laevis oocytes for the functional characterization of heterologously expressed membrane proteins. Cell Physiol Biochem. 2000;10(1–2):1–12. doi: 10.1159/000016341. [DOI] [PubMed] [Google Scholar]
- Wang S, Poon K, Oswald RE, Chuang H-H. Distinct modulations of human capsaicin receptor by protons and magnesium through different domains. J Biol Chem. 2010;285(15):11547–11556. doi: 10.1074/jbc.M109.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Faria LC, Mody I. Low ethanol concentrations selectively augment the tonic inhibition mediated by delta subunit-containing GABAA receptors in hippocampal neurons. J Neurosci. 2004;24(38):8379–8382. doi: 10.1523/JNEUROSCI.2040-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil A, Moore SE, Waite NJ, Randall A, Gunthorpe MJ. Conservation of functional and pharmacological properties in the distantly related temperature sensors TRVP1 and TRPM8. Mol Pharmacol. 2005;68(2):518–527. doi: 10.1124/mol.105.012146. [DOI] [PubMed] [Google Scholar]
- Weiss DR, Levitt M. Can morphing methods predict intermediate structures? J Mol Biol. 2009;385(2):665–674. doi: 10.1016/j.jmb.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JM, Simon SA, Reinhart PH. The activation mechanism of rat vanilloid receptor 1 by capsaicin involves the pore domain and differs from the activation by either acid or heat. Proc Natl Acad Sci USA. 2000;97(25):13889–13894. doi: 10.1073/pnas.230146497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick MJ, Mihic SJ, Ueno S, Mascia MP, Trudell JR, Brozowski SJ, Ye Q, Harrison NL, Harris RA. Mutations of gamma-aminobutyric acid and glycine receptors change alcohol cutoff: evidence for an alcohol receptor? Proc Natl Acad Sci USA. 1998;95(11):6504–6509. doi: 10.1073/pnas.95.11.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakura T, Mihic SJ, Harris RA. Amino acid volume and hydropathy of a transmembrane site determine glycine and anesthetic sensitivity of glycine receptors. J Biol Chem. 1999;274(33):23006–23012. doi: 10.1074/jbc.274.33.23006. [DOI] [PubMed] [Google Scholar]
- Yao L, Fan P, Jiang Z, Gordon A, Mochly-Rosen D, Diamond I. Dopamine and ethanol cause translocation of epsilonPKC associated with epsilonRACK: cross-talk between cAMP-dependent protein kinase A and protein kinase C signaling pathways. Mol Pharmacol. 2008;73(4):1105–1112. doi: 10.1124/mol.107.042580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Koltchine VV, Mihic SJ, Mascia MP, Wick MJ, Finn SE, Harrison NL, Harris RA. Enhancement of glycine receptor function by ethanol is inversely correlated with molecular volume at position alpha267. J Biol Chem. 1998;273(6):3314–3319. doi: 10.1074/jbc.273.6.3314. [DOI] [PubMed] [Google Scholar]
- Yi BA, Lin YF, Jan YN, Jan LY. Yeast screen for constitutively active mutant G protein-activated potassium channels. Neuron. 2001;29(3):657–667. doi: 10.1016/s0896-6273(01)00241-0. [DOI] [PubMed] [Google Scholar]
- Zhu F, Hummer G. Gating transition of pentameric ligand-gated ion channels. Biophys J. 2009;97(9):2456–2463. doi: 10.1016/j.bpj.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt PM, Petersson J, Andersson DA, Chuang H, Sørgård M, Di Marzo V, Julius D, Högestatt ED. Vanilloid receptors on sensory nerves mediate the vasodilator action of anandamide. Nature. 1999;400(6743):452–457. doi: 10.1038/22761. [DOI] [PubMed] [Google Scholar]