Abstract
The metabolism of phosphatidylinositol-4,5-bisphosphate (PIP2) changed during the culture period of the thermoacidophilic red alga Galdieria sulphuraria. Seven days after inoculation, the amount of PIP2 in the cells was 910 ± 100 pmol g−1 fresh weight; by 12 d, PIP2 levels increased to 1200 ± 150 pmol g−1 fresh weight. In vitro assays indicated that phosphatidylinositol monophosphate (PIP) kinase specific activity increased from 75 to 230 pmol min−1 mg−1 protein between d 7 and 12. When G. sulphuraria cells were osmostimulated, transient increases of up to 4-fold could be observed in inositol-1,4,5-trisphosphate (IP3) levels within 90 s, regardless of the age of the cells. In d-12 cells, the increase in IP3 was preceded by a transient increase of up to 5-fold in specific PIP kinase activity, whereas no such increase was detected after osmostimulation of d-7 cells. The increase in PIP kinase activity before IP3 signaling in d-12 cells indicates that there is an additional pathway for regulation of phosphoinositide metabolism after stimulation other than an initial activation of phospholipase C. Also, the rapid activation of PIP2 biosynthesis in cells with already-high PIP2 levels suggests that the PIP2 present was not available for signal transduction. By comparing the response of the cells at d 7 and 12, we have identified two potentially distinct pools of PIP2.
The unicellular thermoacidophilic red alga Galdieria sulphuraria occurs in hot, acidic, volcanic springs (up to 55°C and pH < 2.0; Merola et al., 1981). The algae grow in the springs and colonize surrounding rocks under a silica sinter layer of 2 to 3 mm (Smith and Brock, 1973; Gross et al., 1998). Frequently submerged and exposed to hot, acidic steam from the boiling springs, the rocks provide very unstable conditions for the growth of microorganisms, and the algae are subject to frequent desiccation. Under the heterotrophic conditions used for cell culture (Gross and Schnarrenberger, 1995), the algae exhibit a simple succession of growth stages with the carbon source as a limiting factor for growth (Gross and Schnarrenberger, 1995). The ability of G. sulphuraria to adapt to changing environmental factors in nature (Gross et al., 1998) requires mechanisms to perceive and respond to environmental cues and stimuli.
In both plants and animals, the phosphoinositide pathway is involved in the perception and transduction of external stimuli. In vitro lipid phosphorylation with microsomal membranes from G. sulphuraria has been shown to yield primarily polyphosphoinositides (Gross and Boss, 1993), and in vitro lipid phosphorylation profiles of stationary-phase G. sulphuraria are similar to those of Neurospora or rat liver (Gross and Boss, 1993). Based on this information, we wanted to investigate whether phosphoinositides played a role in signaling in G. sulphuraria.
It had been shown in several systems that the metabolism of phosphoinositides changes with growth or senescence (Heim and Wagner, 1986; Falkenau et al., 1987; Borochov et al., 1994). An increase in specific PIP kinase activity in plasma membranes with senescence had been reported in wilting petunia petals (Borochov et al., 1994). However, the physiological role for an increased PIP2 synthesis in senescing petals was not clear. In Catharanthus roseus suspension cultures, changes in the levels of radiolabeled phosphoinositides with culture age have been observed, leading the authors to suggest a role for phosphoinositides in the regulation of cell proliferation (Heim and Wagner, 1986; Falkenau et al., 1987). These studies imply that the responsive state of the cells will change with age. To our knowledge, there has been no systematic comparison of signal transduction pathways between different stages of growth or development.
To study phosphoinositide signaling in G. sulphuraria, we first had to characterize the phosphoinositide metabolism in the alga. Microsomal membranes were prepared at various times during the culture period and compared with regard to their ability to phosphorylate lipids in vitro. These data suggested changes in the prevalence of phosphoinositides in the cells during the culture period. In our study, we determined that PIP kinase specific activity and PIP2 levels of G. sulphuraria cells were different between d 7 and 12 of the culture period. The aim of this work was to elucidate how these differences in phosphoinositide metabolism affected IP3 signaling after osmostimulation of G. sulphuraria.
In addition to serving as the precursor of the second messengers IP3 and DAG, PIP2 can perform a variety of cellular functions in different locations of the cell. PIP2 can function as a direct affector of proteins (for review, see Toker, 1998), including plasma membrane ATPases (Memon et al., 1989; Memon and Boss, 1990), ion channels (Hilgemann and Ball, 1996; Fan and Makielski, 1997), and the cytoskeletal actin (Fukami et al., 1992; Drøbak et al., 1994; for review, see Janmey, 1994; Shibasaki et al., 1997; Staiger et al., 1997; Sun et al., 1997). It has been difficult to identify and characterize distinct functional pools of polyphosphorylated phosphoinositides within the cell. By comparing levels of PIP2, the specific activity of PIP kinase, and the production of IP3 after mild osmostimulation of G. sulphuraria cells at two different times in the growth cycle (d 7 and 12), we have characterized two different signal transduction pathways. Furthermore, we have gained new insight into the character of the distinct pools of PIP2.
MATERIALS AND METHODS
Cell Culture
Galdieria sulphuraria strain 002 (culture collection of the University of Naples, Italy) was grown at pH 2.0 and 37°C heterotrophically in the dark in 2-L flasks shaking at 100 rpm in liquid medium containing 50 mm Glc, as described by Gross and Schnarrenberger (1995).
In Vivo Labeling
To study the incorporation of [2-3H]myo-inositol into inositol phospholipids, 0.1 to 0.3 g fresh weight of cells from d 7 and 12 was incubated overnight with 0.1 to 10 μCi of [2-3H]myo-inositol in a 2-mL culture. Cells were washed twice in deionized water before lipid extraction. To label polyphosphoinositides with 32Pi, between 0.2 and 0.5 g fresh weight of cells from d 7 and 12 was incubated with 10 μCi of carrier-free 32Pi for 10 min or 2 h in a 2-mL culture. Cells were washed twice in deionized water before extraction of lipids. Cells were lysed with 20% (v/v) TCA and washed in deionized water, and total lipids were extracted according to the method of Cho et al. (1992).
Osmotic Stimulation
Before stimulation, cells were equilibrated overnight in 50 mL of culture medium in 200-mL culture flasks at 26°C in the dark with shaking (150 rpm). Cells were stimulated by the addition of 5 mL of NaCl, KCl, or methyl-Man in conditioned culture medium to final concentrations, as indicated. Conditioned culture medium was obtained immediately before each experiment by centrifuging 100 mL of cell culture for 10 min at 2000g and decanting the medium. NaCl, KCl, and methyl-Man solutions were prepared fresh in conditioned medium for each experiment and adjusted to 26°C before use. Methyl-Man was used as an osmotically active sugar derivative, because unlike sorbitol or mannitol, it is not taken up and metabolized by G. sulphuraria (W. Gross, personal communication).
Preparation of Microsomes
Cells from 50-mL cultures (0.5–1 g fresh weight) were harvested by centrifugation at 2,500g for 30 s and homogenized in 20 mL of ice-cold buffer (250 mm Suc, 3 mm EDTA, 2 mm EGTA, 14 mm β-mercaptoethanol, 2 mm DTT, and 50 mm Tris-HCl, pH 7.4) with 0.1 g of polyvinylpolypyrrolidone using a blender (VirTis Co., Gardiner, NY) and glass beads. For time-course experiments, the times indicated denote the initiation of homogenization. Arabidopsis vegetative tissue and whole rat liver were ground in the same buffer using a VirTis blender and rotating blades. Microsomal membranes were prepared by centrifuging the homogenate for 15 min at 2,500g and then centrifuging the 2,500g supernatant for 60 min at 41,000g. The 41,000g pellets were resuspended in 30 mm Tris, pH 6.5, containing 15 mm MgCl2. Protein was estimated using the Bradford assay (Bio-Rad) with BSA as a standard.
Lipid Kinase Assays
PI kinase and PIP kinase activities were assayed as described previously (Cho and Boss, 1995) using 20 μg of microsomal membrane protein per assay in 50 μL of reaction mixture containing 30 mm Tris-HCl, pH 6.5, 7.5 mm MgCl2, 1 mm sodium molybdate, 0.01% (v/v) Triton X-100, and 0.9 mm [γ-32P]ATP (0.2 μCi/nmol). Reactions were incubated for 10 min at room temperature with intermittent mixing. For assays containing exogenous substrate, PI or PIP presolubilized in 1% (v/v) Triton X-100 was added to give a final concentration of 25 μg of lipid in 0.1% (v/v) Triton X-100 per reaction. After incubation, inositol phospholipids were extracted using an acid-extraction method (Cho et al., 1992). The DAG kinase assay contained 0.01% (v/v) 3-[(cholamidopropyl)dimethylammonio]-1-propanesulfonic acid instead of Triton X-100. To quantify DAG levels, lipids were extracted from equal amounts (500 μg of microsomal protein) of microsomes from G. sulphuraria, Arabidopsis, or rat liver. The extracted lipids were incubated with 0.9 mm [γ-32P]ATP (0.2 μCi/nmol) and 1 unit per assay of a recombinant Escherichia coli DAG kinase (Calbiochem) under the conditions used to measure endogenous DAG kinase activity.
Separation of Phospholipids
Lipids were separated by TLC on LK5D silica-gel plates (Whatman) using a CHCl3:methanol:NH4OH:H2O (86:76:6:16) solvent system (Cho and Boss, 1995). The 32P-labeled phospholipids were quantified using a scanner (system 500, Bioscan, Inc., Washington, DC). 32P-labeled phosphoinositides were hydrolyzed when the samples were incubated on the TLC plate with 0.5 m KOH for 10 min before development, indicating that they were not phosphosphingolipids (data not shown). To distinguish between 4- and 3-phosphorylated PIP, lipids were separated in the presence of boric acid, as described previously by Walsh et al. (1991). Under the conditions used for the in vitro lipid phosphorylation assays, no PI-3-P could be detected (data not shown).
Quantification of IP3 and PIP2 Contents
For IP3 measurements after stimulation, cells were harvested by centrifugation at 2000g for 10 to 30 s in preweighed test tubes. The supernatant was discarded and cells were immediately frozen in liquid N2. The times indicated denote the times the cells were placed in liquid N2. The frozen cells were weighed, and 500 μL of ice-cold 20% (v/v) PCA was added to each sample. After a 20-min incubation on ice, precipitated proteins were pelleted by centrifugation at 2000g for 15 min at 4°C. For IP3 assays, the supernatant was transferred to a clean tube and adjusted to pH 7.5 using ice-cold 1.5 m KOH, 60 mm Hepes containing universal pH indicator dye (Fisher Scientific). The neutralized samples were assayed for IP3 content using the [3H]IP3 receptor-binding assay (Amersham). Assays were carried out at 4°C according to the manufacturer's instructions using 50 μL of sample per assay.
For PIP2 mass measurements, whole-cell PCA precipitate was washed twice with ice-cold deionized water, and lipids were extracted using the acidic CHCl3:methanol extraction method according to Cho et al. (1992). The extracted lipids were hydrolyzed by adding 1 mL of 1 m KOH and heating to 100°C for 15 min. After hydrolysis, samples were adjusted to pH 7.5 with 20% (v/v) PCA containing universal pH indicator dye. Fatty acids were removed by washing twice with 1-butanol:petroleum ether (5:1, v/v). A 500-μL sample of the aqueous phase was lyophilized, resuspended in 110 μL of deionized water, and assayed for IP3 as described above.
Expression and Purification of Inositol Polyphosphate 5-Phosphatase I
To rule out the possibility that inositol phosphate metabolites in G. sulphuraria samples other than IP3 affected the displacement of [3H]IP3 in the IP3 receptor-binding assay, aliquots from G. sulphuraria samples were pretreated with a recombinant human inositol polyphosphate 5-phosphatase I. The recombinant protein was induced for 3 h by the addition of isopropyl-β-d-thiogalactosidase (0.5 mm final concentration). Bacterial cells expressing the His-tagged phosphatase were lysed by sonication and resuspended in 50 mm sodium phosphate, pH 8.0, and 300 mm NaCl. The recombinant protein was purified by metal-affinity chromatography on a nickel-nitrilotriacetic acid agarose resin (Qiagen, Dusseldorf, Germany) and eluted from the column with the same buffer containing 50 to 500 mm imidazole. The activity of purified fractions was tested on commercially available IP3. Samples from G. sulphuraria osmostimulated for 90 s were treated with active or heat-denatured phosphatase at room temperature or at 37°C and assayed for IP3 content. The phosphatase pretreatment eliminated the IP3 from G. sulphuraria samples. Heat-denatured phosphatase had no effect. The human inositol polyphosphate 5-phosphatase I cDNA (Auethavekiat et al., 1997) was a gift from Dr. Phil Majerus (Washington University School of Medicine, St. Louis, MO).
Presentation of Data
Products of in vitro phosphorylation that did not migrate on TLC (origin) were excluded from the quantification of phosphorylated phospholipids presented in Figure 1B. The data shown in Figure 4 were calculated as the percentage change of the osmostimulated samples over the conditionedmedium controls at each time point measured. Experiments presented in Figure 4 were repeated two (A + B) or four times (C + D), samples were assayed in duplicate, and values were averaged from these values before calculation of the percentage change to allow the comparison of changes between different experiments.
Figure 1.
A, Microsomal membranes prepared from G. sulphuraria at different times during the culture period were incubated for 10 min with [γ-32P]ATP. Lipids were extracted under acidic conditions and separated by TLC in a basic solvent. Characteristic patterns of lipid phosphorylation are evident. After 5 to 7 d in culture, PA decreased and PIP2 increased. The autoradiograph is from a representative experiment. The experiment was repeated four times with similar results. B, The in vitro phosphorylation products from the experiment shown in A were quantified with a Bioscan scanner. Vertical bars indicate the duplicate range. Counts at the origin were not included in the quantification.
Figure 4.
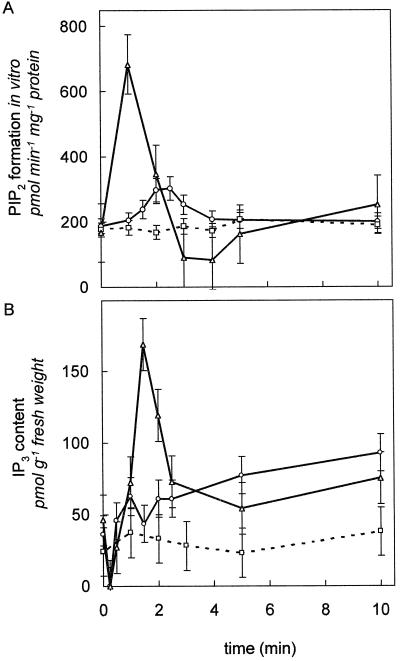
G. sulphuraria cultures were osmostimulated by the addition of different concentrations of KCl (○) or methyl-Man (□) in conditioned medium. The effects of the stimulation on in vitro PIP2 formation by microsomes from 7- and 12-d-old cells are shown in A and B, respectively. A transient increase in PIP2 formation in vitro was observed only in 12-d-old cells. Data represent the means of 6 values from three experiments. Vertical bars indicate the range. C and D, In vivo levels of IP3 were measured in whole cells after the addition of 25 mm KCl in conditioned medium (▵) to the cultures. The effects of both treatments on 7- and 12-d-old cells are illustrated in C and D, respectively. At both stages, a transient decrease in IP3 levels could be observed, followed by a transient increase. Data are the averages of 10 values from five experiments. Vertical bars indicate the range.
Northern Blots
G. sulphuraria total RNA was isolated using an RNeasy (plant) kit (Qiagen). The RNA (25 μg/lane) was fractionated by electrophoresis on formaldehyde-containing agarose gels. It was then transferred to a nylon membrane (Magna Graph, Micron Separations, Inc., Westborough, MA) and cross-linked in UV light. The blots were probed with an Arabidopsis cDNA that was identical to that of the Arabidopsis PIP 5-kinase cDNA (Satterlee and Sussman, 1997) radiolabeled by random priming with [α-32P]dCTP. Hybridizations were carried out at 42°C in hybridization buffer containing 50% (v/v) formamide. The blots were washed sequentially in 1× SSPE (10 mm NaH2PO4, 1 mm EDTA, and 149 mm NaCl) containing 0.1% (w/v) SDS, followed by a final wash in 0.1× SSPE and 0.1% (w/v) SDS at 55°C. Autoradiography was carried out using Kodak X-Omat autoradiography film.
RESULTS
Lipid Phosphorylation Changes with the Time in Culture
To detect lipid-mediated signaling events in response to a stimulus, basal levels of key phospholipids (PA, PIP, and PIP2) had to be characterized. When [2-3H]myo-inositol or 32Pi was added to G. sulphuraria cultures at d 7 or 12 of the growth period, only trace amounts of radiolabeled phosphoinositides could be detected. Because of the low level of incorporation of the radiolabel into phospholipids and to avoid the difficulties in the identification of comigrating phospholipids after in vivo labeling, we conducted in vitro lipid phosphorylation assays to study the phosphoinositide metabolism of G. sulphuraria. Microsomal membranes were isolated from G. sulphuraria cells at 5, 7, 10, and 12 d after transfer and incubated with [γ-32P]ATP. The changes in the lipid-phosphorylation profile with the culture age are illustrated in Figure 1A. In vitro lipid-phosphorylation products were quantified using a scanner (Bioscan) (Fig. 1B). Between 5 to 7 d and 12 d of the 20-d culture period, PA decreased from about 40% of the total 32P-labeled phospholipid to less than 5%. In contrast, at the same time PIP2 increased from about 1% at d 7 to 9% by d 12 (Fig. 1B). The most prevalent phospholipid formed in vitro during the growth period was PIP, with values between 62% and 83% of the total phosphorylated lipid.
The decrease in PA formation during the culture period of G. sulphuraria could have resulted from decreases in either endogenous DAG or DAG kinase activity. To distinguish between these possibilities, DAG levels in the microsomal preparations were compared. Quantitative in vitro phosphorylation assays were carried out using 1 unit per assay of a recombinant E. coli DAG kinase and the extracted lipids from equal amounts of microsomal protein from 7- and 12-d-old G. sulphuraria cells and, for comparison, rat liver and Arabidopsis as the phosphorylation substrates. Approximately equal amounts of PA were formed in vitro by the E. coli enzyme from all of the samples (Table I), indicating that microsomal DAG levels were similar. Based on these data, we conclude that the described differences in PA formation by the isolated microsomes resulted from changes in DAG kinase activity during the culture period of G. sulphuraria. The data are summarized in Table I.
Table I.
DAG content and DAG kinase specific activity was measured in microsomes prepared from G. sulphuraria, Arabidopsis, and rat liver
| Source | Relative DAG Content | Endogenous DAG Kinase Activity | PA Formed by Endogenous DAG Kinase |
|---|---|---|---|
| % relative to Arabidopsis | pmol min−1 mg−1 protein | % of total phosphorylated lipid | |
| Arabidopsis | 100 ± 20 | 2100 ± 200 | 75 ± 10 |
| G. sulphuraria | |||
| d 7 | 117 ± 23 | 1280 ± 150 | 40 ± 5 |
| d 12 | 119 ± 24 | 515 ± 75 | 20 ± 2 |
| Rat liver | 93 ± 19 | 680 ± 85 | 20 ± 7 |
The relative DAG content in microsomes from each species was determined by incubating total lipids extracted from 500 μg of microsomal protein with [γ-32P]ATP and 1 unit per assay of a recombinant E. coli DAG kinase. The amount of PA formed in vitro by the E. coli enzyme is given relative to the amount formed with Arabidopsis microsomes. Values are averaged from three independent experiments.
PIP2 Biosynthesis and PIP2 Levels Increase with Time in Culture
Microsomes prepared from d-7 cells showed 3-fold lower specific PIP kinase activity than those from d-12 cells when excess PIP was added to the in vitro assay (Fig. 2A). These data indicate that the specific activity of the microsomal PIP kinase increased between d 7 and 12 of the culture period. To determine whether levels of the reaction product, PIP2, consistently increased between d 7 and 12, we performed mass measurements of whole-cell PIP2. In cells from 7 d after transfer, the PIP2 was 25% lower (910 ± 100 pmol g−1 fresh weight, n = 3) than that of the cells assayed at 12 d after transfer (1200 ± 150 pmol g−1 fresh weight, n = 3). Whereas there was increased PIP2 biosynthesis based on both in vivo and in vitro data, we have no indication of increased PIP2 hydrolysis, because there were never significant differences in basal IP3 levels between cells from d 7 and 12 of the culture period (both at 42 ± 30 pmol g−1 fresh weight, n = 6 each).
Figure 2.
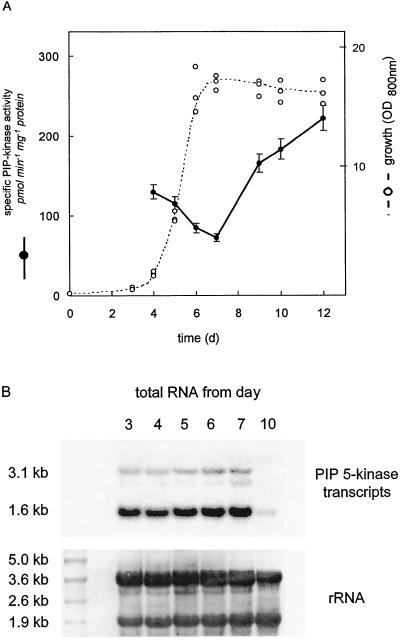
A, Specific activity of PIP kinase was assayed in vitro in microsomes prepared at the indicated times during the culture period of G. sulphuraria. The activity increased from 75 pmol min−1 mg−1 protein on d 7 to 230 pmol min−1 mg−1 protein on d 12. Data are from one representative experiment. The experiment was repeated twice and the results were similar. B, Total RNA was prepared at different times during the culture period, separated by gel electrophoresis, blotted onto a nylon membrane, and probed with a full-length PIP 5-kinase cDNA. The top panel shows the autoradiograph of the northern blot. Two transcripts were detected (3.1 and 1.6 kb). PIP 5-kinase mRNA levels did not appear to correlate with PIP kinase specific activity (A). The bottom panel shows the blot stained with methylene blue to illustrate equal loading of total RNA.
Regulation of PIP Kinase Activity
Northern-blot experiments were performed to investigate whether the changes in microsomal PIP kinase specific activity during the culture period were correlated with changes in PIP 5-kinase mRNA levels. Total RNA was prepared from G. sulphuraria at different times during the culture period and probed with a full-length PIP 5-kinase cDNA clone from Arabidopsis. Two transcripts could be detected (1.6 and 3.1 kb; Fig. 2B, top), which corresponded in size to PIP 5-kinase transcripts obtained with Arabidopsis total RNA (data not shown). During the culture period, PIP 5-kinase mRNA levels (Fig. 2B, top) did not change in a pattern similar to that of the changes in PIP kinase activity (Fig. 2A). In fact, at d 10, when the specific microsomal PIP kinase activity was already high, only very low levels of PIP 5-kinase mRNA were detected. These data imply that the increase in PIP kinase specific activity in the late-stationary-phase cells resulted from posttranslational regulation of the enzyme, from a stimulation of a PIP kinase-activating factor, or from a similar regulatory mechanism affecting enzyme activity. Alternatively, kinase activity and mRNA levels detected with our probe may not be correlated, implying the existence of significantly distinct PIP kinase isoforms.
Phosphoinositide Metabolism of G. sulphurariaChanges in Response to the Addition of Solutions to the Culture Medium
G. sulphuraria cells were very sensitive to the addition of solutions to the culture medium. Because the solutions used for osmostimulation were prepared with conditioned medium, before studying the effects of osmostimulation we had to characterize the effects of adding conditioned medium alone. Figure 3 illustrates changes in PIP2 formation and IP3 levels of d-12 cells that were untreated, treated with conditioned medium, or osmostimulated by the addition of 25 mm KCl in conditioned medium. The basal PIP2 formation from endogenous substrate for untreated cells was 200 ± 30 pmol min−1 mg−1 protein (Fig. 3A, dashed line). When d-12 cells were treated with conditioned medium, there was a transient 50% increase of in vitro PIP2 formation. For a comparison, the transient increase of in vitro PIP2 formation in response to stimulation of d-12 cells with 25 mm KCl in conditioned medium is also shown. Although the changes of in vitro PIP2 formation in microsomes from d-12 cells resulting from conditioned-medium treatment were small compared with those observed after osmostimulation, they were reproducible. Therefore, controls with conditioned medium were used for all time-course experiments to correct for the described effects. Conditioned medium had no effect on the specific PIP kinase activity of d-7 cells (data not shown).
Figure 3.
G. sulphuraria 12-d cells were sensitive to the addition of solutions to the culture medium. In vitro PIP2 formation (A) and IP3 levels (B) were measured in nontreated cells (□), in cells treated with conditioned medium (○), and in cells osmostimulated by 25 mm KCl in conditioned medium (▵). Conditioned medium did not affect PIP2 formation in microsomes from d-7 cells (data not shown). Values are the averages of duplicate assays from a representative experiment. Vertical bars indicate the duplicate range. The experiments were repeated twice and the trends were similar.
In d-12 cells, IP3 levels also changed transiently in response to the addition of conditioned medium (Fig. 3B). In untreated cells, no change in IP3 levels could be observed during an experiment (Fig. 3B, dashed line). However, within 10 s of the addition of conditioned medium, IP3 levels decreased below the limit of detection. The cells recovered by about 30 s. In d-7 cells, no effect of conditioned medium on IP3 levels was observed, but data were not taken from the 10-s time point, so an initial decrease in IP3 levels might have been missed. However, as with the d-12 cells, by 30 s the IP3 levels were not significantly different from those in the unstimulated control (data not shown). For a comparison, the effects of osmostimulation of d-12 cells with 25 mm KCl in conditioned medium are shown in Figure 3B.
Hypertonic Stimulation Results in a Transient Increase in IP3
Large differences in PIP kinase specific activity were detected between G. sulphuraria cells from 7 and 12 d after transfer (compare Fig. 2A). Because the ability of the cells to synthesize PIP2 was so different, we sought to determine whether there would be differences in their ability to produce IP3 in response to a stimulus. Cells from 7 and 12 d after transfer were stimulated by adding KCl, NaCl, or methyl-Man in conditioned culture medium (Fig. 4). For a better comparison, the data shown in Figure 4 are reported relative to the conditioned-medium control at each time point. Because methyl-Man gave identical responses as equal osmolal concentrations of KCl or NaCl, salt effects can be ruled out.
With both d-7 and -12 cells, osmostimulation evoked an initial decrease in IP3 levels, followed by an increase of up to 4-fold in cellular IP3 levels by 90 s (Fig. 4, C and D). The maximum peak IP3 level was 150 ± 30 pmol g−1 fresh weight (n = 5). After 15 min, the IP3 in both cell types returned to control values (data not shown). When cells from d 7 or 12 were stimulated with different concentrations of methyl-Man, the increase in IP3 at 90 s was proportional to the concentration of the osmostimulus applied up to 100 mosmol (Fig. 5).
Figure 5.
In both d-7 (○) and d-12 (□) cells, the production of IP3 increased with the osmostimulus. IP3 levels were quantified in G. sulphuraria cultures stimulated for 90 s with increasing concentrations of methyl-Man (5, 10, 50, and 100 mm final concentration). Data are the averages of four values from two experiments.
An Increase in PIP2 Biosynthesis Precedes the Increase in IP3 in Osmostimulated d-12 Cells
To investigate whether the IP3 production affected PIP2 biosynthesis, lipid-phosphorylation assays were performed with microsomes prepared at different times after osmostimulation. Surprisingly, when d-12 cells were stimulated, there was an increase of up to 5-fold in microsomal PIP kinase activity (Fig. 4B) within 60 s. This increase in PIP2 biosynthesis significantly preceded the maximum increase in IP3 production measured at 90 s. Between 1 and 3 min, in vitro PIP2 formation decreased to about 50% of the initial value (Fig. 4B), and within another 10 min it recovered back to the basal level (data not shown). A similar response to the stimulus was observed when lipid phosphorylation was investigated in the presence of added excess PIP in the in vitro assay (data not shown). The increase in PIP kinase activity was proportional to the concentration of the osmostimulus applied (Fig. 4B). In contrast, with d-7 cells, there was no significant change in PIP2 formation in response to osmostimulation, whether an endogenous (Fig. 4A) or an exogenous substrate was used (data not shown), even though similar changes in IP3 were observed in both cell types (Fig. 4, C and D).
DISCUSSION
There were distinct differences in lipid phosphorylation of microsomes from cells harvested throughout the growth cycle. First, at the transition from the logarithmic to the stationary phase of growth, PA formation decreased. Because microsomal DAG levels were not significantly different between growth stages of G. sulphuraria, the decrease in PA production must have resulted from a decrease in DAG kinase activity. The decreased DAG kinase activity in stationary G. sulphuraria cells may reflect a decreased demand for membrane lipid biosynthesis.
Second, PIP kinase specific activity increased 3-fold between d 7 and 12. The differences in PIP2 formation between these two time points were evident even when excess PIP was added, indicating that the apparent specific activity of the PIP kinase increased from d 7 to 12. Analysis of whole-cell PIP2 content gave consistent results and showed lower PIP2 levels in d-7 than in d-12 cells. The mass measurements indicated that the in vitro phosphoinositide kinase assay was a good measure of PIP2 levels in this system and confirmed the presence of PI-4,5-P2.
Having identified two distinct stages of cell growth, we continued to study changes in phosphoinositide metabolism in response to osmotic stimulation. Regardless of the differences in PIP2 biosynthesis, cells from d 7 and 12 exhibited similar changes in IP3 production after osmotic stimulation. The positive correlation between the concentration of the osmostimulus and the intensity of the short-term signals suggests that the stimulation did not immediately saturate the responsive capacity of the cells. For this study, we used changes in osmolarity of the culture medium of less than 10% and as low as 2% and detected significant effects on phosphoinositide metabolism. Previous studies of osmotic stress and phospholipid metabolism have used severalfold increases in osmolarity of the culture medium as a stimulus (Einspahr et al., 1988a, 1988b; Cho et al., 1993; Dove et al., 1997; Kearns et al., 1998; Mikami et al., 1998). By using mild osmostimulation, we were able to bypass difficulties arising from the complexity of parallel signaling events and gained insight into potentially distinct pools of PIP2.
In 12-d-old G. sulphuraria cells, hypertonic stimulation resulted in a rapid and transient increase in microsomal PIP kinase specific activity, which preceded and overlapped with an increase in IP3. Transient activation of PIP kinase was detected only in d-12 cells. This was surprising because nonstimulated G. sulphuraria cells from d 7 had lower whole-cell PIP2 content and lower microsomal PIP kinase specific activity in vitro, and we had anticipated a greater need for PIP2 biosynthesis in d-7 but not in d-12 cells. However, with the d-7 cells there was no significant increase in microsomal PIP kinase specific activity upon stimulation.
Our data suggest that the d-7 cells had sufficient PIP2 to produce the initial increase in IP3, whereas in the d-12 cells the newly synthesized PIP2 was the primary source of the IP3 formed. Estimated from the amount of IP3 produced after stimulation (approximately 150 ± 30 pmol g−1 fresh weight, n = 5) and the total PIP2 present in the cells before stimulation (910 ± 100 pmol g−1 fresh weight on d 7 and 1200 ± 150 pmol g−1 fresh weight on d 12), between 12% and 16% of the total cellular PIP2 is turned over during the generation of an IP3 signal in G. sulphuraria. The size of the signaling pool, therefore, is similar to that of animal cells, in which 10% to 20% of the cellular PIP2 is turned over during IP3 signaling (Fisher and Agranoff, 1986; Gross and Boss, 1993). Our data imply that the majority of PIP2 present in 12-d-old G. sulphuraria cells was not available for the production of IP3 and that a new PIP2 pool had to be established before IP3 signaling could occur.
The PIP2 present before stimulation in 12-d-old G. sulphuraria may be bound up in PIP2-binding proteins of the plasma membrane or involved in regulation of the actin cytoskeleton. A shift in cellular PIP2 toward a bound state would stabilize filamentous actin (Drøbak et al., 1994; Shibasaki et al., 1997; Staiger et al., 1997), as the cells enter a resting stage, to maintain cellular integrity under the aggressive culture conditions the cells are exposed to in nature (Gross et al., 1998). The physiological status of the cells affects the early signaling cascade. In d-12 G. sulphuraria, the initial step of the cascade is the activation of PIP kinase. A similar effect has been described by Lassing and Lindberg (1990) in platelets after thrombin stimulation, in which phosphoinositide kinases are activated and cause an increase in PIP2 biosynthesis before phospholipase C activation.
The data presented here remind us that the phosphoinositide pathway, like other metabolic pathways, is not a linear set of reactions but rather involves the complex regulation of concerted enzymatic events. An added complexity with this pathway is that the inositol phospholipids bind to many cellular proteins (Memon et al., 1989; Memon and Boss, 1990; Fukami et al., 1992; Drøbak et al., 1994; Hilgemann and Ball, 1996; Fan and Makielski, 1997; for review, see Janmey, 1994; Shibasaki et al., 1997; Staiger et al., 1997; Sun et al., 1997), including enzymes involved in their own trafficking (Kauffmann-Zeh et al., 1995; for review, see Cockroft, 1998; Kearns et al., 1998) and biosynthesis (Stevenson et al., 1998). These lipid-protein complexes define potentially unique subcellular domains or pools that facilitate and optimize different cellular functions identified for PIP2 (for review, see Toker, 1998). By comparing the signal transduction of cells at different stages of growth, we have now begun to characterize selective domains and to dissect functional pools of PIP2.
ACKNOWLEDGMENTS
We thank Dr. Phil Majerus (Washington University School of Medicine) for the inositol polyphosphate 5-phosphatase I cDNA and Dr. Bjørn Drøbak (John Innes Institute, Norwich, UK) for helpful discussion.
Abbreviations:
- DAG
diacylglycerol
- IP3
inositol-1,4,5-trisphosphate
- PA
phosphatidic acid
- PCA
perchloric acid
- PI
phosphatidylinositol
- PIP
phosphatidylinositol monophosphate
- PIP2
phosphatidylinositol-4,5-bisphosphate
Footnotes
This research was supported by the National Aeronautics and Space Administration (grant no. NAGW-4984 to W.F.B.) and a Deutscher Akademischer Austauschdienst fellowship (HSP III to I.H.) financed by the German Federal Ministry of Education, Science, Research, and Technology.
LITERATURE CITED
- Auethavekiat V, Abrams CS, Majerus PW. Phosphorylation of platelet pleckstrin activates inositol polyphosphate 5-phosphatase I. J Biol Chem. 1997;272:1786–1790. doi: 10.1074/jbc.272.3.1786. [DOI] [PubMed] [Google Scholar]
- Borochov A, Cho MH, Boss WF. Plasma membrane lipid metabolism of petunia petals during senescence. Physiol Plant. 1994;90:279–284. [Google Scholar]
- Cho MH, Boss WF (1995) Transmembrane signaling and phosphoinositides. In DW Galbraith, HJ Bohnert, DP Bourque, eds, Methods in Plant Cell Biology: Part A. Academic Press, New York, pp 543–554 [DOI] [PubMed]
- Cho MH, Chen Q, Okpodu CM, Boss WF. Separating and detecting inositol phospholipids. LC-GC. 1992;10:464–468. [Google Scholar]
- Cho MH, Shears SB, Boss WF. Changes in phosphatidylinositol metabolism in response to hyperosmotic stress in Daucus carotaL. cells grown in suspension culture. Plant Physiol. 1993;103:637–647. doi: 10.1104/pp.103.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockroft S. Phosphatidylinositol transfer proteins: a requirement in signal transduction and vesicle traffic. BioEssays. 1998;20:423–432. doi: 10.1002/(SICI)1521-1878(199805)20:5<423::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol 3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Drøbak BK, Watkins PA, Valenta R, Dove SK, Lloyd CW, Staiger CJ. Inhibition of plant plasma membrane phosphoinositide phospholipase C by the actin binding protein, profilin. Plant J. 1994;6:389–400. [Google Scholar]
- Einspahr KJ, Maeda M, Thompson GA. Concurrent changes in Dunaliella salinaultrastructure and membrane phospholipid metabolism after hyperosmotic shock. J Cell Biol. 1988a;107:529–538. doi: 10.1083/jcb.107.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einspahr KJ, Peeler TC, Thompson GA. Rapid changes in polyphosphoinositide metabolism associated with the response of Dunaliella salinato hypoosmotic shock. J Biol Chem. 1988b;263:5775–5779. [PubMed] [Google Scholar]
- Falkenau C, Heim S, Wagner KG. Effect of cytokinins on the phospholipid phosphorylation of the suspension cultured Catharanthus roseuscells. Plant Sci. 1987;50:173–178. [Google Scholar]
- Fan Z, Makielski JC. Anionic phospholipids activate ATP-sensitive potassium channels. J Biol Chem. 1997;272:5388–5395. doi: 10.1074/jbc.272.9.5388. [DOI] [PubMed] [Google Scholar]
- Fisher SK, Agranoff BW (1986) Phosphoinositide turnover in the CNS and in neural-related tissues. In JW Putney, Jr, ed, Phosphoinositides and Receptor Mechanisms. Alan R. Liss, New York, pp 219–243
- Fukami K, Furuhashi K, Inagaki M, Endo T, Hatano S, Takenawa T. Requirement of phosphatidylinositol 4,5-bisphosphate for α-actinin function. Nature. 1992;359:150–152. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- Gross W, Boss WF. Inositol phospholipids and signal transduction. In: Verma DPS, editor. Control of Plant Gene Expression. Boca Raton, FL: CRC Press; 1993. pp. 17–32. [Google Scholar]
- Gross W, Küver J, Tischendorf G, Bouchaala N, Büsch W. Cryptoendolithic growth of the red alga Galdieria sulphurariain volcanic areas. Eur J Phycol. 1998;33:25–31. [Google Scholar]
- Gross W, Schnarrenberger C. Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol. 1995;36:633–638. [Google Scholar]
- Heim S, Wagner KG. Evidence of phosphorylated phosphatidylinositols in the growth cycle of suspension cultured plant cells. Biochem Biophys Res Commun. 1986;134:1175–1182. doi: 10.1016/0006-291x(86)90374-8. [DOI] [PubMed] [Google Scholar]
- Hilgemann DW, Ball R. Regulation of cardiac Na+, Ca2+ exchange and KATP potassium channels by PIP2. Science. 1996;273:956–959. doi: 10.1126/science.273.5277.956. [DOI] [PubMed] [Google Scholar]
- Janmey P. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- Kauffmann-Zeh A, Thomas GMH, Ball A, Prosser S, Cunningham E, Cockroft S, Hsuan JJ. Requirement for phosphatidylinositol transfer protein in epidermal growth factor signalling. Science. 1995;268:1188–1190. doi: 10.1126/science.7761838. [DOI] [PubMed] [Google Scholar]
- Kearns MA, Monks DE, Fang M, Rivas MP, Courtney PD, Chen J, Prestwich GD, Theibert AB, Dewey RE, Bankaitis VA. Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypasses the requirement for an essential phosphatidylinositol transfer protein in yeast. EMBO J. 1998;17:4004–4017. doi: 10.1093/emboj/17.14.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Polyphosphoinositide synthesis in platelets stimulated with low concentrations of thrombin is enhanced before the activation of phospholipase C. FEBS Lett. 1990;262:231–233. doi: 10.1016/0014-5793(90)80197-q. [DOI] [PubMed] [Google Scholar]
- Memon AR, Boss WF. Rapid light-induced changes in phosphoinositide kinases and H+-ATPase in plasma membranes of sunflower hypocotyls. J Biol Chem. 1990;265:14817–14821. [PubMed] [Google Scholar]
- Memon AR, Chen Q, Boss WF. Inositol phospholipids activate plasma membrane ATPase in plants. Biochem Biophys Res Commun. 1989;162:1295–1301. doi: 10.1016/0006-291x(89)90814-0. [DOI] [PubMed] [Google Scholar]
- Merola A, Castaldo R, DeLuca P, Gambardella R, Musachio A, Taddei R. Revision of Cyanidium caldarium: three species of acidophilic algae. G Bot Ital. 1981;115:189–195. [Google Scholar]
- Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;15:563–568. doi: 10.1046/j.1365-313x.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- Satterlee JS, Sussman MR. An Arabidopsis phosphatidylinositol 4-phosphate 5-kinase homolog with seven novel repeats rich in aromatic and glycine residues (accession no. AF019380) (PGR 97-150) Plant Physiol. 1997;115:864. [Google Scholar]
- Shibasaki Y, Ishihara H, Kizuki N, Asano T, Oka Y, Yazaki Y. Massive actin polymerization induced by phosphatidylinositol 4-phosphate 5-kinase in vivo. J Biol Chem. 1997;272:7578–7581. doi: 10.1074/jbc.272.12.7578. [DOI] [PubMed] [Google Scholar]
- Smith DW, Brock TD. The water relations of the alga Cyanidium caldariumin soil. J Gen Microbiol. 1973;79:219–231. [Google Scholar]
- Staiger CJ, Gibbon BC, Kovar DR, Zonia LE. Profilin and actin-depolymerizing factor: modulators of actin organization in plants. Trends Plant Sci. 1997;7:275–281. [Google Scholar]
- Stevenson JM, Perera IY, Boss WF. A phosphatidylinositol 4-kinase pleckstrin homology (PH) domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem. 1998;273:22761–22767. doi: 10.1074/jbc.273.35.22761. [DOI] [PubMed] [Google Scholar]
- Sun H, Lin K, Yin HL. Gelsolin modulates phospholipase C activity in vivothrough phospholipid binding. J Cell Biol. 1997;138:811–820. doi: 10.1083/jcb.138.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Curr Opin Cell Biol. 1998;10:254–261. doi: 10.1016/s0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- Walsh JP, Caldwell KK, Majerus PW. Formation of phosphatidylinositol 3-phosphate by isomerization from phosphatidylinositol 4-phosphate. Proc Natl Acad Sci USA. 1991;88:9184–9187. doi: 10.1073/pnas.88.20.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]