Abstract
Polar body formation is an essential step in forming haploid eggs from diploid oocytes. This process involves completion of a highly asymmetric cytokinesis that results in a large egg and two small polar bodies. Unlike mitotic contractile rings, polar body contractile rings assemble over one spindle pole so that the spindle must move through the contractile ring before cytokinesis. During time-lapse imaging of C. elegans meiosis, the contractile ring moved downward along the length of the spindle and completed scission at the midpoint of the spindle, even when spindle length or rate of ring movement was increased. Patches of myosin heavy chain and dynamic furrowing of the plasma membrane over the entire embryo suggested that global cortical contraction forces the meiotic spindle and overlying membrane out through the contractile ring center. Consistent with this model, depletion of myosin phosphatase increased the velocity of ring movement along the length of the spindle. Global dynamic furrowing, which was restricted to anaphase I and II, was dependent on myosin II, the anaphase promoting complex and separase, but did not require cortical contact by the spindle. Large cortical patches of myosin during metaphase I and II indicated that myosin was already in the active form before activation of separase. To identify the signal at the midpoint of the anaphase spindle that induces scission, we depleted two proteins that mark the exact midpoint of the spindle during late anaphase, CYK-4 and ZEN-4. Depletion of either protein resulted in the unexpected phenotype of initial ingression of a polar body ring with twice the diameter of wild type. This phenotype revealed a novel mechanism for minimizing polar body size. Proteins at the spindle midpoint are required for initial ring ingression to occur close to the membrane-proximal spindle pole.
Keywords: Polar Body, Meiosis, Anaphase, Centralspindlin, Cytokinesis, C. elegans
Introduction
In the oocytes of most animals, half of the homologous chromosomes are eliminated by extrusion into a first polar body during meiosis I and half of the remaining sister chromatids are expelled into a second polar body during meiosis II. Polar body failure results in polyploid embryos and embryonic lethality. Polar body extrusion and mitotic cleavage in C. elegans both require non-muscle myosin II (Shelton et al., 1999) suggesting similar mechanisms. However, the geometric relationship between a mitotic spindle and mitotic contractile ring differs dramatically from the relative positions of a female meiotic spindle and a polar body contractile ring. Mitotic contractile rings assemble either between spindle poles or over the spindle midzone (Bringmann and Hyman, 2005) ensuring that cleavage occurs between separated sister chromatids when the ring contracts in diameter. In contrast, polar body contractile rings assemble on the surface of the oocyte over the extreme end of the spindle pole contacting the cortex (Ma et al., 2006; Maro and Verlhac, 2002; Pielak et al., 2004). If the polar body contractile ring simply contracted in diameter, both sets of segregating chromosomes would remain in the oocyte. Successful extrusion of chromosomes into a polar body thus requires that the spindle move outward through the contractile ring or that the contractile ring move inward down the length of the spindle (Pielak et al., 2004; Zhang et al., 2008).
The mechanism that moves the spindle relative to the polar body contractile ring has not been demonstrated in any species. One hypothesis is that global contraction of myosin throughout the cortex might drive membrane out through the hole in the center of the cortical actomyosin ring. In this model, one pole of the meiotic spindle is anchored to the cortex in the center of the myosin-free hole within the contractile ring, global contraction then forces the spindle through the hole. However, other types of membrane protrusion are generated by actin-polymerization (Small et al., 2002; Bugyi et al., 2008; Mellor, 2010) rather than by global contraction, and cortical pulling mechanisms move the spindle through the contractile ring in budding yeast (Moore and Cooper, 2010). Distinguishing between these possibilities has been stymied by the fact that inhibition of myosin contractility with small molecule inhibitors results in disassembly of the polar body contractile ring (Deng et al., 2007) thus making it impossible to test whether myosin contractility is required to move the spindle through the polar body contractile ring.
A second vital question is how polar body extrusion is coordinated with the cell cycle to ensure that it does not occur before anaphase. One model is that the rho-GEF, ECT-2, becomes activated by transfer to the spindle midzone during anaphase. ECT-2 then activates myosin II through rho kinase and simultaneously activates formin-dependent actin polymerization (Piekny and Mains, 2002). However in several species, constitutively active myosin allows normal mitotic cytokinesis (Dean and Spudich, 2006; Uyeda and Spudich, 1993). Thus it is not clear whether inactivation of myosin during metaphase contributes to ensuring that polar body extrusion or mitotic cleavage occurs after anaphase. A more likely timing mechanism for polar body extrusion is suggested by the finding that separase is required for polar body formation in both mouse (Kudo et al., 2006) and C. elegans (Bembenek et al., 2007) since separase activity is inhibited by securin during metaphase and becomes active when securin is targeted for destruction by the anaphase promoting complex/cyclosome (APC/C). The mechanism by which separase induces polar body formation, however, is not known.
We approached these questions using time-lapse imaging of living C. elegans meiotic embryos where we could unambiguously track the movements of the meiotic spindle, contractile ring and plasma membrane. The C. elegans metaphase I spindle has a steady state pole-to-pole length of 7.7 μm and associates with the cortex in a parallel orientation (McNally et al., 2006). After activation of the APC/C, the spindle shortens to 4.5 μm in length before rotating to a perpendicular orientation relative to the cortex (McNally et al., 2006). Homolog separation and further shortening result in an early anaphase spindle that is 2.8 μm in length with the chromosomes at the ends (McNally et al., 2006). The spindle then narrows and lengthens (McNally and McNally, 2005). While it is possible that spindle shortening might contribute to the formation of a small polar body, the precise timing of polar body membrane pulling away from the eggshell near the spindle and scission relative to the spindle shortening cycle has not been investigated previously. Here, we investigate the roles of actomyosin and the centralspindlin complex during polar body formation. Our results suggest a role for global actomyosin contractility in movement of the spindle through the polar body contractile ring. Global contractility was dependent on the cell cycle and actomyosin, but not spindle contact at the cortex. Further, we demonstrate a role for the centralspindlin components CYK-4 and ZEN-4 in regulating the initial diameter of the polar body contractile ring. Thus we have revealed a previously unrecognized mechanism that contributes to minimizing polar body size.
Results
The velocity of polar body ring ingression is doubled by hyperactivating myosin contraction
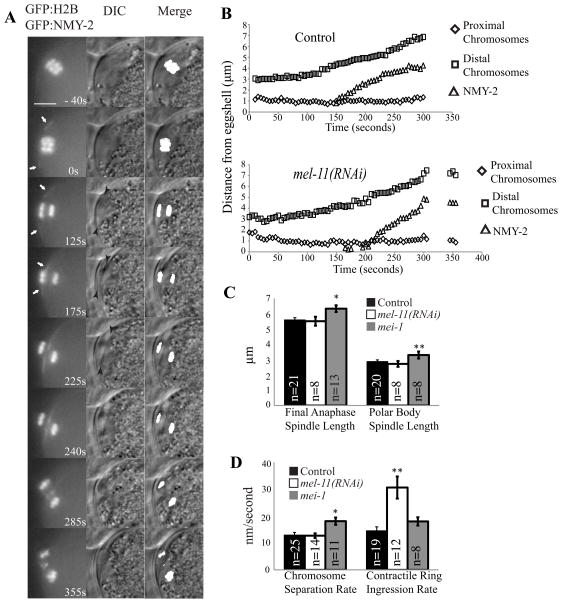
Previous studies (Dorn et al., 2010; Ma et al., 2006; Zhang et al., 2008) have not simultaneously filmed the contractile ring, chromosomes and a stationary reference point, thus the velocity of the ring moving down the spindle or the velocity of the spindle moving outward through the ring have never been reported. We used the C. elegans eggshell as a stationary reference point. Although the eggshell is formed by sequential depositon of discrete layers during meiosis, the initial chitinous layer forms during prometaphase I in response to fertilization (Marcello and Singson, 2010) and can be imaged by differential interference contrast (DIC) microscopy. We simultaneously filmed the stationary eggshell (by DIC), GFP:NMY-2 (non-muscle myosin), which marks the contractile ring, and GFP:histone H2B, which labels the chromosomes during polar body formation in C. elegans meiotic embryos in utero. When viewed in cross section, the contractile ring first became visible during spindle rotation as two lines of NMY-2 (Fig. 1A, 0 s arrows and Video 1). During early anaphase, chromosomes moved to the ends of the spindle (Fig. S1) so that the membrane-proximal chromosomes were closely opposed to the plasma membrane, which was closely opposed to the stationary eggshell (Fig. 1A, 125 s). The membrane-proximal chromosomes remained stationary relative to the eggshell as the contractile ring and associated membrane both constricted in diameter and also moved inward down the length of the elongating spindle (Fig. 1A, 175 s, and Videos 1 and 2). Thus the velocity of contractile ring movement away from the eggshell and proximal chromosomes corresponds to the velocity of ring movement down the spindle (Fig. 1B and 1D). Here we define the movement of the ring down the length of the spindle as “ingression” to distinguish it from constriction (the reduction in diameter of the ring).
Figure 1. Hyperactivation of myosin contractility by depletion of myosin phosphatase doubles the velocity of polar body ring ingression down the length of the spindle.
(A) Images of GFP:histone and GFP:NMY-2 fluorescence and DIC within a meiotic embryo are shown from a representative time-lapse sequence of chromosome separation and contractile ring movement during polar body formation in meiosis I (to = completion of spindle rotation). For the merge column, GFP:histone pixels above an arbitrary threshold were converted to white, then merged with the DIC image to show chromosome placement. White arrows in column one highlight the contractile ring labeled with GFP:NMY-2. Black arrowheads in column two highlight the membrane ingression away from the eggshell. Scale bar, 5 μm. (B) Chromosome and contractile ring movements during polar body formation, measured as distance from the eggshell. Proximal chromosomes are the chromosomes closest to the cortex, which are extruded into the polar body (to = completion of spindle rotation). Apparent delayed ingression in mel-11(RNAi) in this example was not reproducible (see Table 1). (C) Spindle length at completion of anaphase and length of the spindle within the polar body at scission. (D) Rate of movement of chromosomes away from each other during anaphase and rate of movement of the GFP:NMY-2-labeled contractile ring away from the eggshell and proximal chromosomes in control, mel-11(RNAi) and mei-1(ct103) embryos (*p < 0.01; **p < 0.005; all others p > 0.05 ).
Previous attempts to ask whether myosin contractility drives polar body ring ingression along the length of the spindle have been hampered by the fact that the polar body contractile ring disperses or fills in when treated with small molecule inhibitors of myosin II (Deng et al., 2007) and because myosin is required for migration of the MI spindle to the cortex (Schuh and Ellenberg, 2008) and rotation of the MII spindle (Matson et al., 2006) in mouse oocytes. To circumvent these problems, we tested whether hyperactivation of myosin contractility by depleting the myosin phosphatase regulatory subunit, MEL-11, (and thus increasing myosin regulatory light chain phosphorylation) might increase the rate of polar body ring ingression as it does the rate of mitotic cleavage furrow constriction (Piekny and Mains, 2002). Indeed the rate of polar body ring ingression was doubled in mel-11(RNAi) embryos (Fig. 1B and 1D) whereas other aspects of spindle and chromosome movement were unaffected (Figs. 1C and D and S2). This result strongly suggests that polar body ring ingression is driven by myosin contraction. If the spindle was pulled through the contractile ring by a microtubule motor as in budding yeast mitosis (Moore et al., 2009), hyperactivation of myosin would not increase the velocity of movement.
Polar body extrusion is accompanied by global, myosin-dependent furrowing of the plasma membrane
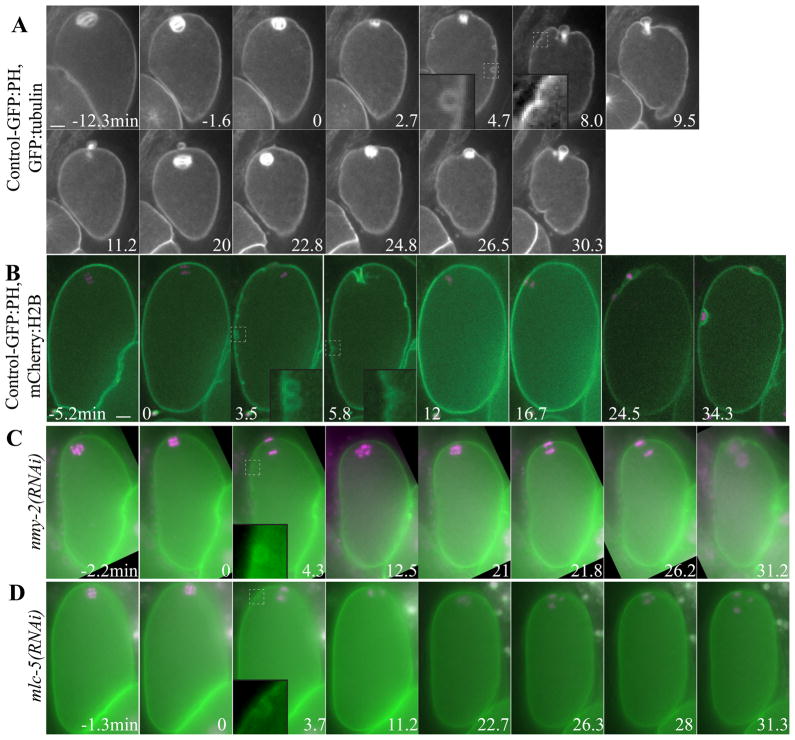
One possible model explaining how myosin contraction could drive movement of the ring down the spindle is that global cortical contractility of the entire embryo cortex “squeezes” out the polar body through the contractile ring, the center of which is void of actin in C. elegans (Dorn et al., 2010) and void of myosin in C. elegans and other species (Shelton et al., 1999; Pielak et al., 2003; Maddox et al., 2007). Evidence for global contractility around the entire oocyte/embryo during polar body extrusion, however, has not been reported in any species. Large cortical myosin patches and transient membrane furrowing have been reported as evidence of global contractility in polarizing C. elegans embryos after completion of meiosis (Chartier et al., 2011; Cowan and Hyman, 2004; Munro et al., 2004). To test for global contractility during polar body formation, we filmed meiotic embryos expressing GFP:tubulin and the plasma membrane marker, GFP:PH PLCδ1 (Audhya et al., 2005). After the meiotic spindle rotated (Fig. 2A, 0 min; Video 3) and shortened (Fig. 2A, 2.7 min), cortical granule exocytosis (CGE) was observed as round pockets of plasma membrane (Fig. 2A, 4.7 min) (Bembenek et al., 2007). During or shortly after CGE, ring ingression around the spindle was visible (Fig. 2A, 4.7 min; Table 1) and then dynamic furrowing of the entire membrane was visible (Fig. 2A, 8–9.5 min; Table 1). After or during completion of polar body extrusion, determined by spindle breakdown and retraction of the membrane around the central spindle, dynamic furrowing ceased (Fig. 2A, 11.2–20 min; Table 1). Dynamic furrowing re-initiated during anaphase II (Fig. 2A, 22.8–30.3 min; Table I) without a visible second wave of CGE. Anaphase II dynamic furrowing continued through pseudocleavage (Fig. S3B), suggesting that anaphase II dynamic furrowing is the same as that previously described during polarization. Time-lapse imaging of GFP:tubulin, GFP:PH embryos allowed analysis of the timing of CGE and furrowing relative to the spindle cycle (Table 1) whereas imaging of embryos with mCherry:histone-labeled chromosomes and GFP:PH-labeled plasma membrane (Fig. 2B) allowed determination of the timing of CGE and dynamic furrowing relative to anaphase chromosome separation (Table 2). This is the first quantitative documentation of global membrane furrowing that corresponds temporally with polar body extrusion.
Figure 2. Global, myosin-dependent membrane furrowing occurs during extrusion of the first and second polar body.
Images from representative time-lapse sequences of (A) wild-type embryo expressing GFP:PH and GFP:tubulin showing rotation (−1.6–0 min), spindle shortening (2.7 min), CGE, spindle elongation and ingression of the membrane next to the spindle (4.7 min) and dynamic furrowing and polar body formation (8–9.5 min) in meiosis I. Then the membrane relaxes (11.2–20 min) until the spindle rotates again (22.8 min) and dynamic furrowing resumes and a polar body forms (26.5–30.3 min) in meiosis II (to = completion of spindle rotation). (B) Wild-type embryo with mCherry:histone and GFP:PH showing chromosomes separating perpendicular to the membrane (0 min), CGE (3.5 min), early membrane furrowing near the spindle (3.5 min), dynamic furrowing (5.8 min) and polar body completion and relaxation of the membrane after meiosis I (5.8–12 min). Dynamic furrowing resumes during anaphase II (24.5 min). (C) nmy-2(RNAi) embryo and (D) mlc-5(RNAi) embryo. Insets (5 μm × 5 μm) show CGE (4.7 min in A, 3.5 min in E, 4.3 min in C, 3.7 min in D) or plasma membrane furrows (8 min in A, 5.8 min in B) magnified from the dashed box in the original image. to = completion of spindle rotation. Scale bar, 5 μm.
Table 1.
Wild-type timing of spindle (GFP:tubulin) and cortical (GFP:PH) events in meiosis I and meiosis II.
| Average Time (min) | SEM | Minimum | Maximum | N | ||
|---|---|---|---|---|---|---|
| Meiosis I | Spindle Rotated | 0 | ||||
| Spindle Shortened | 1.8 | 0.12 | 1.2 | 2.8 | 13 | |
| CGE Starts | 3.0 | 0.20 | 2.2 | 4.0 | 8 | |
| Ring Ingression Visible | 3.6 | 0.16 | 2.8 | 4.8 | 13 | |
| Global Furrowing Begins | 4.1 | 0.29 | 3.2 | 6.7 | 11 | |
| CGE Complete | 4.4 | 0.32 | 3.4 | 6.2 | 8 | |
| Global Furrowing Stops | 7.1 | 0.51 | 5.8 | 10.0 | 9 | |
| Polar Body Complete | 7.2 | 0.39 | 4.9 | 10.5 | 14 | |
| Meiosis II | Spindle Rotated | 0 | ||||
| Spindle Shortened | 1.9 | 0.20 | 1.4 | 2.3 | 6 | |
| Global Furrowing Begins | 1.5 | 0.97 | −2.3 | 4.8 | 8 | |
| Ring Ingression Visible | 3.3 | 0.51 | 0.4 | 14.3 | 9 | |
| Meiosis I to Meiosis II | Rotation to Rotation | 20.4 | 1.62 | 14.4 | 26.7 | 7 |
Table 2.
Wild-type and tba-1(RNAi) timing (in min ± SEM) of chromosome (mCherry:histone) and cortical (GFP:PH) events in meiosis I and meiosis II.
| Wild Type (min) | tba-1(RNAi) (min) | ||
|---|---|---|---|
| Meiosis I | Spindle Rotated | −0.78 ± 0.14 (n=6) | N/A |
| Chromosomes Separate | 0 | 0 | |
| CGE Starts | 1.9 ± 0.44 (n=6) | 2.6 ± 0.27 (n=6) | |
| Ring Ingression Visible | 3.3 ± 0.42 (n=7) | N/A | |
| CGE Complete | 4.3 ± 0.29 (n=6) | 4.0 ± 0.30 (n=6) | |
| Global Furrowing Begins | 4.8 ± 0.63 (n=7) | 4.8 ± 0.39 (n=6) | |
| Global Furrowing Stops | 8.2 ± 0.96 (n=7) | 6.2 ± 0.64 (n=7) | |
| Polar Body Complete | 7.8 ± 0.95 (n=7) | N/A | |
| Meiosis II | Spindle Rotated | −0.69 ± 0.25 (n=8) | N/A |
| Chromosomes Separate | 0 | 0 | |
| Ring Ingression Visible | 0.88 ± 0.59 (n=7) | N/A | |
| Global Furrowing Begins | 0.97 ± 1.2 (n=6) | 0.27 ± 1.2 (n=10) | |
| Meiosis I | Distance From Cortex (μm) | 1.0 ± 0.15 (n=6) | 8.19 ± 0.48 (n=8) |
| Meiosis II | Distance From Cortex (μm) | 1.2 ± 0.21 (n=6) | 8.38 ± 0.46 (n=11) |
To test if dynamic furrowing during meiotic anaphase is due to actomyosin contractility, we knocked down the myosin II heavy chain, NMY-2, using RNAi. nmy-2(RNAi) embryos in strains expressing mCherry:histone and GFP:PH completed spindle rotation (Fig. 2C, 0 min), CGE (Fig. 2C, 4.3 min; n = 7/7 embryos) and anaphase I and II (Fig. 2C, 4.3 min and 21.8–26.2 min; n = 8/8 embryos) normally, but dynamic furrowing (n = 7/8 embryos) and polar body extrusion (n = 8/8 embryos) failed (Fig. 2C), suggesting that dynamic furrowing is due to actomyosin contractility. RNAi knockdown of the recently described myosin essential light chain, MLC-5 (Gally et al., 2009), also eliminated dynamic furrowing in meiotic embryos (Fig. 2D; n = 10/11 embryos) and caused polar body extrusion failure (n = 11/11 embryos), while CGE was normal (Fig. 2D, 3.7 min; n = 11/11 embryos) and anaphase I and II both occurred (Fig. 3D, 3.7–11.2 min and 26.3–31.3 min). Together, these results demonstrate that dynamic furrowing is dependent on actomyosin contractility.
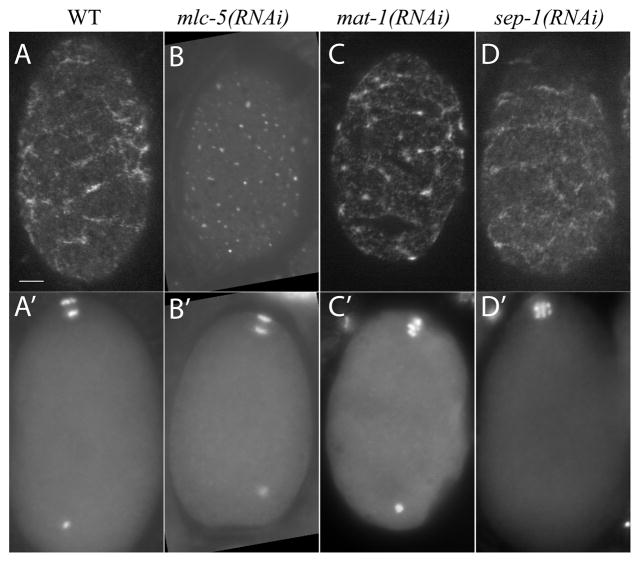
Figure 3. Myosin heavy chain forms large cortical patches indicative of contractility during metaphase and anaphase.
Surface plane of fixed GFP:NMY-2 embryos in early anaphase I (A, B and D) or arrested in metaphase I (C), fed (A) control (L4440) (B) mlc-5, (C) mat-1 or (D) sep-1 RNAi bacterial strains. Staging was determined by DAPI stained chromosome configuration in a different focal plane (A′-D′). The extra DAPI spot is the sperm DNA. Large patches of GFP:NMY-2 were converted to small puncta in embryos depleted of myosin essential light chain (MLC-5) suggesting that large patches are generated by active contraction. Large patches were present on APC/C-depleted and separase-depleted embryos. Scale bar, 5 μm.
GFP fusions and antibody staining of one-cell embryos after meiosis have shown a cortical meshwork of actin and myosin on the surface of the entire embryo (Guo and Kemphues, 1996; Munro et al., 2004). To determine if this meshwork is present during polar body extrusion in meiosis, we imaged embryos with GFP:histone and GFP:NMY-2 or GFP:NMY-2 only. Both live and fixed images showed a meshwork of large patches of GFP:NMY-2 on the surface of early anaphase meiotic embryos (Fig. 3, A and A’; n = 14 MI, n = 10 MII). GFP:NMY-2 appeared as small puncta (0.7 μm average diameter; n = 44 embryos; Fig. 3B) in mlc-5(RNAi) embryos during meiotic anaphase instead of larger patches seen in wild-type embryos throughout meiosis (1.9 μm average diameter; n = 27 embryos; Fig. 3A). Because myosin essential light chain is required for myosin heavy chain contraction (Szent-Gyorgyi et al., 1999), this result further suggests that active, contractile myosin is present around the entire embryo cortex during polar body extrusion, an observation that has not previously been reported in any species.
The APC/C and separase are required to initiate global furrowing and polar body extrusion but are not required for myosin activation
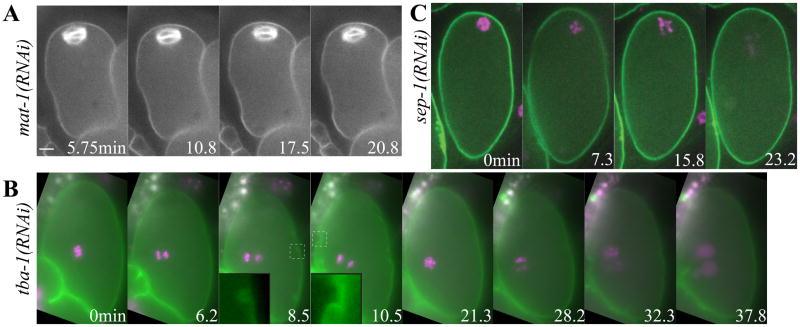
To address the question of how global furrowing and polar body extrusion are repressed during metaphase I, activated in anaphase I, repressed again in metaphase II, then re-activated during anaphase II, we filmed anaphase promoting complex/cyclosome (APC/C)-depleted and separase-depleted embryos. In embryos depleted of the APC/C subunit MAT-1, spindles maintained a constant length of 10.7 μm (n = 24 spindles), did not rotate (6/6 embryos), as previously described (Ellefson and McNally, 2009), and dynamic furrowing of the plasma membrane did not occur (6/6 embryos) over a period of at least 20 min (Fig. 4A). In contrast, wild-type spindles shorten to 5.1 μm by the time of spindle rotation, which occurs 8.7–9.7 min after spermatheca exit (Yang et al., 2005). The APC/C, however, does not appear to activate myosin contractility directly, as large GFP:NMY-2 patches were present on mat-1(RNAi) metaphase-arrested embryos (n = 27; Fig. 3C).
Figure 4. Global cortical contraction requires the APC/C and separase, but does not require contact of the spindle or chromosomes with the cortex.
Images from representative time-lapse sequences of (A) mat-1(RNAi) metaphase-arrested embryo showing a long, parallel spindle and a non-furrowing membrane (to = exit from spermatheca). (B) tba-1(RNAi) embryo from a worm expressing GFP:PH and mCherry:histone showing anaphase I (6min) and anaphase II (28.2 min) far from the membrane. CGE (8.5 min), anaphase I dynamic furrowing (10.5 min) and anaphase II dynamic furrowing (28.2–32.3 min) occurred with wild-type kinetics (to = start of chromosome separation). (C) sep-1(RNAi) embryos from worms expressing GFP:PH and mCherry:histone showing metaphase (0 min) and abnormal anaphase (15.8 min) (to= completion of spindle rotation) but no furrowing. Insets (5 μm × 5 μm) show CGE or dynamic furrows magnified from the dashed box in the original image. Scale bar, 5 μm.
Because mat-1(RNAi) spindles do not rotate, the failure to initiate dynamic furrowing might be caused by the failure of one spindle pole or one set of chromosomes to come into close contact with the cortex. To test whether chromosome or spindle contact with the cortex is required to initiate dynamic furrowing, we looked at chromosomes (mCherry:histone) and the plasma membrane (GFP:PH) in tba-1(RNAi) embryos in which tubulin-dependent translocation to the cortex does not occur (Yang et al., 2003). In these embryos, the meiotic chromosomes remained in the center of the embryo (Fig. 4B and Video 4), but CGE (Fig. 4B, 8.5 min) and dynamic furrowing still occurred shortly after chromosome separation in both meiosis I (Fig. 4B, 10.5 min) and meiosis II (Fig. 4B, 32.3 min; Table 2). This result indicates that neither spindle nor chromosome contact at the cortex are the cue for membrane furrowing during meiosis, and the APC/C must activate furrowing by another mechanism. In addition, a centrally located spindle appears to attract a deep furrow (Fig. 4B and Video 4), suggesting the meiotic central spindle may attract the membrane during polar body formation.
One conserved target of the APC/C is cyclin B, whose destruction inactivates CDK-1 (Murray, 2004; Pesin and Orr-Weaver, 2007). Mimicking cyclin B proteolysis by small molecule inhibition of CDK-1 induces cytokinesis in vertebrate cells arrested in metaphase (Potapova et al., 2006), suggesting that cyclin B degradation is sufficient to initiate cytokinesis. Small molecule inhibition of CDK-1 in metaphase I-arrested, mat-1(RNAi) C. elegans meiotic embryos induced spindle shortening from a length of 10.7 μm (n = 24) to 5.1 μm (n = 15; p < 0.0001) (Fig. S4) as previously described (Ellefson and McNally, 2011), however, dynamic furrowing did not occur over a period of at least 20 min of imaging (n = 11/11 embryos), suggesting that furrowing is not prevented by CDK-1 activity as it is in vertebrate mitotic cells.
Another conserved target of the APC/C is securin, which inhibits separase until securin is degraded through ubiquitination by the APC/C (Nasmyth, 2002). SEP-1, C. elegans separase, is required for homolog separation (Siomos et al., 2001), CGE and polar body extrusion (Bembenek et al., 2007) but the mechanism by which separase induces polar body formation is not known. RNAi knockdown of SEP-1 prevented dynamic furrowing during meiotic anaphase (Fig. 4C; n = 10/10 embryos). This is the first time that separase has been linked to global contractility in any system. SEP-1 depletion also reduced the extent of anaphase chromosome separation (sep-1(RNAi) 4.0 μm; n = 9 vs. control 5.6 μm; n = 8), prevented CGE in 5/9 embryos and prevented polar body extrusion in 10/10 embryos, consistent with previous studies (Bembenek et al., 2007; Siomos et al., 2001). The presence of CGE and chromosome segregation in some embryos may represent incomplete RNAi knockdown. Alternatively, Siomos et al. (2001) showed that the anaphase-like chromosome movement in sep-1(RNAi) does not necessarily reflect homolog separation in anaphase I, as homologs travelled to the same side of the spindle. Large patches of GFP:NMY-2, indicative of contractile myosin, were present on the cortex of sep-1(RNAi) embryos during meiosis (Fig. 3D; n = 17/20), suggesting that separase does not activate furrowing by activating myosin contractility. Global contractility may be important for moving the spindle through the polar body ring, but we next wanted to investigate how scission of the contractile ring is specified to ensure proper physical segregation of half of the meiotic chromosomes into the polar body.
Polar body ring scission occurs at the exact midpoint of the anaphase spindle
A relationship between meiotic spindle length and polar body size has been suggested previously (McNally et al., 2006; Verlhac et al., 2000) but not definitively demonstrated, in part because mutants with long spindles have other defects that might be the cause of the large polar bodies. Our quantitative analysis of wild-type polar body ring ingression revealed that scission occurs at the exact midpoint of the anaphase spindle when the spindle reaches its longest length (Figs. 1, A–C and S2B). Scission still occurred at the exact midpoint when the velocity of ring ingression was doubled in mel-11(RNAi) embryos or when the final length of the elongating spindle was increased in a mutant [mei-1(ct46ct103) (Clandinin and Mains, 1993)] of katanin, a microtubule-severing protein known to regulate meiotic spindle length (McNally et al., 2006) (Figs. 1, B–D and S2B). These results suggest that some molecule at the midpoint of the spindle might activate scission and that the final length of the anaphase spindle must contribute to determining polar body size.
Passenger proteins are distributed along the entire length of the C. elegans meiotic anaphase spindle (Dumont et al., 2010) and thus are unlikely to induce scission at the spindle midpoint. In contrast, the kinesin-6 family member, ZEN-4, localizes in a discrete stripe at the C. elegans meiotic spindle midpoint (Dumont et al., 2010; Powers et al., 1998) making it a prime candidate for the molecular marker identifying the spindle midpoint.
Centralspindlin ensures that the polar body ring ingresses close to the spindle
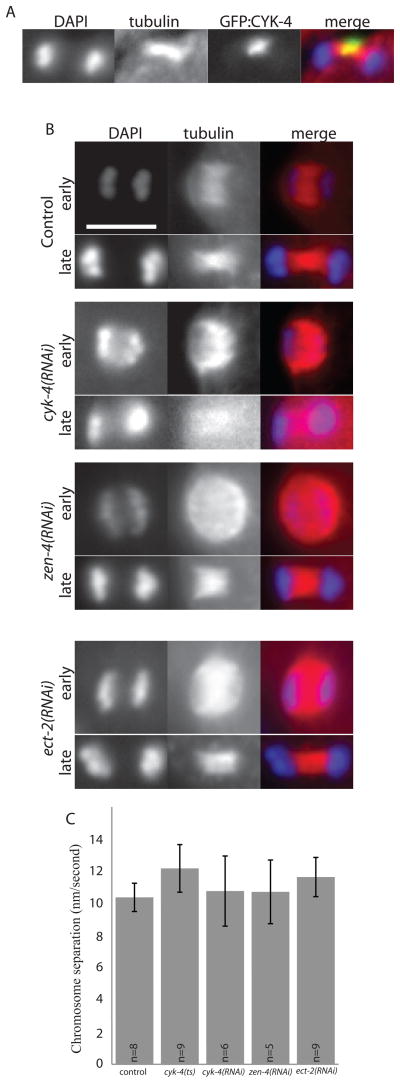
ZEN-4 forms the conserved centralspindlin complex with the GTPase-activating protein (GAP), CYK-4 (Glotzer, 2009), and sometimes associates with the conserved rhoGEF, ECT-2 (Glotzer, 2009). All three of these proteins are required for mitotic cytokinesis (Jantsch-Plunger et al., 2000; Powers et al., 1998) and polar body formation (Sonnichsen et al., 2005) but the exact role in polar body formation has not been reported. To test if CYK-4, like ZEN-4, localizes to the center of the meiotic spindle, we fixed embryos from a strain expressing GFP:CYK-4 and stained for tubulin and DAPI. In 5/5 meiotic spindles observed, GFP:CYK-4 localized to the center of the meiotic anaphase spindle, only co-localizing with the central region of microtubules (Fig. 5A). Unlike mitotic central spindles, meiotic central spindles treated with cyk-4(RNAi) were intact in both early and late anaphase (Fig. 5B). Because the meiotic spindle is intact and chromosome separation still occurred when we knocked down ZEN-4 (n=9), CYK-4 (n=8) or ECT-2 (n=5) (Fig. 5B and C), it is unlikely that cytokinesis failure in meiosis is due to lack of a central spindle, as is thought to be the case in mitosis.
Figure 5. Meiotic spindle microtubule structure is unaffected by loss of centralspindlin.
(A) shows central spindle localization of GFP:CYK-4 on a fixed meiotic anaphase spindle, stained to show tubulin (red) and chromosomes (DAPI, blue). (B) shows meiotic anaphase spindles fixed and stained to show tubulin (red) and chromosomes (DAPI, blue) in early and late anaphase under conditions of control RNAi, cyk-4(RNAi), zen-4(RNAi) or ect-2(RNAi), as labeled. Scale bar, 5 μm. (C) shows anaphase chromosome separation rates for control or centralspindlin-depleted embryos (all, p > 0.1).
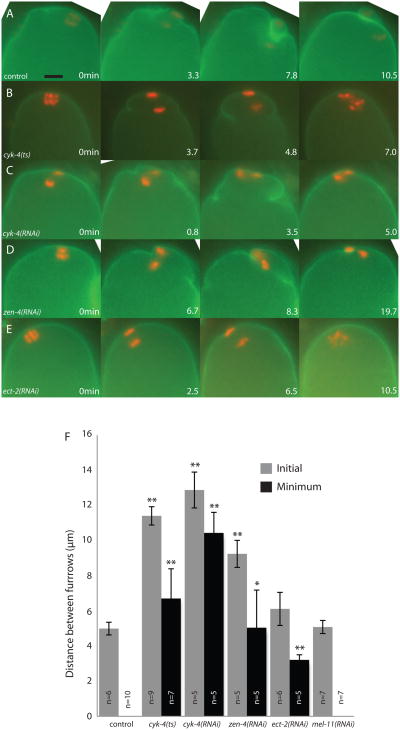
Live imaging of meiotic embryos expressing GFP:PH and mCherry:histone revealed a completely novel phenotype when components of centralspindlin were depleted. While the contractile ring ingresses directly around the meiotic chromosomes anchored at the cortex in wild-type meiotic anaphase (Fig. 6A and S5A), depletion of ZEN-4 or CYK-4 with feeding RNAi resulted in extremely wide ingressions, far from the meiotic chromosomes and spindle, with an initial furrow to furrow distance that was twice the width of wild type (Figs. 6, C, D and F and S5, C and D; Video 5). In addition these furrows regressed, causing polar body failure in both cyk-4(RNAi) (n = 7) and zen-4(RNAi) (n = 5) (Fig. 6 and S5; Video 5). To ensure that the wide contractile ring phenotype was not due to partial depletion, we examined the meiotic phenotype of a cyk-4(ts) mutant, cyk-4(or749ts). This mutant blocks completion of cytokinesis in mitotic embryos (Canman et al., 2008). We did not see any marked difference between this mutant and cyk-4(RNAi) embryos (Figs. 6 and S5), suggesting that the wide contractile ring is due to complete loss of CYK-4.
Figure 6. CYK-4 and ZEN-4 minimize polar body size by ensuring that the contractile ring ingresses adjacent to the membrane proximal chromosomes.
Images from representative time-lapse sequences of embryos progressing through anaphase I of meiosis expressing GFP:PH and mCherry:histone. (A) A wild-type control embryo shows contractile ring ingression tightly around the anaphase spindle and polar body completion, while (B) cyk-4(or749ts), (C) cyk-4(RNAi) and (D) zen-4(RNAi) show wide furrows moving toward the anaphase spindle between the separating chromosomes. The wide furrows regress, allowing the chromosomes to rejoin. (E) ect-2(RNAi) results in little or no ingression of a contractile ring, resulting in polar body failure. (F) The distance between the furrows nearest to the meiotic spindle when they first appear (Initial = grey bars) and when they are in closest proximity (Minimum = black bars). A distance of 0 μm represents touching of the furrows. (*p < 0.005; **p < 0.001) (to = start of chromosome separation). Scale bar, 5 μm.
Inhibition of ECT-2 with RNAi showed a completely different phenotype from the other components of centralspindlin. In 3/9 ect-2(RNAi) embryos, furrows were not present at all (Fig. 6E and F and S5E; Video 6) and in the remaining embryos the furrow-to-furrow distance was more similar to that of wild-type embryos; however, the furrows did not ingress along the spindle as far as in wild-type. The range of phenotypes observed in ect-2(RNAi) embryos likely reflects different degrees of protein depletion. Since none of the ect-2(RNAi) embryos exhibited the wide contractile ring phenotype seen in cyk-4(RNAi) and zen-4(RNAi) embryos, these results demonstrate that CYK-4 and ZEN-4 have an additional function that is discrete from targeting ECT-2 (Glotzer, 2009). The wide contractile ring in zen-4(RNAi) and cyk-4(RNAi) embryos indicates that centralspindlin ensures that ring ingression occurs close to the membrane proximal chromosomes in wild type, thus minimizing the size of the polar body.
Discussion
Global contraction model for polar body formation
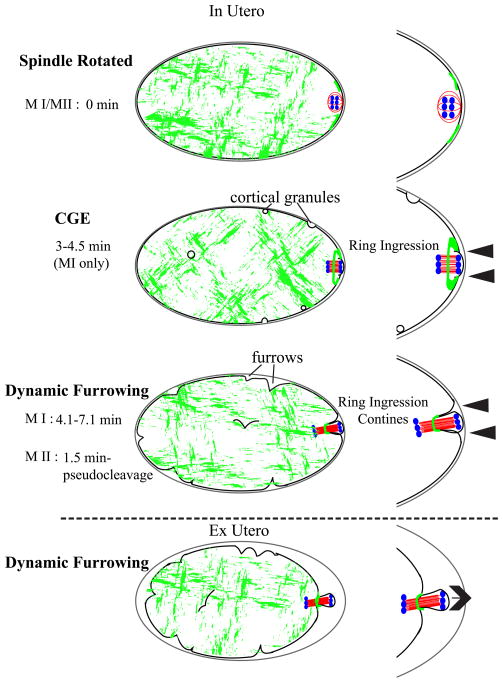
Our data support the hypothesis that global contraction pushes the meiotic spindle through the contractile ring. Polar body extrusion in mouse (Deng and Li, 2009) and clam (Pielak et al., 2004) initiates as an outward protrusion of the plasma membrane that occurs as the cortex-proximal spindle pole moves outward through the myosin-free hole in the center of the contractile ring. Our time-lapse imaging of GFP:NMY-2 and GFP:PH in C. elegans meiotic embryos filmed under in utero osmotic conditions revealed that the plasma membrane over the cortex-proximal spindle pole is pressed against the eggshell, most likely by turgor. The contractile ring and associated plasma membrane then move inward, away from the eggshell, down the length of the spindle (Fig. 7). Both the outward membrane protrusion observed in oocytes of organisms without eggshells and the inward movement of the contractile ring in C. elegans could be explained by a common mechanism in which actomyosin contraction of the entire embryo pushes membrane and cytoplasm out through the less rigid, myosin-free hole in the contractile ring (Fig. 7). In the case of C. elegans, the presence of a membrane closely-opposed to the rigid eggshell paired with outward force of the polar body pushing through the contractile would convert the resulting outward force into inward pushing of the contractile ring. However, hyperosmotic conditions resulting from dissecting embryos from the worm may allow the membrane to pull further away from the eggshell, resulting in what looks like a polar body being pushed through a stationary contractile ring (Fig. 7, ex utero). The finding that depletion of MEL-11 increases the rate of movement of the contractile ring down the length of the spindle strongly supports this model. Further support of a global contraction mechanism comes from the observation of large GFP:myosin patches and dynamic furrowing on the surface of the entire embryo. Ingression of the contractile ring was observed just before dynamic furrowing of the rest of the embryo, a pattern similar to that reported for cortical granule exocystosis, which initiates at the spindle then progresses away from the spindle (Bembenek et al., 2007). Another explanation for ingression of the contractile ring occurring before global furrowing is that less force may be required for initial extrusion through a weaker point of the cortex (within the contractile ring), than for global furrowing.
Figure 7. Global contraction model for polar body formation.
After APC/C activation, the spindle rotates to contact the cortex pole-first. There is an actomyosin ring on the surface of the embryo with a more flexible actomyosin-free center through which the spindle is pushed. Force is generated by global actomyosin contractility and the spindle midzone confers specificity of scission of the actomyosin contractile ring. In utero, it appears that the contractile ring is moving inward along the spindle (arrowheads), while in hyperosmotic conditions ex utero, the polar body appears to be pushed out (arrow) while the ring remains stationary. Grey-eggshell, black-membrane/cortex, red-microtubules, green-myosin, blue-chromosomes.
Cell cycle regulation of polar body formation
In metaphase-arrested mouse oocytes, the contractile ring forms on the cortex but protrusion of membrane through the ring does not occur until the oocyte is induced to proceed into anaphase (Deng and Li, 2009). Likewise, polar body contractile ring ingression and global dynamic furrowing in C. elegans occurred only during anaphase I and anaphase II in wild-type embryos and did not occur in APC/C-depleted, metaphase I-arrested embryos. In budding yeast (Thornton and Toczyski, 2003) and Drosophila embryos (Oliveira et al., 2010), there are only two essential substrates for the APC/C, cyclin B and securin. Ubiquitin-dependent proteolysis of cyclin B leads to inactivation of the cyclin-dependent kinase, CDK-1. Mimicking cyclin B degradation with the CDK-1 inhibitor, purvalanol A did not induce dynamic membrane furrowing or polar body extrusion in metaphase-arrested embryos, in contrast with vertebrate mitotic cells (Potapova et al., 2006). Proteolysis of securin leads to activation of separase, which cleaves cohesin allowing anaphase chromosome separation (Nasmyth, 2002). Separase has been shown to be required for polar body formation in C. elegans (Bembenek et al., 2007) and mouse (Kudo et al., 2006). In C. elegans, separase is required for secretion of cortical granules (Bembenek et al., 2007) which we found initiates just before dynamic furrowing. In this study we found that separase is required for dynamic furrowing around the entire meiotic embryo, but not for large myosin patches. These results support a model in which cortical actomyosin is already contractile at metaphase (indicated by large myosin patches in metaphase-arrested embryos), then APC/C activation of separase drives CGE which allows the contractile actomyosin to pull the plasma membrane away from the eggshell generating dynamic furrowing. Dynamic furrowing squeezes the cytoplasm and membrane out through the hole in the contractile ring.
The role of centralspindlin in minimizing polar body size
If the spindle is squeezed through the contractile ring by a global force, it is necessary for the contractile ring to complete scission before the entire spindle is inside the polar body, but not before it passes the first set of chromosomes. This would ensure that exactly one set of chromosomes is disposed of. In this study, the contractile ring consistently bisected the meiotic spindle after spindle elongation, even when the velocity of ring ingression was increased by RNAi of MEL-11, indicating that a signal from the central spindle might trigger completion of scission. This signal may position the contractile ring, holding it in place until scission has completed or, as suggested in frog oocytes (Zhang et al., 2008), the central spindle signal might increase contractility when the contractile ring reaches the central spindle, allowing rapid scission at that point. In a simplistic model, we would expect depletion of the central spindle signal to cause ingression of the contractile ring past the midpoint of the spindle. Depletion of CYK-4 or ZEN-4 had the unexpected phenotype of initial ingression much further away from the membrane proximal chromosomes than in wild-type. This novel phenotype revealed a completely new mechanism for minimizing polar body size, minimizing initial contractile ring diameter, but precluded a clear interpretation about the central spindle signal.
A relationship between meiotic spindle length and polar body size has been suggested previously from analysis of mos mutant mice (Choi et al., 1996; Verlhac et al., 2000) and katanin mutant C. elegans (McNally et al., 2006) but the relationship between these measurements was previously unclear because of the defective spindle organization observed in both cases. We found that the wild-type meiotic spindle is perfectly bisected, indicating that the length of the spindle is one determinant of the size of the polar body. Other determinants include asymmetric spindle positioning and anillin-dependent shaping of the polar body (Dorn et al., 2010) which occurs after the ring has reached the midpoint of the spindle.
Materials and Methods
C. elegans strains
In these studies, wild type was one of several integrated transgenic strains. The integrated GFP:NMY-2, GFP:histone strain was made by crossing zuIs45 (nmy-2::NMY-2::GFP) (Nance et al., 2003) males with GFP:histone (AZ212) (Praitis et al., 2001) hermaphrodites. We also used the following integrated strains: GFP:PHPLCδ1, GFP:tubulin (OD73) (Maddox et al., 2007) and GFP:PH PLCδ1, mCherry:histone (OD95) (Green et al., 2008). The GFP:CYK-4 strain used was MG110 from (Jantsch-Plunger et al., 2000). The mei-1 experiments were done using the mei-1(ct46ct103) mutant (Clandinin and Mains, 1993) crossed into the GFP:NMY-2, GFP:histone strain above. The CYK-4 GAP mutant used was cyk-4(or749ts) from (Canman et al., 2008).
In utero filming
Adult hermaphrodites were anesthetized as described previously (Kirby et al., 1990; McCarter et al., 1999) and placed on a thin agarose pad and covered with a coverslip. Imaging was carried out, as described previously (Yang et al., 2003), with four different microscopes. 1: Microphot SA (Nikon, Tokyo, Japan) equipped with a 60x PlanApo 1.4 objective and a charge-coupled device ([CCD], Qimaging Retiga Exi Fast 1394 camera). Excitation light from an HBO100 light source was attenuated with a heat and UV reflecting “hot mirror” (Chroma Technology, Bellows Falls, VT) and a 25% transmission neutral density filter. A GFP long pass filter set (Omega Optical, Brattleboro, VT) was used. Excitation light was shuttered with a Sutter shutter controlled by a Sutter Lambda 10-3 controller and IVision software (BioVision Technologies, Exton, PA) (Fig. 1A; Videos 1 and 2). 2: Olympus IX71 inverted microscope with a U-RFL-T mercury burner power supply and 60x PlanApo N.A. 1.42 objective and equipped with a Hamamatsu Orca R2 C10600-10B digital CCD camera. A GFP long pass filter set (Omega Optical, Brattleboro, VT) was used. Excitation light was shuttered with a Sutter shutter controlled by a Sutter Lambda 10-3 controller and Metamorph (MDS Analytical Technologies, Sunnyvale, CA) software (Figs. 2, C and D; 3; 4B; 5; 6; S3; S4 and S5; Videos 4, 5 and 6). An Olympus DSU (disk scanning unit) on this microscope was used only for Figs. 3, 5 and S3A. 3: A Marianas Real Time Confocal SDC Workstation spinning disc confocal microscope (Intelligent Imaging Innovations) employing a CSU-X1 Yokogawa scan head and a modified Cascade QuantEM 512SC EMCCD camera. Objective lenses were either a Zeiss 63x NA 1.4 or Zeiss 100x NA 1.46 oil immersion. Images were acquired using the SlideBook 5.0 software (Intelligent Imaging Innovations) (Figs. 2, A and B; 4, A and C; and S3B; Video 3). 4: A Perkin-Elmer UltraView spinning disk confocal microscope equipped with an Olympus 100X Plan Apo 1.35 objective, a 488 nm Argon laser, Hamamatsu Orca ER CCD and Slidebook acquisition software (Fig. S1). 1×1 binning was used for all live imaging. All live filming was done at 23–25° C with images captured at 5-, 10- or 15-s intervals.
Osmotic sensitivity
Siomos et al. (2001) reported that separase-depleted mitotic embryos, dissected from their mothers, are osmotically sensitive and that any mitotic cleavage defect in sep-1(RNAi) embryos could be rescued by relieving mechanical pressure on the embryo. The osmotic and mechanical sensitivity of sep-1(RNAi) embryos is due to an eggshell defect caused by failure to secrete cortical granules at anaphase I (Bembeneck et al., 2007). Wild-type meiosis I embryos are osmotically sensitive because they have not yet secreted these cortical granules required to generate an impermeable eggshell. Culturing meiosis I embryos ex utero thus requires osmotic support medium (Bembenek et al., 2007; Dumont et al., 2010; Monen et al., 2005). All of our studies were conducted in utero. Because progression of wild-type embryos through meiosis I and II occurred normally, we infer that wild-type C. elegans adults maintain the osmolality of the fluid in the uterus so that osmotically-sensitive embryos can develop normally before secretion of the final components of the eggshell. Thus differences between wild-type and sep-1(RNAi) embryos are unlikely to be due to the osmotic sensitivity of sep-1(RNAi) embryos.
RNAi by feeding
Feeding RNAi bacterial clones for mel-11(RNAi), cyk-4(RNAi), zen-4(RNAi), ect-2(RNAi), mlc-5(RNAi), nmy-2(RNAi) and mat-1(RNAi) were from the Ahringer feeding library (Kamath et al., 2003). For tba-1(RNAi) we used the feeding clone from Yang et al. (2003). The sep-1(RNAi) feeding clone was made using the EcoRI-XhoI restriction fragment from the cDNA clone yk429h5 in the L4440 plasmid in HT115 cells. Feeding was carried out as described previously (Timmons et al., 2001). 24 hour feeding was used for mel-11(RNAi), cyk-4(RNAi), zen-4(RNAi), ect-2(RNAi), mat-1(RNAi), sep-1(RNAi) and tba-1(RNAi) and 30–36 hour feeding was used for mlc-5(RNAi) and nmy-2(RNAi) to allow effective knockdown. 24 hour feeding was performed on L4 larvae and 30–36 hour RNAi was performed on approximately L3 larvae. Thus, all of our analyses were on embryos within adult worms of similar stage, usually containing a single row of embryos. Hatch rates were counted for all feeding strains and resulted in increased embryonic lethality from the control feeding strain (vector only).
Purvalanol A Injection
mat-1(RNAi) GFP:PH, GFP:tubulin worms were injected using standard injection methods (Mello et al., 1991) placed into a drop of mineral oil on a desicated agarose pad on a cover slip. Using a glass needle, 100 mM purvalanol A (Tocris) in DMSO was injected into the uterus through the vulva or by puncturing the surrounding body wall. The worms were quickly recovered in M9 buffer and mounted onto a 3% agarose pad on a glass slide for filming. The time between injection and filming was 3–5 min.
Immunostaining
Immunostaining in Figs. 2 and S3A was done using standard freeze-fracture methods followed by methanol and paraformaldehyde (PFA) fixation. Embryos adhered to polylysine-coated slides were then incubated with primary antibodies. The same method, but without PFA was used for immunostaining shown in Fig. 5. Anti-tubulin DM1α (Sigma-Aldrich, St. Louis, MO) was used at 1:200 diluted in phosphate-buffered saline with 0.05% Tween 20. All secondary antibodies (Invitrogen, Carlsbad, CA) and 4,6-Diamidino-2-phenylindole (DAPI) were diluted 1:200 in PBS-T. All immunostained embryos were imaged using microscope 2.
Measurements and Quantification
Contractile ring and chromosome movements (Figs. 1 and S2) were measured using iVision software (Biovision). All measurements of tba-1(RNAi) embryos and wild-type GFP:PH, mCherry:histone embryos were done in Metamorph software (MDS Analytical Technologies). Spindle length in mat-1(RNAi) (with or without purvalanol A) embryos was measured using Metamorph software. Timing of contractility and polar body formation were done using iVision, Metamorph or ImageJ (NIH) software.
When using the eggshell as a stationary point for measurements of contractile ring and chromosome movements, we used an overlaid spot to ensure that the point of the eggshell used for measurement did not move from frame to frame except after occasional refocusing
Supplementary Material
Time-lapse sequence of GFP:NMY-2 and GFP:histone fluorescence during meiosis in a wild-type worm corresponds to Fig. 1A. Images were captured at 5-s intervals. Display rate is 5 frames/s.
Time-lapse images of a meiotic spindle labeled with GFP:tubulin (green) and mCherry:histone H2B (red). After spindle rotation and shortening, the arrangement of the spindle microtubules changes so the chromosomes are on the ends of the spindle (10.2–12.8 min) through completion of anaphase. This justifies the use of chromosome position to infer spindle position in Fig. 1. The cell cortex is highlighted for clarity of spindle rotation (to = completion of spindle rotation). Scale bar, 5 μm.
(A) Total duration of chromosome segregation in meiotic anaphase and (B) placement of the GFP:NMY-2-labeled contractile ring on the meiotic spindle at the completion of anaphase as a percentage of spindle length in control, mel-11(RNAi) and mei-1 embryos. All, p > 0.05.
DIC time-lapse sequence during meiosis in a wild-type worm corresponds to Fig. 1A. Images were captured at 5-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH and GFP:tubulin fluorescence during meiosis in a wild-type worm corresponds to Fig. 2A. Images were captured at 10-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH (green) and mCherry:histone (magenta) fluorescence during meiosis in a tba-1(RNAi) worm corresponds to Fig. 4B. Images were captured at 10-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH (green) and mCherry:histone (red) fluorescence during meiosis in a cyk-4(RNAi) worm corresponds to Figs. 6C and S5C. Images were captured at 15-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH (green) and mCherry:histone (red) fluorescence during meiosis in an ect-2(RNAi) worm corresponds to Figs. 6E and S5E. Images were captured at 15-s intervals. Display rate is 5 frames/s.
(A) Image of cortical GFP:NMY-2 at the anterior half of the cell in a fixed early mitotic embryo. (B) Images from a representative time-lapse sequence of wild-type embryos expressing GFP:PH and mCherry:histone showing the end of anaphase II and continuation of dynamic furrowing into pseudocleavage (25.6–33.5 min) during pronuclear migration (*) and pronuclear meeting (36.8 min) (to = completion of spindle rotation). Scale bar, 5 μm.
Images from a representative time-lapse sequence of a GFP:tubulin; GFP:PH mat-1(RNAi) metaphase-arrested embryo exposed to the CDK-1 inhibitor, purvalanol A (to = start of imaging). Scale bar, 5 μm.
Images from representative time-lapse sequences of embryos progressing through anaphase II of meiosis expressing GFP:PH and mCherry:histone. (A) A wild-type control embryo shows contractile ring ingression tightly around the anaphase spindle and polar body completion, while (B) cyk-4(or749ts), (C) cyk-4(RNAi) and (D) zen-4(RNAi) show wide furrows moving toward the anaphase spindle between the separating chromosomes, which regress, allowing the chromosomes to rejoin. (E) ect-2(RNAi) results in little or no ingression of a contractile ring, resulting in polar body failure. (to = start of chromosome separation) Scale bar, 5 μm.
Highlights.
Contractile ring ingression associated with polar body formation is directly driven by myosin contractility
Polar body extrusion is accompanied by global, APC/C- and myosin-dependent furrowing of the plasma membrane
Centralspindlin minimizes polar body size by ensuring that the polar body ring ingresses close to the spindle.
Acknowledgments
We thank Ed Munro, Julie Canman, Michael Glotzer, Karen Oegema, John White, Karen McNally and the Caenorhabditis Genetics Center for strains and Yuji Kohara for cDNA clones. We thank Lesilee Rose for advice and use of facilities throughout this work and Dan Starr, Karen McNally and Marina Ellefson for critical reading of the manuscript
This work was supported by National Institute of General Medical Sciences grant 1R01GM-079421 (to F.J.M.).
Abbreviations
- APC/C
anaphase promoting complex/cyclosome
- CGE
cortical granule exocytosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–79. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek JN, Richie CT, Squirrell JM, Campbell JM, Eliceiri KW, Poteryaev D, Spang A, Golden A, White JG. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 2007;134:3837–48. doi: 10.1242/dev.011361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–4. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 2008;322:1543–6. doi: 10.1126/science.1163086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier NT, Salazar Ospina DP, Benkemoun L, Mayer M, Grill SW, Maddox AS, Labbe JC. PAR-4/LKB1 mobilizes nonmuscle myosin through anillin to regulate C. elegans embryonic polarization and cytokinesis. Curr Biol. 2011;21:259–69. doi: 10.1016/j.cub.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Mains PE. Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics. 1993;134:199–210. doi: 10.1093/genetics/134.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–53. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Deng M, Li R. Sperm chromatin-induced ectopic polar body extrusion in mouse eggs after ICSI and delayed egg activation. PLoS One. 2009;4:e7171. doi: 10.1371/journal.pone.0007171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Suraneni P, Schultz RM, Li R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev Cell. 2007;12:301–8. doi: 10.1016/j.devcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Dorn JF, Zhang L, Paradis V, Edoh-Bedi D, Jusu S, Maddox PS, Maddox AS. Actomyosin tube formation in polar body cytokinesis requires Anillin in C. elegans. Curr Biol. 2010;20:2046–51. doi: 10.1016/j.cub.2010.10.030. [DOI] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol. 2010;12:894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson ML, McNally FJ. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2722–30. doi: 10.1091/mbc.E08-12-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson ML, McNally FJ. CDK-1 inhibits meiotic spindle shortening and dynein-dependent spindle rotation in C. elegans. J Cell Biol. 2011;193:1229–44. doi: 10.1083/jcb.201104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally C, Wissler F, Zahreddine H, Quintin S, Landmann F, Labouesse M. Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development. 2009;136:3109–19. doi: 10.1242/dev.039412. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Audhya A, Pozniakovsky A, Dammermann A, Pemble H, Monen J, Portier N, Hyman A, Desai A, Oegema K. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 2008;85:179–218. doi: 10.1016/S0091-679X(08)85009-1. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 1996;382:455–8. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–15. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, Ellenberg J, de Boer P, Nasmyth K. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–46. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Ma C, Benink HA, Cheng D, Montplaisir V, Wang L, Xi Y, Zheng PP, Bement WM, Liu XJ. Cdc42 activation couples spindle positioning to first polar body formation in oocyte maturation. Curr Biol. 2006;16:214–20. doi: 10.1016/j.cub.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox AS, Lewellyn L, Desai A, Oegema K. Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev Cell. 2007;12:827–35. doi: 10.1016/j.devcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Marcello MR, Singson A. Fertilization and the oocyte-to-embryo transition in C. elegans. BMB Rep. 2010;43:389–99. doi: 10.5483/bmbrep.2010.43.6.389. [DOI] [PubMed] [Google Scholar]
- Maro B, Verlhac MH. Polar body formation: new rules for asymmetric divisions. Nat Cell Biol. 2002;4:E281–3. doi: 10.1038/ncb1202-e281. [DOI] [PubMed] [Google Scholar]
- Matson S, Markoulaki S, Ducibella T. Antagonists of myosin light chain kinase and of myosin II inhibit specific events of egg activation in fertilized mouse eggs. Biology of Reproduction. 2006;74:169–176. doi: 10.1095/biolreprod.105.046409. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–28. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–91. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev Biol. 2005;282:218–30. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–55. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- Moore JK, Cooper JA. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21:283–9. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009;66:546–55. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–24. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–34. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Nance J, Munro EM, Priess JR. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–50. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–65. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat Cell Biol. 2010;12:185–92. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 2007;3:e202. doi: 10.1371/journal.pgen.0030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny AJ, Mains PE. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J Cell Sci. 2002;115:2271–82. doi: 10.1242/jcs.115.11.2271. [DOI] [PubMed] [Google Scholar]
- Pielak RM, Gaysinskaya VA, Cohen WD. Cytoskeletal events preceding polar body formation in activated Spisula eggs. Biol Bull. 2003;205:192–3. doi: 10.2307/1543247. [DOI] [PubMed] [Google Scholar]
- Pielak RM, Gaysinskaya VA, Cohen WD. Formation and function of the polar body contractile ring in Spisula. Dev Biol. 2004;269:421–32. doi: 10.1016/j.ydbio.2004.01.033. [DOI] [PubMed] [Google Scholar]
- Potapova TA, Daum JR, Pittman BD, Hudson JR, Jones TN, Satinover DL, Stukenberg PT, Gorbsky GJ. The reversibility of mitotic exit in vertebrate cells. Nature. 2006;440:954–8. doi: 10.1038/nature04652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–6. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–92. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Shelton CA, Carter JC, Ellis GC, Bowerman B. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J Cell Biol. 1999;146:439–51. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomos MF, Badrinath A, Pasierbek P, Livingstone D, White J, Glotzer M, Nasmyth K. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr Biol. 2001;11:1825–35. doi: 10.1016/s0960-9822(01)00588-7. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi AG, Kalabokis VN, Perreault-Micale CL. Regulation by molluscan myosins. Mol Cell Biochem. 1999;190:55–62. [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol. 2003;5:1090–4. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Uyeda TQ, Spudich JA. A functional recombinant myosin II lacking a regulatory light chain-binding site. Science. 1993;262:1867–70. doi: 10.1126/science.8266074. [DOI] [PubMed] [Google Scholar]
- Verlhac MH, Lefebvre C, Guillaud P, Rassinier P, Maro B. Asymmetric division in mouse oocytes: with or without Mos. Current Biology. 2000;10:1303–1306. doi: 10.1016/s0960-9822(00)00753-3. [DOI] [PubMed] [Google Scholar]
- Yang HY, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol. 2005;169:447–57. doi: 10.1083/jcb.200411132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, McNally K, McNally FJ. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev Biol. 2003;260:245–59. doi: 10.1016/s0012-1606(03)00216-1. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ma C, Miller AL, Katbi HA, Bement WM, Liu XJ. Polar body emission requires a RhoA contractile ring and Cdc42-mediated membrane protrusion. Dev Cell. 2008;15:386–400. doi: 10.1016/j.devcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse sequence of GFP:NMY-2 and GFP:histone fluorescence during meiosis in a wild-type worm corresponds to Fig. 1A. Images were captured at 5-s intervals. Display rate is 5 frames/s.
Time-lapse images of a meiotic spindle labeled with GFP:tubulin (green) and mCherry:histone H2B (red). After spindle rotation and shortening, the arrangement of the spindle microtubules changes so the chromosomes are on the ends of the spindle (10.2–12.8 min) through completion of anaphase. This justifies the use of chromosome position to infer spindle position in Fig. 1. The cell cortex is highlighted for clarity of spindle rotation (to = completion of spindle rotation). Scale bar, 5 μm.
(A) Total duration of chromosome segregation in meiotic anaphase and (B) placement of the GFP:NMY-2-labeled contractile ring on the meiotic spindle at the completion of anaphase as a percentage of spindle length in control, mel-11(RNAi) and mei-1 embryos. All, p > 0.05.
DIC time-lapse sequence during meiosis in a wild-type worm corresponds to Fig. 1A. Images were captured at 5-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH and GFP:tubulin fluorescence during meiosis in a wild-type worm corresponds to Fig. 2A. Images were captured at 10-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH (green) and mCherry:histone (magenta) fluorescence during meiosis in a tba-1(RNAi) worm corresponds to Fig. 4B. Images were captured at 10-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH (green) and mCherry:histone (red) fluorescence during meiosis in a cyk-4(RNAi) worm corresponds to Figs. 6C and S5C. Images were captured at 15-s intervals. Display rate is 5 frames/s.
Time-lapse sequence of GFP:PH (green) and mCherry:histone (red) fluorescence during meiosis in an ect-2(RNAi) worm corresponds to Figs. 6E and S5E. Images were captured at 15-s intervals. Display rate is 5 frames/s.
(A) Image of cortical GFP:NMY-2 at the anterior half of the cell in a fixed early mitotic embryo. (B) Images from a representative time-lapse sequence of wild-type embryos expressing GFP:PH and mCherry:histone showing the end of anaphase II and continuation of dynamic furrowing into pseudocleavage (25.6–33.5 min) during pronuclear migration (*) and pronuclear meeting (36.8 min) (to = completion of spindle rotation). Scale bar, 5 μm.
Images from a representative time-lapse sequence of a GFP:tubulin; GFP:PH mat-1(RNAi) metaphase-arrested embryo exposed to the CDK-1 inhibitor, purvalanol A (to = start of imaging). Scale bar, 5 μm.
Images from representative time-lapse sequences of embryos progressing through anaphase II of meiosis expressing GFP:PH and mCherry:histone. (A) A wild-type control embryo shows contractile ring ingression tightly around the anaphase spindle and polar body completion, while (B) cyk-4(or749ts), (C) cyk-4(RNAi) and (D) zen-4(RNAi) show wide furrows moving toward the anaphase spindle between the separating chromosomes, which regress, allowing the chromosomes to rejoin. (E) ect-2(RNAi) results in little or no ingression of a contractile ring, resulting in polar body failure. (to = start of chromosome separation) Scale bar, 5 μm.