Abstract
BACKGROUND
The genetic alterations responsible for an adverse outcome in most patients with acute myeloid leukemia (AML) are unknown.
METHODS
Using massively parallel DNA sequencing, we identified a somatic mutation in DNMT3A, encoding a DNA methyltransferase, in the genome of cells from a patient with AML with a normal karyotype. We sequenced the exons of DNMT3A in 280 additional patients with de novo AML to define recurring mutations.
RESULTS
A total of 62 of 281 patients (22.1%) had mutations in DNMT3A that were predicted to affect translation. We identified 18 different missense mutations, the most common of which was predicted to affect amino acid R882 (in 37 patients). We also identified six frameshift, six nonsense, and three splice-site mutations and a 1.5-Mbp deletion encompassing DNMT3A. These mutations were highly enriched in the group of patients with an intermediate-risk cytogenetic profile (56 of 166 patients, or 33.7%) but were absent in all 79 patients with a favorable-risk cytogenetic profile (P<0.001 for both comparisons). The median overall survival among patients with DNMT3A mutations was significantly shorter than that among patients without such mutations (12.3 months vs. 41.1 months, P<0.001). DNMT3A mutations were associated with adverse outcomes among patients with an intermediate-risk cytogenetic profile or FLT3 mutations, regardless of age, and were independently associated with a poor outcome in Cox proportional-hazards analysis.
CONCLUSIONS
DNMT3A mutations are highly recurrent in patients with de novo AML with an intermediate-risk cytogenetic profile and are independently associated with a poor outcome. (Funded by the National Institutes of Health and others.)
Whole-genome sequencing is an unbiased approach for discovering somatic variations in cancer genomes. We recently reported the DNA sequence and analysis of the genomes of two patients with acute myeloid leukemia (AML) with a normal karyotype.1,2 We did not find new recurring mutations in the first study but did observe a recurrent mutation in IDH1, encoding isocitrate dehydrogenase 1, in the second study.2 Subsequent work has confirmed and extended this finding, showing that mutations in IDH1 and related gene IDH2 are highly recurrent in patients with an intermediate-risk cytogenetic profile (20 to 30% frequency) and are associated with a poor prognosis in some subgroups of patients.3-5 Improvements in sequencing techniques prompted us to reevaluate the first case with deeper sequence coverage, during which we discovered a frameshift mutation in the DNA methyltransferase gene DNMT3A.
The human genes DNMT1, DNMT3A, and DNMT3B encode DNA methyltransferases, enzymes that catalyze the addition of a methyl group to the cytosine residue of CpG dinucleotides. Clusters of CpG dinucleotides (islands) are concentrated in regions upstream of genes; increased methylation of these CpG islands is often associated with reduced expression of the downstream gene. Aberrant DNA methylation has long been hypothesized to contribute to the pathogenesis of cancer.6-9 Although cancer genomes tend to be globally hypomethylated, as compared with normal tissues, hypermethylation of CpG islands in the promoters of tumor-suppressor genes is common in many tumors.
Genomewide methylation profiling of AML samples with the use of microarrays has shown that subgroups of samples have similar methylation patterns.10 Samples obtained from patients with myelodysplastic syndromes have a pattern of aberrant hypermethylation, as compared with that of de novo AML, which is defined as newly diagnosed disease with no history of chemotherapy or radiation therapy and no history of a myelodysplastic syndrome or a myeloproliferative neoplasm.11 DNA methyltransferase inhibitors are widely used to treat patients with AML and myelodysplastic syndromes, although the response rate is low, the response is unpredictable in individual patients, and it is not clearly associated with methylation status.12-14 In addition, in a recent study,15 higher expression of miR-29b, a microRNA that targets DNMT3A messenger RNA (mRNA), in AML blasts was associated with an improved clinical response to the DNA methyltransferase inhibitor decitabine.
After discovering a frameshift mutation in DNMT3A with whole-genome sequencing, we conducted a study to determine whether DNMT3A is recurrently mutated in AML samples and whether DNMT3A mutations are associated with poor survival.
METHODS
We have described the clinical features of the proband previously.1 The patient’s next of kin consented to the sequencing of her tumor and skin genomes on the basis of a protocol that was approved by the institutional review board and that explicitly described the privacy issues of whole-genome sequencing. We have also described the methods that we used for sequencing, determining whether mutations were recurrent, expression analysis, and outcomes analysis.2 Methods that are used in whole-genome analysis of 5-methylcytosine content and methylated DNA immunoprecipitation-chip (MeDIP-chip) analysis are described in the Supplementary Appendix, available with the full text of this article at NEJM.org.
RESULTS
IDENTIFICATION OF DNMT3A MUTATIONS
We previously sequenced the AML genome of a sample obtained from Patient 933124 and obtained short single-end reads, yielding 98 billion base pairs of sequence and 91.2% diploid coverage of the genome.1 In this study, we obtained 116.4 billion base pairs with paired-end reads from the genome of the relapsed tumor, yielding 99.6% diploid coverage of the genome. The improved sequence coverage with paired-end reads and improved mutation calling algorithms allowed us to identify several nonsynonymous tier 1 mutations that we did not detect in our initial sequencing effort, including a 1-base-pair deletion in DNMT3A. We subsequently verified that this mutation was also present in the dominant clone of the original AML sample obtained at presentation (see Supplementary Appendix for details). This deletion causes a frameshift in the codon encoding lysine at amino acid position 723, which predicts the synthesis of 56 novel amino acids (i.e., amino acids that are not encoded by nonmutated DNMT3A at positions including and beyond amino acid position 723), followed by an in-frame, premature stop codon.
DNMT3A MUTATIONS IN AML
We sequenced the exons of DNMT3A and DNMT3L (DNMT3L forms heterodimers with both DNMT3A and its homologue, DNMT3B) by polymerase-chain-reaction (PCR) amplification and Sanger sequencing of samples from 188 patients with de novo AML, which were banked at Washington University. (Primer sequences are provided in the Supplementary Appendix.) All 188 tumors had matched normal DNA samples available for analysis. We obtained 94 additional AML samples from the Cancer and Leukemia Group B (CALBG); matched normal tissue was not available for these samples. The characteristics of all 282 patients, the treatments they received, and the mutations found are provided in the Supplementary Appendix. Although these patients were not treated uniformly, the prevalence of common AML mutations among them was typical, and the clinical outcomes were similar to those in other series of patients that have been described recently.
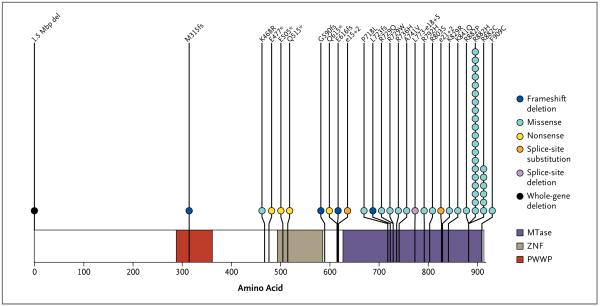
We amplified and sequenced all 24 exons of DNMT3A for 281 of the 282 tumor samples. Of these samples, 62 (22.1%) had mutations predicting translational consequences; 5 of these 62 tumors had two independent mutations, with none of these mutations predicted to affect amino acid position 882. We determined that all mutations in the tumors obtained from Washington University were somatic events by virtue of their absence in DNA sequences obtained from matched skin samples; a summary of all confirmed somatic mutations is shown in Figure 1. We observed seven novel sequence changes in DNMT3A in the CALGB sample set but could not determine whether they were somatic changes without matched DNA from unaffected tissue. (Putative somatic CALGB mutations are listed in Fig. 1 in the Supplementary Appendix.) None of the novel variants that were detected in the CALGB tumors have previously been identified as single-nucleotide polymorphisms. We identified 18 different missense mutations across both sets of tumors. The most common missense mutations are predicted to affect amino acid R882 (in 37 tumors) (Table 2 in the Supplementary Appendix). A total of 27 tumors had the R882H variant, 7 had R882C, 2 had R882P, and 1 had R882S. We also observed six frameshift, six nonsense, and three splice-site mutations and a 1.5-Mb deletion that included DNMT3A and eight other genes (Fig. 2 in the Supplementary Appendix). We observed no somatic mutations in the coding exons of DNMT3L in 188 samples that were tested.
Figure 1. DNMT3A Mutations in 188 Patients with Acute Myeloid Leukemia (AML).
Shown are data from samples banked at Washington University that were obtained from 188 patients with AML. All mutations that are shown were confirmed to be somatic. The locations of the PWWP domain (characterized by the presence of a highly conserved proline–tryptophan–tryptophan–proline motif), the ADD (ATRX, DNMT3, and DNMT3L)-type zinc finger (ZNF) domain, and the methyltransferase (MTase) domain are shown. Each patient with a DNMT3A mutation is designated with a circle. The deletion that is noted at position 1 is a 1.5-Mbp deletion that includes DNMT3A and part or all of eight other genes (for details, see Fig. 2 in the Supplementary Appendix).
The relationship between DNMT3A mutations and other common mutations in AML genomes is shown in Table 1 and Figure 2. Mutations in FLT3, NPM1, and IDH1 were significantly enriched in samples with DNMT3A mutations. No DNMT3A mutations (and likewise no NPM1 or IDH1/2 mutations) were found in 79 AML samples with cytogenetic findings associated with a favorable outcome (P<0.001). Likewise, none of the 11 patients with structural variations involving 11q23, the location of the mixed-lineage leukemia gene, had DNMT3A mutations. DNMT3A mutations were significantly enriched in 56 of 166 patients with a cytogenetic profile associated with intermediate risk (33.7%, P<0.001), including 44 of 120 patients with a normal cytogenetic profile (36.7%, P<0.001), as well as 20 of 61 patients with the M4 subtype (32.8%, P = 0.04) and 12 of 21 patients with the M5 subtype (57.1%, P<0.001), according to the French–American–British (FAB) classification system. White-cell counts at presentation were significantly higher in patients with R882 mutations than in other patients. The single patient with a FAB M3 morphologic subtype and a DNMT3A mutation (Patient 287), who died 2.2 months after presentation, had a normal cytogenetic profile and did not have the characteristic expression signature of t(15;17) AML.16,17
Table 1.
Clinical Characteristics of 281 Patients with Acute Myeloid Leukemia with or without DNMT3A Mutations.*
| Variable | No DNMT3A Mutation (N = 219) |
R882 DNMT3A Mutation (N = 37) |
Non-R882 DNMT3A Mutation (N = 25) |
Any DNMT3A Mutation (N = 62) |
P Value† |
|---|---|---|---|---|---|
| Age at study entry — yr | 47.8±16.6 | 52.1±13.1 | 54.6±14.8 | 53.1±13.7 | 0.02‡ |
| Race — no. (%)§ | 0.58¶ | ||||
| White | 195 (89) | 34 (92) | 21 (84) | 55 (89) | |
| Black | 18 (8) | 2 (5) | 2 (8) | 4 (6) | |
| Other | 6 (3) | 1 (3) | 2 (8) | 3 (5) | |
| Male sex — no. (%) | 125 (57) | 17 (46) | 11 (44) | 28 (45) | 0.10∥ |
| Bone marrow blasts at diagnosis — % | 69.4±18.6 | 73.5±16.3 | 63.6±18.0 | 69.5±17.6 | 0.96‡ |
| Normal karyotype — no./total no. (%) | 76/216 (35) | 27/36 (75) | 17/25 (68) | 44/61 (72) | <0.001∥ |
| White-cell count at diagnosis — (×10−3/mm3) | <0.001** | ||||
| No. of patients | 142 | 27 | 18 | 45 | |
| Mean | 39.3±65.8 | 78.0±57.3 | 29.8±29.4 | 58.7±53.3 | |
| Median | 11.8 | 72.6 | 19.2 | 46.4 | |
| Cytogenetic risk — no./total no. (%)†† | <0.001¶ | ||||
| Favorable | 79/215 (37) | 0/36 | 0/24 | 0/60 | |
| Intermediate | 110/215 (51) | 34/36 (94) | 22/24 (92) | 56/60 (93) | |
| Adverse | 26/215 (12) | 2/36 (6) | 2/24 (8) | 4/60 (7) | |
| AML subtype — no. (%) | |||||
| M3 | 47 (21) | 1 (3) | 0 | 1 (2) | <0.001∥ |
| M4 | 41 (19) | 14 (38) | 6 (24) | 20 (32) | 0.02∥ |
| M5 | 9 (4) | 9 (24) | 3 (12) | 12 (19) | <0.001¶ |
| Mutation — no./total no. (%) | |||||
| NPM1 | 27/216 (13) | 24/37 (65) | 13/25 (52) | 37/62 (60) | <0.001∥ |
| FLT3 | 48/217 (22) | 17/36 (47) | 8/25 (32) | 25/61 (41) | 0.003∥ |
| IDH1 | 12/218 (6) | 10/37 (27) | 3/25 (12) | 13/62 (21) | <0.001∥ |
| IDH2 | 13/217 (6) | 1/37 (3) | 6/25 (24) | 7/62 (11) | 0.15∥ |
| t(15;17) | 45/219 (21) | 0/37 | 0/25 | 0/62 | <0.001∥ |
| t(8;21) | 12/219 (5) | 0/37 | 0/25 | 0/62 | 0.07¶ |
| inv(16) | 20/219 (9) | 0/37 | 0/25 | 0/62 | 0.01¶ |
Plus–minus values are means ±SD. DNMT denotes DNA methyltransferase.
P values are for the comparisons between no DNMT3A mutations and any DNMT3A mutation.
The P value was calculated by means of Student’s t-test.
Race was self-reported.
The P value was calculated by means of Fisher’s exact test.
The P value was calculated by means of Pearson’s chi-square test.
The P value was calculated by means of the Wilcoxon test.
The Cancer and Leukemia Group B classification system was used to stratify risk on the basis of cytogenetic findings.16
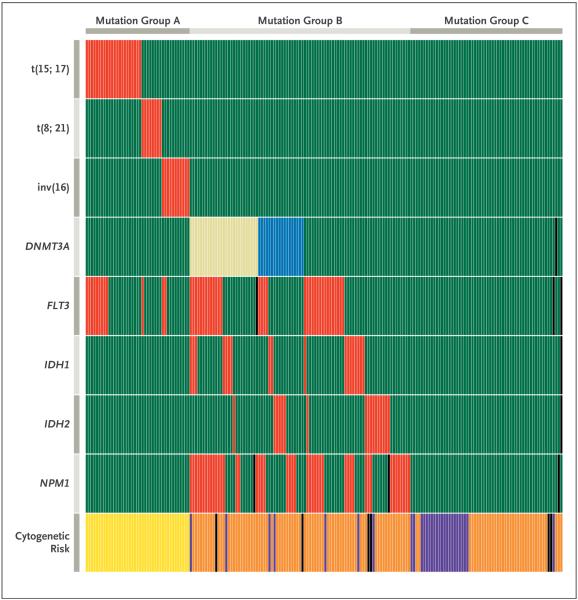
Figure 2. Relationship between DNMT3A Mutations and Other Common Mutations in 188 Patients with Acute Myeloid Leukemia (AML).
Shown is the mutation status of each of 188 patients with AML on the basis of sequencing results shown in Figure 1. Red indicates the presence of the specified mutation in the designated patient, and green the absence of the mutation. For DNMT3A mutations, the presence of any R882 variant is shown in tan, and all other DNMT3A mutations are shown in blue. DNMT3A, NPM1, and IDH1/2 mutations were not detected in any sample with t(15;17), t(8;21), complex t(8;21), or inv(16). In samples with DNMT3A mutations, mutations in FLT3 (either internal tandem duplications or mutations at amino acid position D835), NPM1, and IDH1 were significantly enriched. Cytogenetic risk groups are shown at the bottom of the graph, with yellow indicating favorable risk, orange intermediate risk, and purple adverse risk. Black boxes indicate that no data were available for that sample. Mutation groups as defined by these data are designated at the top of the graph. Samples in mutation group A have cytogenetic findings associated with favorable risk and have only FLT3 mutations among the five commonly mutated AML genes. Samples in mutation group B are highly enriched for mutations in the five commonly mutated AML genes that are associated with intermediate risk. Samples in mutation group C have no mutations in any of the five common AML genes, suggesting that a different set of mutations may contribute to the pathogenesis in this group.
We obtained expression array data from 180 of the 188 samples obtained from Washington University (GeneChip Human Genome U133 Plus 2.0 Array, Affymetrix). We identified the expression of genes encoding DNA methyltransferases in AML samples and in normal hematopoietic cells with these data (Fig. 3 in the Supplementary Appendix). DNMT3A was expressed in all 180 AML samples and in normal human CD34+ bone marrow cells. Its expression decreased with terminal myeloid differentiation. We observed no difference in the levels of DNMT3A expression between AML samples with DNMT3A mutations and those without DNMT3A mutations. Likewise, DNMT3B and DNMT1 were both highly expressed in AML samples and normal CD34+ cells. The single DNMT3L probe set on the expression array revealed no expression of DNMT3L in any sample; however, spliced DNMT3L was detected with transcriptome sequencing of AML cells from several patients (data not shown).
To determine whether the mutant DNMT3A alleles are expressed, we amplified the target regions from 21 primary tumor samples from bone marrow using reverse-transcriptase PCR and then sequenced the amplicons using the Roche 454-Titanium platform to obtain deep read counts (Table 3 and Fig. 4 in the Supplementary Appendix). For most of the samples, the proportion of complementary DNA (cDNA) sequence reads containing the variant allele was about 50%, suggesting that the variant and wild-type alleles are expressed at nearly equal levels. One nonsense mutation (E477*) and one frameshift mutation (M315fs) were not detected in the cDNA samples, suggesting that mRNAs carrying these variants are subject to nonsense-mediated decay. There was no evidence of homozygous loss of DNMT3A expression for any of the five samples carrying two mutations. We did not detect variant cDNAs carrying the missense allele (A741V) or the nonsense allele (E477*) in the sample from Subject 246634, although we readily detected cDNA from the nonmutated allele in this sample (Table 3 in the Supplementary Appendix). We suspect that A741V and E477* were on the same allele and thus were equally affected by nonsense-mediated decay.
PATTERNS OF MUTATIONS
We have recently sequenced the genomes of 38 cytogenetically normal samples of de novo AML, with an average of more than 25 times genome coverage obtained for each tumor and matched normal skin genome. Of these genomes, 11 have mutations in DNMT3A, but no mutations were detected in DNMT3L, DNMT1, or DNMT3B (data not shown). We identified with high confidence somatic single-nucleotide variants in each genome and determined that the total number of these variants was not significantly influenced by DNMT3A mutation status (Fig. 5 in the Supplementary Appendix). Furthermore, the types of mutations detected in each genome (C-to-T, C-to-G, C-to-A, etc.) were not associated with DNMT3A mutation status (Table 4 and Fig. 6 in the Supplementary Appendix).
DNA METHYLATION
To determine whether the 5-methylcytosine content was altered in AML genomes with DNMT3A mutations, we hydrolyzed genomic DNA derived from the bone marrow of patients with AML to nucleoside monophosphates and assayed 5-methyl-2′-deoxycytidine 5′-monophosphate using liquid chromatography–tandem mass spectrometry (see the Supplementary Appendix). The mean (±SD) 5-methylcytosine content of each genome with a DNMT3A mutation was virtually identical to that of AML genomes without DNMT3A mutations (wild type, 4.89±0.57%; any DNMT3A mutation, 4.77±0.50%; P = 0.45) (Table 5 and Fig. 7 in the Supplementary Appendix).
We determined the methylation patterns of five AML genomes with the R882H mutation and five matched AML genomes (i.e., with the same FAB subtypes and myeloblast percentages) with nonmutated DNMT3A, using MeDIP-chip analysis. Nearly all the methylated regions were similar among the 10 samples (Fig. 8A in the Supplementary Appendix), with only a small number of exceptions (Table 6 and Fig. 8B and 9 in the Supplementary Appendix). A total of 182 genomic regions had significantly different methylation levels (at specific genomic locations) associated with DNMT3A mutation status. All 182 regions had significantly reduced methylation on average in the mutant genomes. There was no consistent correlation between any of the differentially methylated regions and altered expression of “nearest neighbor” genes (data not shown).
GENE EXPRESSION AND DNMT3A MUTATIONS
We performed unsupervised clustering, using expression data from 180 of the 188 samples banked at Washington University (Fig. 10 in the Supplementary Appendix). Although multiple expression clusters were clearly identified, none were clearly defined by DNMT3A mutation status. Similar results were obtained with the subgroup of 76 arrays from AML samples with a normal cytogenetic profile (Fig. 11 in the Supplementary Appendix).
CLINICAL OUTCOME AND DNMT3A MUTATIONS
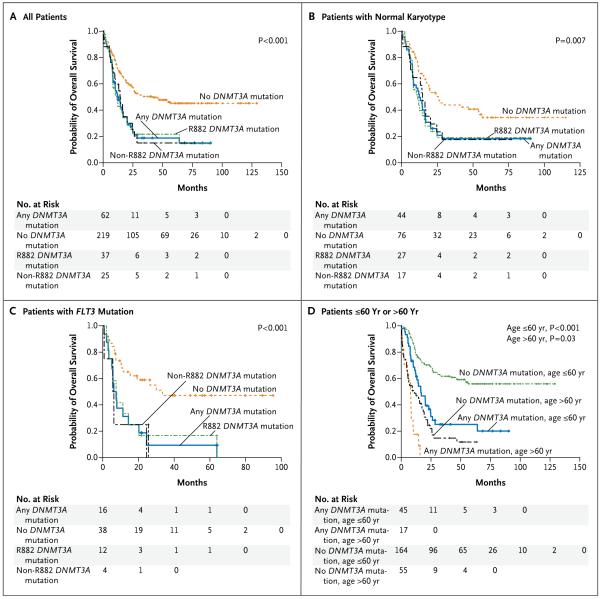
We determined event-free and overall survival for all 281 patients with AML for whom DNMT3A mutation status was known. The two metrics were very similar, so only overall survival is reported. Patients with DNMT3A mutations had significantly worse survival (Fig. 3A). The presence of a DNMT3A mutation was also associated with worse survival among patients with a normal cytogenetic profile (P = 0.007) (Fig. 3B) and those with an intermediate-risk profile (P = 0.006) (Fig. 12 in the Supplementary Appendix). When patients with FLT3 internal tandem duplication mutations were classified according to DNMT3A status, those carrying a DNMT3A mutation had a significantly worse outcome (P<0.001) (Fig. 3C). Finally, DNMT3A mutations were associated with an adverse outcome for patients regardless of age (P<0.001 for ≤60 years and P = 0.03 for >60 years), although patients with DNMT3A mutations who were older than 60 years had an extremely poor outcome (Fig. 3D). Data on DNMT3A mutations and overall survival among patients with all combinations of common AML mutations (NPM1, IDH1/2, and FLT3) are provided in Figures 13, 14, and 15 in the Supplementary Appendix; Cox proportional-hazards models showed that the variables that were independently associated with overall survival were DNMT3A mutations, FLT3 mutations, and age.
Figure 3. Overall Survival among Patients with Acute Myeloid Leukemia (AML) with DNMT3A Mutations.
Panel A shows the overall survival among all 281 patients, including 62 with a DNMT3A mutation and 219 without a DNMT3A mutation. Panel B shows the overall survival of 120 patients with normal karyotypes, including 44 with a DNMT3A mutation and 76 without a DNMT3A mutation. Panel C shows the overall survival of 54 patients with an FLT3 internal tandem duplication mutation, including 16 with a DNMT3A mutation and 38 without a DNMT3A mutation. Panel D shows the overall survival of all 281 patients stratified according to DNMT3A mutation status and age; 72 patients were over the age of 60 years and 209 were 60 years of age or younger. All P values, which are for the comparison between any DNMT3A mutation and no mutation, were calculated by means of the log-rank test.
DISCUSSION
DNMT3A mutations are recurrent in patients with AML and are associated with poor event-free and overall survival, independently of age and the presence of FLT3 or NPM1 mutations and regardless of the type of mutation or genetic location. This finding strongly suggests that DNMT3A mutations are probably relevant to the pathogenesis of AML. DNMT3A mutations do not cause genomic instability (since most genomes with mutations have a normal cytogenetic profile and an unaltered number of total mutations), do not alter total 5-methylcytosine content or global patterns of methylation, and do not dramatically alter gene expression. Currently, the only clue regarding the pathogenetic mechanism is strong selection against DNMT3A mutations (and also IDH1/2 and NPM1 mutations) in patients who have a favorable-risk cytogenetic profile, suggesting a biologic relationship that is not random (Fig. 2).
In patients with AML, there are two major classes of DNMT3A mutations. The first class is the highly recurrent set of mutations at R882, as recently described in 3 of 74 AML samples tested by Yamashita et al.18 (The low prevalence of this variant in that study could reflect a different sample population or a lower sensitivity of mutation detection.) The second class is represented by all the other mutations in this gene. The locations of mutations in DNMT3A are similar to those of DNMT3B mutations associated with the immunodeficiency, centromere instability, and facial anomalies (ICF) syndrome.19,20
Recurrent mutations at a single amino acid position suggest a gain-of-function mechanism, although widely divergent mutations at many positions in a gene generally suggest loss of function, a pattern seen for many classic tumor-suppressor genes (e.g., TP53 and BRCA1). Indeed, several of the non-R882 mutations clearly cause loss of function in DNMT3A. A gain-of-function property that is induced by the R882 mutation is not yet apparent.
The MeDIP-chip experiments revealed a significant reduction in DNA methylation at 182 genomic locations, suggesting that R882 mutations may act in a dominant-negative fashion to reduce the methyltransferase activity of the enzyme. This hypothesis is supported by the fact that all R882 mutations are heterozygous and by observations that this mutation reduces methyltransferase activity in vitro.18,21 Both the R882H and R882C mutations are caused by a C-to-T transition at a CpG dinucleotide (R882H on the noncoding strand and R882C on the coding strand), suggesting that these mutations may be caused by the deamination of methylcytosine on either strand of this CpG dinucleotide.22 Many DNMT3A mutations are predicted to cause changes in the DNA binding groove of DNMT3A, and some are predicted to change its interaction with DNMT3L (Fig. 16 in the Supplementary Appendix). However, it is possible that R882 mutations alter functions of DNMT3A that are not yet fully understood, including its ability to bind to other proteins involved in transcriptional regulation and localization to chromatin regions containing methylated DNA.23-26 In any case, all DNMT3A mutations are associated with poor overall survival, suggesting that they have an important common effect on the potential of AML cells to cause lethal disease.
The association between DNMT3A mutations and mutations in the four other most commonly mutated genes in AML (FLT3, NPM1, IDH1, and IDH2) is evident, as shown in Figure 2. A large proportion of patients with an intermediate-risk cytogenetic profile had mutations in one or more of these genes (mutation group B in Fig. 2). However, many patients with an intermediate- or adverse-risk cytogenetic profile had no mutations in any of these genes (mutation group C), which was not a random finding (P<0.001). Patients in mutation group C had outcomes similar to those of patients in mutation group B with one or two mutations (Fig. 17 in the Supplementary Appendix) but may have a unique set of driver mutations. These may include some of the less commonly mutated AML genes (e.g., CEBPA, RUNX1, NRAS, and KRAS), although whole-genome sequencing may ultimately be required to identify the key mutations in this group.
Remarkably, no DNMT3A mutations were found in the group of 79 patients with a favorable-risk cytogenetic profile (Table 1 and Fig. 2), which includes patients with t(15;17), t(8;21), and inv(16). Mutations in NPM1, IDH1, and IDH2 were not detected in these patients either, a finding that is consistent with data reported in other studies.4,27-30 The virtual exclusion of mutations in these four genes in patients with a favorable-risk profile is not random and may reflect the leukemogenic properties of the fusion proteins created by these chromosomal rearrangements. The PML-RARA fusion protein, which is created by t(15;17), physically interacts with DNMT3A, and AML-ETO, which is created by t(8;21), interacts with DNMT1; both fusion proteins alter the methylation of specific promoters.31-33 Both PML and DNMT3A regulate telomere function, and all-trans retinoic acid, which is part of the therapy for patients with t(15;17), down-regulates DNMT3A expression.34,35 Together, these data suggest that DNMT3A mutations and the favorable-risk fusion oncogenes (e.g., PML-RARA and AML-ETO) may not be found in the same AML genomes because they both act to alter the function of DNA methyltransferases and are therefore redundant. However, the outcomes for patients with DNMT3A mutations and those with a favorable-risk cytogenetic profile are dramatically different, for reasons that are currently unclear.
DNMT3A mutations do not change 5-methylcytosine content in AML genomes, and the R882H mutation appears to minimally perturb the methylation of CpG islands. We did not detect changes in DNA methylation that were directly correlated with local changes in gene expression. These data might suggest that DNMT3A mutations do not directly affect the cytosine methyltransferase properties of DNMT3A. However, a recent study has suggested that DNMT3A may alter the methylation of nonpromoter-associated CpG regions, affecting gene expression indirectly.36 Many further experiments will be required to define the precise mechanisms by which these mutations act.
The discovery of highly recurrent mutations in DNMT3A may provide a new tool for the classification of intermediate-risk AML. If these data are reproduced in other series, clinical trials designed to assess the effects of early intensification of treatment in patients with DNMT3A mutations may be warranted. In our small series, allogeneic transplantation provided a significant benefit for patients with DNMT3A mutations (Fig. 18 in the Supplementary Appendix). The careful analysis of many additional AML samples with unbiased genomic methods may reveal changes in methylation or expression that help to define the mechanisms of action of DNMT3A mutations.
Supplementary Material
Acknowledgments
Supported by grants from the National Center for Research Resources (UL1 RR024992), the National Institutes of Health (P41RR000954 and UL1 RR024992, to Dr. Townsend; PO1 CA101937, to Dr. Ley; and U54 HG003079, to Dr. Wilson), and the Barnes–Jewish Hospital Foundation (00335-0505-02, to Dr. Ley).
We thank Nancy Reidelberger for administrative support; Todd Hepler, William Schroeder, Justin Lolofie, Scott Abbott, Shawn Leonard, Ken Swanson, Indraniel Das, and Michael Kiwala for their contributions to the Laboratory Information Management System; Gary Stiehr, Richard Wohlstadter, Matt Weil, and Kelly Fallon for information-technology support; Jin Shao for statistical support; Clara Bloomfield, Michael Caligiuri, and James Vardiman for providing the AML samples from the CALGB Leukemia Bank; the Washington University Cancer Genome Initiative for support; Alvin J. Siteman for supporting the sequencing of the original AML genome that is further described in this study; and our patients.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–55. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–43. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 5.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 7.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome — components and functional correlates. Genes Dev. 2006;20:3215–31. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 8.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2009;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa ME, Skrabanek L, Li Y, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–58. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cashen AF, Schiller GJ, O’Donnell MR, DiPersio JF. Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J Clin Oncol. 2010;28:556–61. doi: 10.1200/JCO.2009.23.9178. [DOI] [PubMed] [Google Scholar]
- 13.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a Study of the Cancer and Leukemia Group B. J Clin Oncol. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 14.Fandy TE, Herman JG, Kerns P, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114:2764–73. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum W, Garzon R, Klisovic RB, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A. 2010;107:7473–8. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 17.Payton JE, Grieselhuber NR, Chang LW, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. 2009;119:1714–26. doi: 10.1172/JCI38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita Y, Yuan J, Suetake I, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–31. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 19.Ehrlich M, Sanchez C, Shao C, et al. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity. 2008;41:253–71. doi: 10.1080/08916930802024202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang YL, Rigolet M, Bourc’his D, et al. DNMT3B mutations and DNA methylation defect define two types of ICF syndrome. Hum Mutat. 2005;25:56–63. doi: 10.1002/humu.20113. [DOI] [PubMed] [Google Scholar]
- 21.Gowher H, Loutchanwoot P, Vorobjeva O, et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J Mol Biol. 2006;357:928–41. doi: 10.1016/j.jmb.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Rideout WM, III, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–90. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 23.Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–99. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 24.Wang YA, Kamarova Y, Shen KC, et al. DNA methyltransferase-3a interacts with p53 and represses p53-mediated gene expression. Cancer Biol Ther. 2005;4:1138–43. doi: 10.4161/cbt.4.10.2073. [DOI] [PubMed] [Google Scholar]
- 25.Brenner C, Deplus R, Didelot C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–46. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]; N Engl J Med. 2005;352:740. Erratum. [Google Scholar]
- 28.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–20. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 29.Chou WC, Hou HA, Chen CY, et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood. 2010;115:2749–54. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- 30.Boissel N, Nibourel O, Renneville A, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association Group. J Clin Oncol. 2010;28:3717–23. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 31.Di Croce L, Raker VA, Corsaro M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 32.Liu S, Shen T, Huynh L, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–84. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 33.Fazi F, Zardo G, Gelmetti V, et al. Heterochromatic gene repression of the retinoic acid pathway in acute myeloid leukemia. Blood. 2007;109:4432–40. doi: 10.1182/blood-2006-09-045781. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–24. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 35.Fazi F, Travaglini L, Carotti D, et al. Retinoic acid targets DNA-methyltransferases and histone deacetylases during APL blast differentiation in vitro and in vivo. Oncogene. 2005;24:1820–30. doi: 10.1038/sj.onc.1208286. [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Coskun V, Tao J, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–8. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.