Abstract
Patients vary in their responses to drug therapy, and some of that variability is genetically-determined. This review outlines general approaches used to identify genetic variation that influences drug response. Examples from specific therapeutic areas are presented: cholesterol management, arrhythmias, heart failure, hypertension, warfarin anticoagulation, and anti-platelet agents. A brief view of potential pathways to implementation is presented.
Keywords: genetics, pharmacogenetics, pharmacogenomics, drug therapy
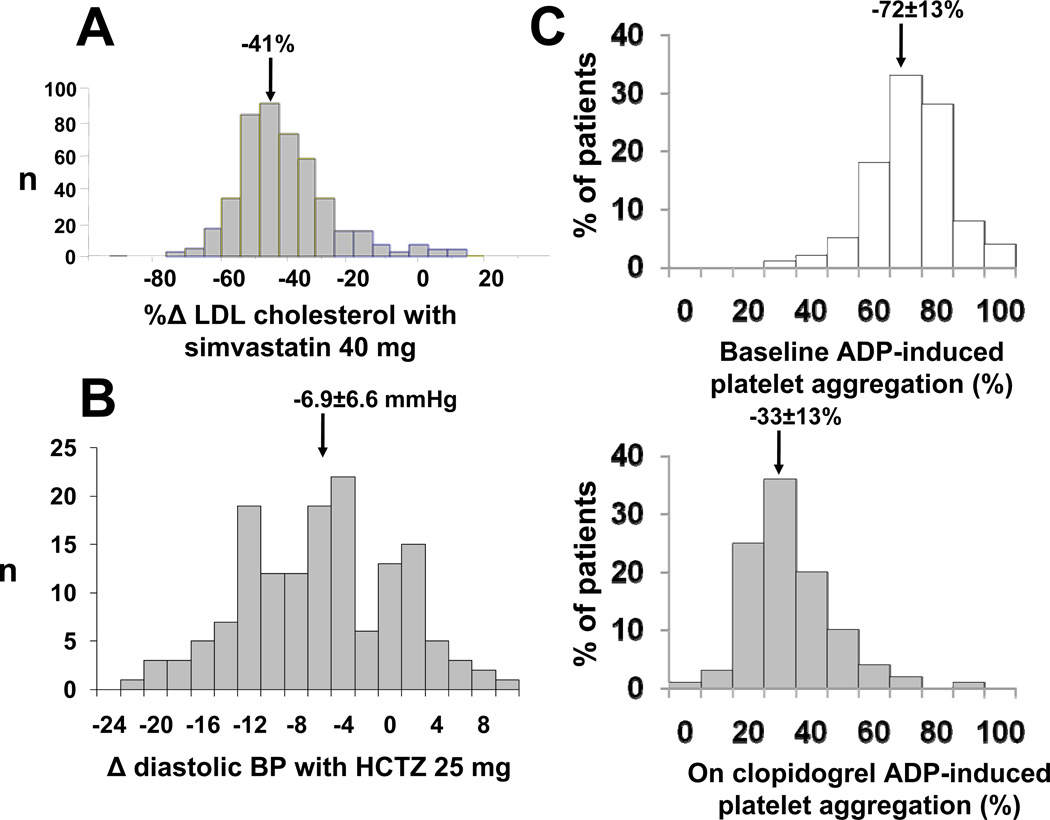
Americans spent $234,000,000,000 on drug therapy in 2008, and cardiovascular drugs are among the most widely-used.1 While large randomized clinical trials unequivocally demonstrate population benefits with many of these agents, individual patients display striking variability in response: variability in efficacy and serious adverse effects continue to plague therapy. Figure 1 shows examples of interindividual variability in response to common cardiovascular therapies. These data highlight the idea that use of single drug doses ignores the possibility – indeed the near-inevitability – that individuals vary in response.
Figure 1. Variability in response to cardiovascular therapies.
A. Variability in LDL cholesterol lowering in response to simvastatin 40 mg/day for 6 weeks. The drug is highly effective, judged by mean response, but there is substantial variability with some subjects displaying no response and others LDL cholesterol lowering of up to 80%. Adapted by permission from Simon et al.156 B. Variability in decrease in diastolic blood pressure in response to hydrochlorothiazide (HCTZ) in 147 African-American subjects. As in panel A, the drug is effective, judged by mean response, but there is substantial variability. C. The top panel shows variability in ADP-induced platelet aggregation at baseline, and the bottom panel after clopidogrel. As in the other examples, there is a very obvious drug effect, but substantial variability across the population. Adapted by permission from Shuldiner et al.9
There are many sources of variability in response to drug therapy, such as noncompliance and unrecognized drug interactions. This review focuses on genetic mechanisms contributing to variability in response to cardiovascular drug therapy. An introductory section discusses principles of drug action and approaches to identifying genetic contributors to drug action, followed by sections examining the relationship between genetic variation and outcomes of treatment with commonly used cardiovascular drugs. The conclusion includes a discussion of the current status of implementing this new knowledge in drug prescribing.
INTRODUCTION – BROAD PRINCIPLES
Principles of Drug Action
Drugs interact with specific receptors in the circulation, on the cell surface, or inside cells, to exert their beneficial and detrimental effects. Variability in response to drug therapy can reflect either variability in the amount of drug delivered to receptor sites (pharmacokinetic factors) or variability in response at equivalent drug concentrations (pharmacodynamic factors). Describing the molecular basis of these processes is the critical first step to evaluating the extent to which their genetic variation leads to variable drug responses.
Pharmacokinetics
This encompasses the processes of absorption, distribution, metabolism, and elimination. The most widely-studied of these processes is metabolism, most often accomplished by members of the cytochrome P450 (CYP) superfamily. CYP-mediated biotransformation generally results in metabolites that are more polar than the parent drug, and thus more readily excreted. Metabolites may display no pharmacologic activity, similar pharmacologic activity to the parent drug, or different pharmacologic activity from the parent drug. In some cases, the parent drug is inactive (a “prodrug”) and requires bioactivation by metabolism to exert its pharmacologic effects; clopidogrel is one example. A second common pathway of drug metabolism is conjugation with specific side groups, such as methyl, acetyl, or glucuronide moieties; this is accomplished by specific transferases (e.g. methyltransferases, acetyltransferases, etc.).
The processes of absorption, distribution, and elimination may be passive or may depend on expression or function of specific drug transport molecules. Thus, for example, renal or biliary excretion of a drug or drug metabolite often involves active uptake of drug by specific transport molecules into the renal or biliary epithelium, followed by drug excretion of drug into the urine or bile. Similarly, absorption from the gastrointestinal tract may involve active uptake into enterocytes followed by excretion into the portal circulation or back into the gut by specific drug uptake and efflux molecules.
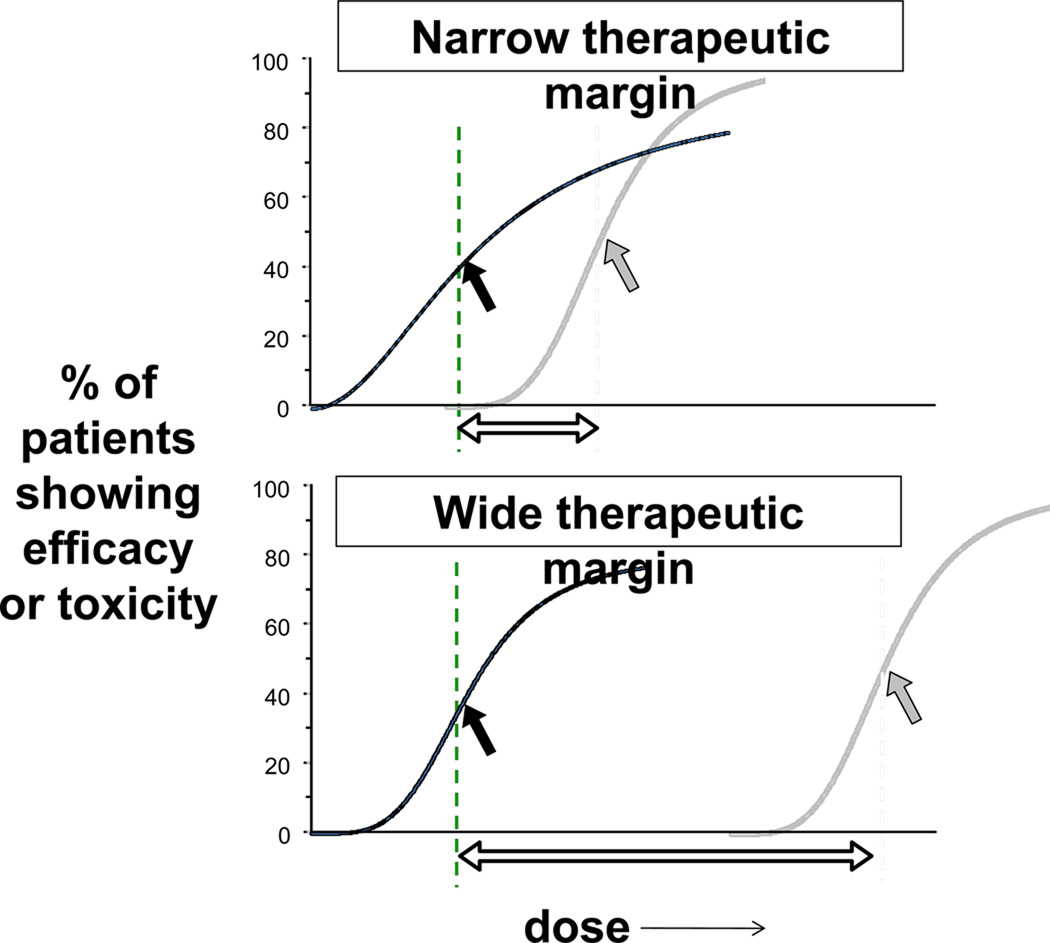
Variation in these processes, by genetic factors, drug interactions, or generalized disease of organ function (e.g. liver or kidney disease) may alter pharmacokinetics, and hence drug response. The functional consequences of such variation are drug-specific. For drugs whose metabolism or elimination is dependent on the function of a single CYP or drug transport molecule, variation in that pathway may lead to especially large variability in drug concentrations. This is a situation that has been termed “high risk pharmacokinetics”2 and applies to many commonly used drugs, including clopidogrel, warfarin, and digoxin. Variable efficacy by this mechanism is a particular risk for prodrugs. In the case of drugs inactivated by metabolism, the problem is especially acute when the margin between concentrations required for efficacy and those producing side effects is narrow, thus increasing risk for side effects (Figure 2). On the other hand, altered function of a single CYP or transporter is unlikely to have major consequences for a drug that is eliminated by multiple pathways.
Figure 2. Therapeutic Index.
For any drug, there is a relationship between dose and efficacy (black lines) and a second relationship between dose and toxicity (gray lines). These curves are derived from populations, and efficacy may be incomplete, as indicated in these examples. The arrows on each plot identify the dose at which 50% of the response is seen, and the therapeutic index is indicated by the open arrow at the bottom of each plot. Some drugs considered here, such as warfarin and clopidogrel, have narrow therapeutic indices, while others (beta-blockers, statins) have wider ones. DNA variants may modulate the relationship between efficacy and toxicity in populations and in individuals.
Pharmacodynamic factors
The most straightforward example of genetically-determined variability in drug action despite equivalent drug concentration at the receptor site is functional variation in the gene encoding the drug receptor itself. Thus, coding or regulatory variation in VKORC1, encoding the warfarin target, is a key contributor to variability in warfarin response3 and variants in the beta-adrenergic receptor genes appear to contribute to variability in response to beta-adrenergic receptor blockers (beta-blockers).4
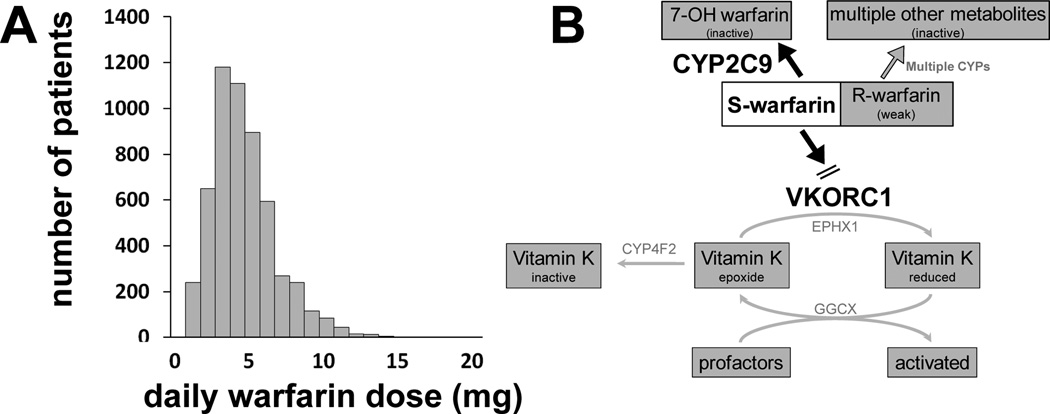
More generally, any physiologic variable that impacts the interaction of drug with its receptor or the downstream effects that this interaction produces can itself modulate drug action. Examples include changes in serum potassium influencing drug-ion channel interactions, or variable catecholamines affecting a drug-adrenergic receptor interaction. This concept extends far beyond single genetic variants and encompasses the idea that drug-receptor interactions occur in a complex biologic milieu5 whose function may be modified by multiple interacting modules, each of which may display genetic variation (Figure 3). Another factor clearly contributing to pharmacodynamic variability is the disease process itself that the drug is targeting.
Figure 3. Multiple genes affecting warfarin dose.
A. Steady state warfarin dosage varies widely. Adapted by permission from Kurnik et al.157 B. As discussed in the text, variants in CYP2C9 and VKORC1 contribute to variable warfarin dosage. Other genes that may also play a role are illustrated here. Pharmacokinetic response genes include other CYPs, although their role is likely minimal. Within the pharmacodynamic pathway, the microsomal epoxide hydroxylase (EPHX1) and γ-glutamyl carboxylase (GGCX) have been associated with warfarin response. Other genes include calumenin, which regulates the carboxylation of coagulation factors and proteins; apolipoprotein E, which facilitates cellular uptake of chylomicrons, the main vehicle of vitamin K transport to the liver; coagulation pathway genes encoding for factors II, VII, IX, and X; and genes that accelerate the inactivation of factor Va and VIIIa (endothelial protein C receptor (PROCR) and protein S (PROS1)).89
Approaches to identifying genetic contributors to variable drug actions
Candidate genes
One intuitively appealing experimental design is to identify a series of genes modulating an important biological process and to then address variability in the biologic process by examining the effects of common variants in such “candidate” genes. In general, it has been difficult to replicate even the most biologically appealing results in candidate gene studies.6 However, studies of candidate variants in pharmacokinetic pathways provide an important exception to this rule: as discussed below, variants in pharmacokinetic pathways involved in variable responses to warfarin, clopidogrel, and simvastatin were first implicated by candidate gene studies based on the drugs’ pharmacokinetics, and these have since been validated in genome-wide association (GWA) studies.7–10 These studies provide further evidence for the very large effects that single gene variants may exert for some drugs.
Unbiased approaches
The GWA paradigm has now been applied in over 800 studies11 to identify common genetic variants modulating disease susceptibility, physiologic traits, and – in a small number of studies – variable drug responses.12 This approach has revealed new biologic mechanisms, but has also been subject to a number of criticisms; interestingly, GWA for drug response phenotypes seems less subject to these drawbacks. For example, while the signals identified in a GWA study may be statistically robust, the odds ratios that they confer are often small, and validated only with very large numbers of subjects; one reasonable explanation is that single variant alleles with major detrimental effects are unlikely to propagate in a large population. However, there may be no survival disadvantage for common polymorphisms modulating drug action; hence, statistically valid associations have emerged in GWA studies of drug response traits that involve relatively small numbers of patients.7–10 A second criticism has been that loci identified by GWA studies explain only small proportions of variability in the traits being studied. However, GWA has explained relatively large proportions of variability compared to studies of traits such as disease susceptibility or physiologic measurements. Thus, for example, adding pharmacogenomic variants to clinical information improves the predictability of warfarin dose from ~20% to 45–60%13 and CYP2C19*2 explains 12% of variability in clopidogrel inhibition of ADP-induced platelet aggregation.9 A third criticism of the GWA approach has been that a “hit” often identifies a genomic locus, but generally leaves open the question of which specific variant, or combinations of variants, within the locus actually exerts the biologic effects to modulate the trait. This has not proven to be the case in pharmacogenetically-based GWA studies conducted to date, although as studies include increasing numbers of patients, such “unexplained” loci may well emerge.
Another potentially powerful approach is to use GWA to examine variability in common “intermediate” phenotypes (endophenotypes) such as electrocardiographic intervals or LDL cholesterol. These studies can readily acquire tens of thousands of subjects and thus identify multiple genomic loci with extremely low P values that contribute to the trait being studied. Such loci, then, can be taken forward as candidates, alone or in combination, that modulate drug response or other “hard” cardiovascular end-points such as myocardial infarction or sudden arrhythmic death. The development of rapid relatively inexpensive large scale sequencing capacity (“next-generation sequencing”) is a logical next step in this area of science.
Combined approaches – moving to systems biology
An intermediate experimental approach, between single candidate gene variant and whole genome approaches, is to consider the possibility that variability in a trait such as drug response is attributable to combinations5 of variants in functionally-linked sets of genes. This approach may be especially appealing to dissect the contributions of variable physiologic perturbations caused by disease to variable drug responses, as in the use of beta-blockers in heart failure14 discussed further below. One experimental approach is to examine associations between large numbers (hundreds or thousands) of variants in such candidate pathways and specific traits, this idea has been applied to examine variability in response to HMG-CoA reductase inhibitors (statins),15 among others.
Another highly promising approach is to combine information on genetic variation with other high dimensional data, such as gene expression profiling in cell lines or specific tissues, to identify networks of genes modulating variable drug response phenotypes.16,17 Studies of drug response in mice or zebrafish with defined genetic backgrounds have also been proposed as approaches to discovery and validation in pharmacogenomics.18,19
DISEASE-SPECFIC CARDIOVASCULAR THERAPIES
Atherosclerosis /statins
HMG CoA reductase (HMGCR) inhibitors (statins) are highly efficacious in the primary and secondary prevention of cardiovascular disease. HMGCR catalyzes the rate limiting step of cholesterol biosynthesis. By attenuating the endogenous production of cholesterol, statins also upregulate expression of the LDL receptor (LDLR) in a variety of tissues. This is the fundamental basis for the clinical effect of statins in lowering plasma LDL cholesterol (LDLC).20
Cholesterol lowering efficacy – in vitro studies
The clinical results described below have been further validated by studies in lymphoblastoid cell lines (LCLs) from individuals with specific variants thought to affect statin response. For example, in vitro statin exposure of LCLs derived from African American L5 carriers showed smaller induction of cell surface LDLR protein compared to non-carriers, a finding consistent with the smaller reductions in plasma LDL-cholesterol with statin treatment seen clinically.21 Similarly, in vitro statin exposure identified a mechanism whereby the HMGCR H7 haplotype modulates response to statins: one of the three SNPs within this haplotype, rs3846662, was associated with differential regulation of statin-induced expression of an HMGCR splice variant lacking exon 13, HMGCR13(−).22 Additional studies found that cellular enrichment of HMGCR13(−) attenuated sensitivity to statin inhibition, demonstrating the likelihood that HMGCR alternative splicing is not only a marker but also a determinant of inter-individual variation in LDL-cholesterol reduction with statin treatment.22 Compared to the 1–2% of variation explained by most gene variants related to drug response, statin-induced HMGCR13(−) expression explained up to 15% of the variation in LDL-cholesterol response to statins.22
Cholesterol lowering efficacy – human studies
The LDL-lowering effect of statins has been unequivocally associated with variation in two key pharmacodynamic candidate genes (HMGCR and LDLR). The H7 haplotype within HMGCR, defined by the presence of three intronic single nucleotide polymorphisms (SNPs), rs17244841, rs3846662, and rs17238540, has been associated with an 11–19% smaller reduction in LDL-cholesterol with statin treatment in multiple independent populations as well as ethnically diverse population based cohorts.15,23–25 The H7 haplotype has been shown to interact with other genetic variants including a second HMGCR haplotype, H2, defined primarily by rs3846662, as well as the LDLR L5 haplotype, defined by six SNPs within the LDLR 3’ UTR. LDL-cholesterol lowering with statin treatment in African Americans who carry multiple copies of these haplotypes is further attenuated compared to those who carry any haplotype alone.21,24,25
Additionally, genetic variants in CYP3A4, that metabolizes simvastatin, atorvastatin and lovastatin, have been associated with variability in statin efficacy. Both a non-synonymous polymorphism (M445T, rs4986910) as well the CYP3A4*4 haplotype have been associated with lower LDL cholesterol levels with atorvastatin.26,27 By contrast, the lipid lowering effect of statins appears to be attenuated in carriers of either a CYP3A4 promoter polymorphism (A290G) or the CYP3A4*1G haplotype.26–28 The A290G variant in CYP3A4 is in high linkage disequilibrium (D’ >0.8) with an intronic variant in CYP3A5.29
Initial application of GWA approaches to the study of statin efficacy did not identify variants at the genome-wide level.30 Subsequently, Bayesian methods have identified CLMN1 as a predictor of LDL-cholesterol lowering efficacy.31 Analysis of genetic determinants of statin efficacy generally relies on single doses used in large clinical trials, so individuals with different dose response curves can appear similar if sampled at only one dose. Analysis of response to multiple doses in individual subjects, accomplished in electronic medical record (EMR) environments, has been proposed as method to refine analysis of statin efficacy in clinical practice.32,33
Predictors of muscle toxicity
The spectrum of statin-related myotoxicity includes stopping the drug for muscle aches to serum creatine kinase (CK) elevations to the very rare occurrence of rhabdomyolysis. Initial studies identified variants in CYP3A534 and SLCO1B1 as potential predictors of myotoxicity.35 A GWA study examined 85 subjects with myotoxicity during high dose (80 mg/day) simvastatin and implicated a SNP in SLCO1B1; resequencing and subsequent replication indicated that a non-synonymous variant, resulting in Val174Ala, conferred an odds ratio for myopathy of 16.9 in homozygotes (95% confidence interval [CI], 4.7–61.1) and 4.5 (2.6–7.7) in heterozygotes.8 Homozygotes made up 2.1% of the study group and heterozygotes 24.9%; this single variant accounted for ~60% of risk. This association has since been replicated in two independent study populations.36,37
Predictors of clinical outcome
A missense SNP, Trp719Arg (rs20455), in KIF6 has been associated with increased risk of coronary artery disease, coronary heart disease and myocardial infarction,38,39 although the mechanisms for the association are unknown and the association is disputed.40 In multiple trials,38,39 statin treatment has been shown to significantly reduce coronary events in carriers of Trp719Arg, and SNPs in high linkage disequilibrium with it, with relative risk reduction of ~30%, while no benefit of statin treatment is reported in non-carriers.
Clinical Utility
SLCO1B1 genotype exerts a large effect on risk for simvastatin myotoxicity, and may be useful in some settings. Current studies identifying genomics of LDL response are a key step to addressing the broader question of whether genomic markers identify subjects developing coronary events during statin therapy.
Antiarrhythmics and drug-induced arrhythmias
Drug-induced long QT syndrome
1–5% of patients exposed to QT-prolonging antiarrhythmic drugs (sotalol, dofetilide, quinidine) develop striking QT interval prolongation and the polymorphic ventricular tachycardia, torsades de pointes (TdP);41 this also occurs with a range of “non-cardiovascular” therapies including some antipsychotics, antibiotics, and methadone.41 Many clinical features of drug-induced TdP (diTdP) resemble those seen in the congenital long QT syndromes, diseases caused by mutations in genes (13 have now been described) encoding ion channels or function-modifying subunits and characterized by incomplete penetrance.42
Small studies have estimated that 10–40% of subjects with diTdP have subclinical congenital long QT syndrome gene mutations.43–45 A study using targeted next-generation sequencing to screen all 13 congenital long QT syndrome disease genes and other arrhythmia susceptibility genes identified rare variants predicted to be deleterious to protein function in 20/31 patients (64.5%).46 Thus, there seems little doubt that congenital long QT syndrome mutations contribute to risk for diTdP, but the extent to which they explain the risk is uncertain. The beta-adrenergic receptor polymorphisms discussed below were not associated with diTdP.47
GWA approaches in electrophysiology
GWA has implicated multiple loci modulating variability in the normal QT interval duration.48–50 These fall into two broad groups. The first includes “obvious” candidates, that encode cardiac ion channels and in which mutations cause the congenital long QT or other arrhythmia susceptibility syndromes. These include SCN5A, encoding the cardiac sodium channel as well as genes encoding a series of cardiac potassium channels (KCNH2, KCNQ1, KCNJ2, KCNE1). The second group includes genes not previously implicated as modulating the QT interval. One of these is NOS1AP, encoding an accessory protein for neuronal nitric oxide synthase and another is GINS3, now implicated as a potential modulator of the extent to which challenge with a QT-prolonging drug (dofetilide) modulates action potential duration in zebrafish.51 The convergence of the zebrafish experiment on the GINS3 locus provides biologic validation for the GWA result, although the underlying physiology remains to be explored. One study suggested that a NOS1AP variant was associated with total and cardiovascular mortality during treatment with dihydropyridine calcium channel blockers.52 Variants in NOS1AP have also been reported to modulate the risk of arrhythmias, at equivalent QT interval durations, in patients with the congenital long QT syndrome,53,54 and to modulate risk for sudden death in the general population.55 To date, the mechanisms whereby NOS1AP variation affects QT remain uncertain; one study suggested that the NOS1AP is a modulator of L-type calcium current.56 Variability in PR and QRS durations have also been analyzed by GWA. As with QT, the results point to previously-implicated genes, as well as to loci previously not implicated in cardiac electrophysiology. One locus, at chromosome 4q25, has consistently been associated with risk of atrial fibrillation (AF).57 Preliminary data have linked 4q25 variants to outcome for ablation58 and of antiarrhythmic drug therapy for AF.59 Candidate gene studies have also suggested a role for beta-adrenergic receptor polymorphisms in rate control during AF.60
Clinical Utility
Identifying patients at risk for long QT-related arrhythmias during drug therapy may become possible as platforms to identify both common and rare variants are increasingly deployed. Early studies suggest that available AF therapies may be less effective in some genetically-defined subsets, and these patients may thus be candidates for alternate approaches.
Hypertension
Pharmacogenomics of the antihypertensive response61
One major focus of candidate gene studies has been two common, nonsynonymous SNPs, resulting in Ser49Gly and Arg389Gly, in the gene encoding the beta1 adrenergic receptor (ADRB1). These variants demonstrate altered biologic function in vitro,62 including enhanced agonist induced adenylyl cyclase activation by Gly49 compared to Ser49 and by Arg389 compared to Gly389. The effects in humans, where multiple haplotypes are the rule, have been more difficult to dissect. Many, but not all, studies have shown that the Arg389Arg genotype, or Ser49/Arg389 haplotype, is associated with the greatest BP lowering.4,63,64 These findings are consistent with beta-blocker response associations in heart failure outcomes discussed below.
A common functional polymorphism resulting in Gly460Trp in the alpha-adducin gene ADD161 has been associated with response to thiazides in some but not all studies. This association led to the development of a novel antihypertensive drug class targeting adducin.65 NEDD4L is also a candidate gene with a documented functional SNP, a role in sodium reabsorption, and several studies have found an association between this SNP and BP response with thiazides.66,67
There has been one GWA study conducted to date examining antihypertensive response, and this revealed an association between thiazide response in African Americans and a locus at chromosome 12q15.68 This association has since been replicated in an independent cohort.69 While no genes in the region are obvious thiazide response genes, an interesting candidate is FRS2, which is involved in fibroblast growth factor signaling which plays a role in vascular smooth muscle cell regulation.
Myocardial infarction, stroke, and death during antihypertensive therapy
The ADD1 Gly460Trp polymorphism has been associated with increased risk of myocardial infarction or stroke during thiazide diuretic treatment.70 However, analyses from both ALLHAT and INVEST were unable to replicate this finding.71,72 In the INVEST cohort, the ADRB1 Ser49/Arg389 haplotype was associated with increased risk of death, and the risk of this allele was offset by treatment with the beta-blocker atenolol compared to the calcium channel blocker (CCB) verapamil.4 A cohort study similarly found significant interactions between SNPs in ADRB1 and risk for myocardial infarction or stroke.
Other studies have suggested potential associations between SNPs in CACNA1C, CACNB2, and KCNMB1 (involved in calcium signaling) and MI or stroke with beta-blockers versus CCBs.73–75 CACNB2 was also one of ten genes that replicated in large GWA studies of hypertension.76 Other reported genetic associations include variable stroke risk by genotype for an MMP3 promoter polymorphism in patients treated with lisinopril,77 an ACE gene polymorphism and differential outcomes by genotype during treatment with an ACE inhibitor78 and different treatment-related outcomes with thiazides and beta-blockers, but not diltiazem, by NEDD4L genotype.66
Clinical Utility
Genetic determinants of BP and long-term outcomes in hypertensive patients are being identified, and their role in choosing among therapies is under active investigation.
Heart Failure
The largest body of literature centers on the genetic associations between ADRB1 SNPs (discussed above) and either beta-blocker mediated improvements in left ventricular ejection fraction (LVEF) or clinical outcomes.4 Some studies have suggested that the Arg389Arg genotype is associated with the greatest improvement in LVEF, but this has not been consistently observed.4 In BEST, a large trial of bucindolol in heart failure, the survival benefit was confined to Arg389Arg patients, while there were no difference for Gly389 carriers on therapy versus those on placebo.79 The sponsor for bucindolol has announced plans for a superiority trial in 3,200 ADRB1 Arg389Arg patients who will be randomized to long-acting metoprolol or bucindolol.80 A population cohort suggested that Arg389Arg patients on high dose beta-blocker had significantly better outcomes than those with the same genotype on no or low dose beta-blocker, also consistent with the findings in BEST.81 However, other studies have not documented an association between Arg389Gly genotype and improved outcome with beta-blockers.82,83
These differences may be explained by whether studies examine outcomes in patients treated with beta-blocker compared to a control (low dose or placebo), or whether they examine outcome by genotype in a cohort of patients all treated with beta-blocker. One interpretation of the data is that Gly389 carriers may be at lower risk of adverse outcomes, and so garner less benefit from beta-blockers than Arg389Arg patients; in this case, beta-blocker treatment benefits Arg389Arg patients, moving their outcomes to that of Gly389 carriers (treated or untreated). This is consistent with the treatment related outcomes in hypertension for this gene, discussed above, where beta-blocker treatment reduced the risk of the Arg389Arg genotype.
The adrenergic signaling pathway includes a number of other genes that have been associated with severity of the heart failure phenotype or with benefits of response to treatment. A 12 nucleotide (4 amino acid) insertion deletion polymorphism in ADRA2C has been shown to lead to a loss of feedback inhibition, resulting in increased norepinephrine, and in combination with the ADRB1 Arg389Arg genotype has been associated with increased risk of heart failure in African Americans.84 A study of metoprolol reported that the combination of ADRB1 Arg389Arg/ ADRA2C Del carrier status led to the greatest improvement in LVEF.85 By contrast, ADRA2C Del carrier status was associated with reduced efficacy (death and hospitalization outcomes) in BEST.86 The difference in effect of ADRA2C genotype of efficacy in these two studies may reflect differences in the pharmacological properties of bucindolol versus metoprolol.
The Leu41Gln variant in GRK5 is common in African-Americans, and has been found to blunt catecholamine-induced responses in vitro and in experimental animals.87 In three reported cohort studies,82,87 carriers of the Leu41 variant had improved outcomes compared to those with Gln41Gln. In addition, Leu41 carriers did not appear to derive benefit from beta-blocker therapy, while Gln41Gln seemed to derive substantial survival benefit.
Clinical Utility
Some subjects with heart failure may derive little benefit from beta-blocker therapy; trials are underway to further address this issue.
Warfarin
Genetics of Warfarin Response
CYP2C9 is largely responsible for the metabolic clearance of (S)-warfarin, the more active of the two warfarin enantiomers. Two non-synonymous reduction-of-function variants in CYP2C9, designated *2 and *3, are clearly associated with lower warfarin dose requirements and increased bleeding risk in Caucasian populations;3,88–90 they are much less prevalent in African Americans, and thus do not have as large a population effect on dose.91
VKORC1 is the warfarin sensitive and rate-limiting enzyme of the vitamin K cycle that recycles the epoxide and quinone form of vitamin K to the reduced non-oxidized form. Multiple linked variants in the VKORC1 promoter have been associated with variability in gene expression and dose requirement:3 rs9934438 in intron 1 (also designated 1173C/T) is as informative as haplotypes for predicting warfarin dose in Caucasians and African Americans. Additional VKORC1 variants and CYP2C9 variants (e.g., CYP2C9*5, *6, *8, and *11 alleles) may improve prediction, particularly in African Americans.92 Rare coding region variants that generate relative or absolute warfarin resistance have also been reported.93
GWA identified variants in CYP2C9, VKORC1, and CYP4F2, now recognized as a vitamin K1 oxidase, as determinants of increased warfarin dose requirements.10,94 Carriers of the CYP4F2 V433M allele (rs2108622) were found to have reduced capacity to metabolize vitamin K1, suggesting that the variant may be associated with elevated hepatic levels of vitamin K, thus explaining the higher dose requirements in those with this variant.95 The effects of CYP4F2 on warfarin dosing have not been consistent across studies.96 Other genes implicated in variable warfarin dosages are highlighted in Figure 3.
Dosing Algorithms
Numerous dosing algorithms have been developed that incorporate both clinical and genetic information to predict warfarin dose requirements. Two serve as useful examples: the International Warfarin Pharmacogenetics Consortium (IWPC) algorithm97 and an algorithm developed by Gage et al.98 Both groups developed models using clinical parameters alone and then developed models with both clinical and genetic information, and both demonstrated clinically meaningful improvements in dose prediction with the addition of genetic data. These models have been externally validated.99,100
Research on dosing algorithms has demonstrated three important findings: (1) The vast majority of models include both clinical parameters and a single VKORC1 SNP plus CYP2C9*2 and CYP2C9*3 variants. (2) Most recent models have similar overall predictive ability, explaining about 40–65% of warfarin dose variability (in mostly Caucasian populations), compared with only about 20% variability for algorithms that use clinical information alone.99–101 Whether the residual variability is genetic or environmental (e.g. due to diet) is under study. (3) They perform substantially better in non-African American populations than they do in African-American populations.92,97,100,101 All of the findings are consistent with the known associations between genetic variants and warfarin dosing across different populations.
Clinical Utility
It is currently unknown if pharmacogenetic-based dosing of warfarin will improve anticoagulation control and clinical outcomes. The largest published, randomized clinical trial to date included only 206 patients and did not demonstrate an advantage of pharmacogenetic dosing on anticoagulation control, despite the fact that the dosing algorithm clearly predicted the final stable dose better than a clinical-only algorithm.102 Another small study that used CYP2C9 only to determine dosing did show some benefit on anticoagulation control and minor bleeding, but suffered from a high drop-out rate.103 An observational study reported a relationship between providing warfarin genetic information to clinicians and reduced hospitalizations.104 Large ongoing and future randomized studies are addressing this issue.
Antiplatelet agents
Pharmacogenomics of Anti-platelet Medications
Dual anti-platelet therapy with aspirin and clopidogrel is the standard of care for prevention of thromboembolic events in patients at high risk of myocardial infarction. However, there is marked inter-individual variation in response to aspirin105–107 and clopidogrel.108–113 Ex vivo quantitative measures of changes in platelet aggregation in response to aspirin and clopidogrel indicate that response is normally distributed in the population (Figure 1). Mounting evidence supports a genetic component to both baseline platelet function and anti-platelet drug response. In healthy Amish families, the heritability (h2) of baseline ADP-stimulated platelet aggregation and changes in response to clopidogrel, and clopidogrel plus aspirin was 0.33 (p=0.005), 0.73 (p<0.001) and 0.83 (p<0.001), respectively;114 the heritability of response to aspirin monotherapy appears to be more modest (h2 = 0.19; p<0.01 for changes in collagen-induced platelet aggregation).115
Genetics of Aspirin Response
Most reports of aspirin response to date have studied one or a few candidate variants, and these have not yielded reproducible results. A meta-analysis116 of 50 polymorphisms in 11 genes reported in 31 studies and a combined sample size of 2834 subjects suggested that the common PLA1/2 polymorphism (Leu59Pro; rs5918) in glycoprotein IIIa (GPIIIa) does confer aspirin resistance (odds ratio in healthy subjects = 2.36, p = 0.009); however, when combining both healthy subjects and those with cardiovascular disease, the odds ratio was 1.14 (p = 0.40). The meta-analysis found no effect on aspirin sensitivity with variants in COX1, GPIa, GP1balpha; GPIIIa, GPVI, FXIII, P2Y1, and P2Y12,.
Genetics of Clopidogrel Response
Clopidogrel is a prodrug, requiring biotransformation in the liver to an active thiol derivative primarily by CYP2C19, CYP3A4/5, CYP1A2, and CYP2B6;117–119 the active metabolite irreversibly binds to platelet ADP P2Y12 receptors. Early candidate gene studies implicated variants in the P2Y12 receptor 120,121 and CYP3A4 122 as potential determinants of clopidogrel resistance; however these studies were not consistently replicated. More recently, the loss-of-function CYP2C19*2 allele (rs4244285) has been reproducibly shown to be associated with a decreased conversion of clopidogrel into its active metabolite,123–125 anti-platelet effect,126–134 and increased risk for cardiovascular events in patients on clopidogrel.135–140 A GWA study identified CYP2C19*2 as the single major genetic determinant of biochemical response to clopidogrel, accounting for approximately 12% of the variation in ADP-stimulated platelet aggregation during drug treatment.9 In a large meta-analysis CYP2C19*2 carriers treated with clopidogrel have an increased risk for major adverse cardiovascular events compared to non-carriers (hazard ratio (HR) 1.55, 95% CI 1.11–2.17 for heterozygotes; HR 1.76, 95% CI 1.24–2.50 for homozygotes), and increased risks of stent thrombosis (HR 2.67, 95% CI 1.69–4.22 for heterozygotes; HR 3.97, 95% CI 1.75–9.02 for homozygotes).139 In Caucasians, African-Americans, and Mexican Americans, CYP2C19*2 is present is 18–33% (2–3% homozygotes), and the allele frequency is higher in Asians. The studies showing a relationship between CYP2C19 genotype and clopidogrel response have been conducted in relatively high risk patients, and thus may not apply to all indications for clopidogrel such as atrial fibrillation, stroke, peripheral artery disease, or chronic stable angina.141,142
There is some evidence that the loss-of-function *3 variant, present in Asians, is also associated with poorer response. Other loss-of-function alleles are rare. By contrast, the gain-of-function *17 variant may be associated with increased response (and thus adverse bleeding outcomes);143 However it is unclear whether the *17 variant has effect that are independent of the *1 allele due to linkage disequilibrium.144
Variants in ABCB1, which is involved in clopidogrel absorption, has been implicated in clopidogrel response,145 but these findings have not been replicated in all studies. A recent study reported that the common decreased function Gln192Arg (rs662) allele in paraoxonase 1 (PON1) may be associated with decreased active metabolite levels, decreased inhibition of ADP-stimulated platelet aggregation, and increased risk for stent thrombosis;146 that study did not find an effect of CYP2C19 and the results await replication by others.
Clinical Utility
Guided by the replicated findings of CYP2C19*2 as a determinant of clopidogrel response in patients undergoing PCI, the FDA approved new labeling of clopidogrel in March 2010. This includes a boxed warning alerting physicians to the genetic findings and suggests alternative anti-platelet therapy in CYP2C19*2 homozygotes. These may include prasugrel147 and the investigational agent ticagrelor148 which are not appreciably affected by CYP2C19 genotype. The idea of increasing clopidogrel dose has also been evaluated149 but not specifically in CYP2C19*2 homozygotes.
SUMMARY AND FUTURE DIRECTIONS
The focus of this brief review has been to emphasize that even for widely-deployed and clinically-familiar therapies, there is substantial variability in response across patients: one size cannot fit all. Substantial work, particularly over the last decade has elucidated genetic contributors to this variability. These include variants in pharmacokinetic and pharmacodynamic pathways, with an increasing view that drugs act in a complex biologic milieu. The technology to allow accumulation of high dimensional datasets, such as genomic, tissue-specific expression, and drug response phenotypes in large groups of patients is enabling for this approach; next-generation sequencing will similarly expand our catalog of variants associated with variable drug responses.
There are substantial barriers to applying pharmacogenomic information to practice (Table 1), and these include challenges in discovery, translation, and implementation. One significant obstacle is determining the level of evidence that is required before a variant is moved into clinical practice. It is an interesting paradox that the ability to personalize therapy – to prescribe with high confidence that specific variant alleles influence the outcome in an individual patient – must rely on analysis of very large datasets, to enable discovery and replication of response in genetically defined subsets. Another set of challenges is logistic: determining genotype at the time drug is prescribed means the practitioner must get the result, know how to act on it, and recontact the patient if needed to change drug or dose. An alternate approach is a preemptive one, in which genotypic information is deposited into an EMR prior to drug prescription. The preemptive scenario has the potential appeal of automated delivery of point of care decision support to practitioners and the ability to acquire and manage large amounts of genomic information that may ultimately be accessible to an EMR system. Thus, advanced information technology must be part of a vision for developing, analyzing, and ultimately applying new genomic data to healthcare.33,150 Indentifying outcomes and analyzing costs are other challenges.
Table 1.
Issues in implementing pharmacogenomics in practice
| Discovery |
| Identifying the contribution of genetic variants to drug outcomes |
| Establishing interactions among genes and between genes and the environment |
| Translation |
| Establishing which genetic variants should be used in which settings |
| Implementation |
| Education |
| Patients |
| Healthcare providers |
| Genotyping |
| At the point of care or preemptively |
| Assay quality |
| Decision support |
| Developing rules |
| Deploying advice in an electronic medical record environment |
| Analyzing outcomes |
| Population and individual health |
| Balancing costs and benefits |
Translating fundamental discovery in genome science to individual patients and populations is a challenge for the field and genotyping for selection of appropriate drug therapy is one of the first ways this is being accomplished. In oncology, tumor genotyping is rapidly becoming standard of care to identify specific mutations that then dictate selection of therapy.151 Pre-prescription germline genotyping is becoming standard of care to reduce the risk of serious adverse reactions to carbamazepine152 and abacavir.153 In cardiovascular therapy, a number of centers are now deploying programs to use CYP2C19 genotypes to guide clopidogrel therapy. NIH is making major investments both in discovery of new pharmacogenomic pathways and how that information can be used to improve healthcare: these include the warfarin trials, and efforts such as the Pharmacogenomics Research Network154 and the electronic Medical Records and Genomics Network.150 The newly-developed NHGRI strategic plan includes an emphasis on clinical implementation of new genomic knowledge.155 Biomedical discovery over the past several decades has included studies to understand sources of individual variability in response to drug therapy. We are clearly entering an era where this knowledge will increasingly be leveraged to improve healthcare.
Acknowledgments
Grant Support: Supported in part by grants from the US National Institutes of Health (U19 HL065962, U01 GM074492, U01 HL105198, U01 HL069757, R01 DK080007)
Non-standard Abbreviations and Acronyms
- ACE
Angiotensin converting enzyme
- AF
Atrial fibrillation
- ALLHAT
The Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial
- CCB
Calcium channel blocker
- CI
Confidence interval
- CK
Creatine kinase
- CYP
cytochrome P450
- EMR
Electronic Medical Record
- GPIIIa
glycoprotein IIIa
- GWA
Genome-Wide Association
- HCTZ
Hydrochlorothiazide
- HMG-CoA
3-hydroxy-3-methylglutaryl-coenzyme A
- HMGCR
HMG-CoA reductase
- INVEST
International Verapamil SR/Trandolapril Study
- IWPC
International Warfarin Pharmacogenetics Consortium
- LCL
Lymphoblastoid cell line
- LDL
Low density lipoprotein
- LDLC
LDL cholesterol
- LDLR
LDL receptor
- LVEF
Left ventricular ejection fraction
- NHGRI
National Human Genome Research Institute
- NIH
National Institutes of Health
- PGRN
Pharmacogenomics Research Network
- SNP
Single nucleotide polymorphism
- UTR
Untranslated region
- VKORC1
Vitamin K epoxide reductase complex subunit 1;
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Dr. Roden receives grant support from NIH for research on antiarrhythmic pharmacogenomics, has been a consultant to Merck, Astellas, Sanofi-Aventis, and Novartis, and receives royalties for a patent describing genetic predictors of drug induced arrhythmias. Dr. Johnson serves as an advisor to Medco. Dr. Kimmel has served as a consultant to Centocor, Pfizer, Merck, and Novartis, and has received research funding from the Aetna Foundation and from Shire, Astra Zeneca, and Bristol Myers Squibb, all unrelated to the subject of this manuscript. Dr. Krauss receives research grants from Merck and Roche and consults for Celera and Genentech. Dr. Shuldiner receives grant support from NIH for research on anti-platelet pharmacogenomics, and is a consultant for Bristol Myer-Squibb/Sanofi-Aventis and United States Diagnostics Standards, Inc. Drs. Medina and Wilke report no potential conflicts.
References
- 1.Gu Q, Dillon CF, Burt VL. Prescription drug use continues to increase: U.S. prescription drug data for 2007–2008. NCHS Data Brief. 2010:1–8. [PubMed] [Google Scholar]
- 2.Roden DM, Stein CM. Clopidogrel and the concept of high risk pharmacokinetics. Circulation. 2009;119:2127–2130. doi: 10.1161/CIRCULATIONAHA.109.865907. [DOI] [PubMed] [Google Scholar]
- 3.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JA, Liggett SB. Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. 2011:366–378. doi: 10.1038/clpt.2010.315. 2011/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilke RA, Reif DM, Moore JH. Combinatorial pharmacogenetics. Nat Rev Drug Discov. 2005;4:911–918. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- 6.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genetics. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 7.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 9.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, Pollin TI, Post W, Parsa A, Mitchell BD, Faraday N, Herzog W, Gurbel PA. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA. Genomewide Association Studies and Assessment of the Risk of Disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 12.Motsinger-Reif AA, Jorgenson E, Relling MV, Kroetz DL, Weinshilboum R, Cox NJ, Roden DM. Genome-wide association studies in pharmacogenomics: successes and lessons. Pharmacogenet Genomics. 2010 doi: 10.1097/FPC.0b013e32833d7b45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The International Warfarin Pharmacogenetics Consortium. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorn GW. Adrenergic Signaling Polymorphisms and Their Impact on Cardiovascular Disease. Physiol Rev. 2010;90:1013–1062. doi: 10.1152/physrev.00001.2010. [DOI] [PubMed] [Google Scholar]
- 15.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–282. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 18.Lusis AJ, Weiss JN. Cardiovascular networks: systems-based approaches to cardiovascular disease. Circulation. 2010;121:157–170. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milan DJ, MacRae CA. Zebrafish genetic models for arrhythmia. Progress in Biophysics and Molecular Biology. 2010;98:301–308. doi: 10.1016/j.pbiomolbio.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangravite LM, Medina MW, Cui J, Pressman S, Smith JD, Rieder MJ, Guo X, Nickerson DA, Rotter JI, Krauss RM. Combined Influence of LDLR and HMGCR Sequence Variation on Lipid-Lowering Response to Simvastatin. Arterioscler Thromb Vasc Biol. 2010 doi: 10.1161/ATVBAHA.110.203273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnelly LA, Doney AS, Dannfald J, Whitley AL, Lang CC, Morris AD, Donnan PT, Palmer CN. A paucimorphic variant in the HMG-CoA reductase gene is associated with lipid-lowering response to statin treatment in diabetes: a GoDARTS study. Pharmacogenet Genomics. 2008;18:1021–1026. doi: 10.1097/FPC.0b013e3283106071. [DOI] [PubMed] [Google Scholar]
- 24.Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI. Variation in the 3-Hydroxyl-3-Methylglutaryl Coenzyme A Reductase Gene Is Associated With Racial Differences in Low-Density Lipoprotein Cholesterol Response to Simvastatin Treatment. Circulation. 2008;117:1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 25.Poduri A, Khullar M, Bahl A, Sehrawat BS, Sharma Y, Talwar KK. Common variants of HMGCR, CETP, APOAI, ABCB1, CYP3A4, and CYP7A1 genes as predictors of lipid-lowering response to atorvastatin therapy. DNA Cell Biol. 2010;29:629–637. doi: 10.1089/dna.2009.1008. [DOI] [PubMed] [Google Scholar]
- 26.Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ. CYP3A4 genotypes and plasma lipoprotein levels before and after treatment with atorvastatin in primary hypercholesterolemia. The American Journal of Cardiology. 2004;93:104–107. doi: 10.1016/j.amjcard.2003.08.078. [DOI] [PubMed] [Google Scholar]
- 27.Wang A, Yu BN, Luo CH, Tan ZR, Zhou G, Wang LS, Zhang W, Li Z, Liu J, Zhou HH. Ile118Val genetic polymorphism of CYP3A4 and its effects on lipid-lowering efficacy of simvastatin in Chinese hyperlipidemic patients. Eur J Clin Pharmacol. 2005;60:843–848. doi: 10.1007/s00228-004-0848-7. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Zhang Lr, Fu Q. CYP3A4polymorphism is associated with lipid-lowering efficacy of atorvastatin but not of simvastatin. Eur J Clin Pharmacol. 2008;64:877–882. doi: 10.1007/s00228-008-0502-x. [DOI] [PubMed] [Google Scholar]
- 29.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genetics. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, Hovingh GK, Kastelein JJP. Comprehensive Whole-Genome and Candidate Gene Analysis for Response to Statin Therapy in the Treating to New Targets (TNT) Cohort / CLINICAL PERSPECTIVE. Circ Cardiovasc Genet. 2009;2:173–181. doi: 10.1161/CIRCGENETICS.108.818062. [DOI] [PubMed] [Google Scholar]
- 31.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT, Ridker PM, Chatterjee A, Rotter JI, Nickerson DA, Stephens M, Krauss RM. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilke RA, Berg RL, Linneman JG, Zhao C, McCarty CA, Krauss RM. Characterization of low-density lipoprotein cholesterol-lowering efficacy for atorvastatin in a population-based DNA biorepository. Basic Clin Pharmacol Toxicol. 2008;103:354–359. doi: 10.1111/j.1742-7843.2008.00291.x. [DOI] [PubMed] [Google Scholar]
- 33.Wilke RA, Xu H, Denny JC, Roden DM, Krauss RM, McCarty CA, Davis RL, Skaar T, Lamba J, Savova G. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther. 2011;89:379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 35.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 36.Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS. The SLCO1B1*5 Genetic Variant Is Associated With Statin-Induced Side Effects. J Am Coll Cardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donnelly LA, Doney ASF, Tavendale R, Lang CC, Pearson ER, Colhoun HM, McCarthy MI, Hattersley AT, Morris AD, Palmer CNA. Common Nonsynonymous Substitutions in SLCO1B1 Predispose to Statin Intolerance in Routinely Treated Individuals With Type 2 Diabetes: A Go-DARTS Study. Clin Pharmacol Ther. 2011;89:210–216. doi: 10.1038/clpt.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iakoubova OA, Robertson M, Tong CH, Rowland CM, Catanese JJ, Blauw GJ, Jukema JW, Murphy MB, Devlin JJ, Ford I, Shepherd J. KIF6 Trp719Arg polymorphism and the effect of statin therapy in elderly patients: results from the PROSPER study. Eur J Cardiovasc Prev Rehabil. 2010;17:455–461. doi: 10.1097/HJR.0b013e328336a0dd. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Iakoubova OA, Shiffman D, Devlin JJ, Forrester JS, Superko HR. KIF6 polymorphism as a predictor of risk of coronary events and of clinical event reduction by statin therapy. Am J Cardiol. 2010;106:994–998. doi: 10.1016/j.amjcard.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 40.Assimes TL, Holm H, Kathiresan S, Reilly MP, Thorleifsson G, Voight BF, Erdmann J, Willenborg C, Vaidya D, Xie C, Patterson CC, Morgan TM, Burnett MS, Li M, Hlatky MA, Knowles JW, Thompson JR, Absher D, Iribarren C, Go A, Fortmann SP, Sidney S, Risch N, Tang H, Myers RM, Berger K, Stoll M, Shah SH, Thorgeirsson G, Andersen K, Havulinna AS, Herrera JE, Faraday N, Kim Y, Kral BG, Mathias RA, Ruczinski I, Suktitipat B, Wilson AF, Yanek LR, Becker LC, Linsel-Nitschke P, Lieb W, Konig IR, Hengstenberg C, Fischer M, Stark K, Reinhard W, Winogradow J, Grassl M, Grosshennig A, Preuss M, Schreiber S, Wichmann HE, Meisinger C, Yee J, Friedlander Y, Do R, Meigs JB, Williams G, Nathan DM, Macrae CA, Qu L, Wilensky RL, Matthai WH, Jr, Qasim AN, Hakonarson H, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser VE, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Martinelli N, Olivieri O, Trabetti E, Malerba G, Pignatti PF, Guiducci C, Mirel D, Parkin M, Hirschhorn JN, Asselta R, Duga S, Musunuru K, Daly MJ, Purcell S, Eifert S, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Ouwehand WH, Deloukas P, Scholz M, Cambien F, Huge A, Scheffold T, Salomaa V, Girelli D, Granger CB, Peltonen L, McKeown PP, Altshuler D, Melander O, Devaney JM, Epstein SE, Rader DJ, Elosua R, Engert JC, Anand SS, Hall AS, Ziegler A, O'Donnell CJ, Spertus JA, Siscovick D, Schwartz SM, Becker D, Thorsteinsdottir U, Stefansson K, Schunkert H, Samani NJ, Quertermous T. Lack of association between the Trp719Arg polymorphism in kinesin-like protein-6 and coronary artery disease in 19 case-control studies. J Am Coll Cardiol. 2010;56:1552–1563. doi: 10.1016/j.jacc.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roden DM. Long QT syndrome: reduced repolarization reserve and the genetic link. J Intern Med. 2006;259:59–69. doi: 10.1111/j.1365-2796.2005.01589.x. [DOI] [PubMed] [Google Scholar]
- 42.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 43.Paulussen AD, Gilissen RA, Armstrong M, Doevendans PA, Verhasselt P, Smeets HJ, Schulze-Bahr E, Haverkamp W, Breithardt G, Cohen N, Aerssens J. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J Mol Med. 2004;82:182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 44.Yang P, Kanki H, Drolet B, Yang T, Wei J, Viswanathan PC, Hohnloser SH, Shimizu W, Schwartz PJ, Stanton MS, Murray KT, Norris K, George ALJ, Roden DM. Allelic variants in Long QT disease genes in patients with drug-associated Torsades de Pointes. Circulation. 2002;105:1943–1948. doi: 10.1161/01.cir.0000014448.19052.4c. [DOI] [PubMed] [Google Scholar]
- 45.Itoh H, Sakaguchi T, Ding WG, Watanabe E, Watanabe I, Nishio Y, Makiyama T, Ohno S, Akao M, Higashi Y, Zenda N, Kubota T, Mori C, Okajima K, Haruna T, Miyamoto A, Kawamura M, Ishida K, Nagaoka I, Oka Y, Nakazawa Y, Yao T, Jo H, Sugimoto Y, Ashihara T, Hayashi H, Ito M, Imoto K, Matsuura H, Horie M. Latent Genetic Backgrounds and Molecular Pathogenesis in Drug-induced Long QT Syndrome. Circ Arrhythmia Electrophysiol. 2009 doi: 10.1161/CIRCEP.109.862649. CIRCEP. [DOI] [PubMed] [Google Scholar]
- 46.Ramirez AH, Shaffer C, Sexton DP, Levy S, George AL, Roden DM. Abstract 12560: Targeted Next Generation Exomic Sequencing of Cardiac Electrophysiology Genes in Cases of Drug-Induced Torsades de Pointes. Circulation. 2010;122:A12560. [Google Scholar]
- 47.Kanki H, Yang P, Xie HG, Kim RB, George AL, Roden DM. Polymorphisms in beta-adrenergic receptor genes in patients with the acquired long QT syndrome. J Cardiovasc Electrophysiol. 2002:252–256. doi: 10.1046/j.1540-8167.2002.00252.x. [DOI] [PubMed] [Google Scholar]
- 48.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O'Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 49.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PIW, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JCM, Hofman A, Heckbert SR, O'Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Ch Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genetics. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WHL, Muhleisen TW, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Muller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis GR, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genetics. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kaab S, Roden DM, Macrae CA. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker ML, Visser LE, Newton-Cheh C, Hofman A, Uitterlinden AG, Witteman JC, Stricker BH. A common NOS1AP genetic polymorphism is associated with increased cardiovascular mortality in users of dihydropyridine calcium channel blockers. Br J Clin Pharmacol. 2009;67:61–67. doi: 10.1111/j.1365-2125.2008.03325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, Greenberg DA, Schwartz PJ, George AL., Jr NOS1AP Is a Genetic Modifier of the Long-QT Syndrome. Circulation. 2009;120:1657–1663. doi: 10.1161/CIRCULATIONAHA.109.879643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomas M, Napolitano C, De GL, Bloise R, Subirana I, Malovini A, Bellazzi R, Arking DE, Marban E, Chakravarti A, Spooner PM, Priori SG. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. J Am Coll Cardiol. 2010;55:2745–2752. doi: 10.1016/j.jacc.2009.12.065. [DOI] [PubMed] [Google Scholar]
- 55.Kao WH, Arking DE, Post W, Rea TD, Sotoodehnia N, Prineas RJ, Bishe B, Doan BQ, Boerwinkle E, Psaty BM, Tomaselli GF, Coresh J, Siscovick DS, Marban E, Spooner PM, Burke GL, Chakravarti A. Genetic Variations in Nitric Oxide Synthase 1 Adaptor Protein Are Associated With Sudden Cardiac Death in US White Community-Based Populations. Circulation. 2009;119:940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang KC, Barth AS, Sasano T, Kizana E, Kashiwakura Y, Zhang Y, Foster DB, Marban E. CAPON modulates cardiac repolarization via neuronal nitric oxide synthase signaling in the heart. Proc Natl Acad Sci U S A. 2008;105:4477–4482. doi: 10.1073/pnas.0709118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, Macrae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 58.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 59.Rowan SB, Estrada JC, Stubblefield T, Kucera G, Carter S, Roden DM, Darbar D. A single nucleotide polymorphism at 4q25 associated with atrial fibrillation modulates symptomatic response to antiarrhythmic drug therapy. Heart Rhythm. 2009;6 PO04-7. [Google Scholar]
- 60.Vaglio JC, Rowan SB, Stubblefield T, Carter S, Roden DM, Darbar D. Genetic and clinical predictors of response to rate control therapy in patients with atrial fibrillation. Circulation. 2008;118:S828. [Google Scholar]
- 61.Johnson JA. Pharmacogenomics of antihypertensive drugs: past, present and future. 2010:487–491. doi: 10.2217/pgs.10.34. 2010/03/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brodde OE. Beta1- and beta2-adrenoceptor polymorphisms and cardiovascular diseases. Fundam Clin Pharmacol. 2008;22:107–125. doi: 10.1111/j.1472-8206.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 63.Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74:44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, Liu ZQ, Tan ZR, Chen XP, Wang LS, Zhou G, Zhou HH. Gly389Arg polymorphism of beta1-adrenergic receptor is associated with the cardiovascular response to metoprolol. Clin Pharmacol Ther. 2003;74:372–379. doi: 10.1016/S0009-9236(03)00224-8. [DOI] [PubMed] [Google Scholar]
- 65.Lanzani C, Citterio L, Glorioso N, Manunta P, Tripodi G, Salvi E, Carpini SD, Ferrandi M, Messaggio E, Staessen JA, Cusi D, Macciardi F, Argiolas G, Valentini G, Ferrari P, Bianchi G. Adducin- and Ouabain-Related Gene Variants Predict the Antihypertensive Activity of Rostafuroxin, Part 2: Clinical Studies. 2010;2 doi: 10.1126/scitranslmed.3001814. 59ra87. [DOI] [PubMed] [Google Scholar]
- 66.Svensson-Farbom P, Wahlstrand B, Almgren P, Dahlberg J, Fava C, Kjeldsen S, Hedner T, Melander O. A functional variant of the NEDD4L gene is associated with beneficial treatment response with beta-blockers and diuretics in hypertensive patients. 2011:388–395. doi: 10.1097/HJH.0b013e3283410390. 2010/11/06. [DOI] [PubMed] [Google Scholar]
- 67.Manunta P, Lavery G, Lanzani C, Braund PS, Simonini M, Bodycote C, Zagato L, li Carpini S, Tantardini C, Brioni E, Bianchi G, Samani NJ. Physiological interaction between alpha-adducin and WNK1-NEDD4L pathways on sodium-related blood pressure regulation. 2008:366–372. doi: 10.1161/HYPERTENSIONAHA.108.113977. 2008/07/02. [DOI] [PubMed] [Google Scholar]
- 68.Turner ST, Bailey KR, Fridley BL, Chapman AB, Schwartz GL, Chai HS, Sicotte H, Kocher JP, Rodin AS, Boerwinkle E. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duarte JD, Tran B, Langaee TY, Chapman AB, Gums JG, Boerwinkle E, Turner ST, Johnson JA. YEATS4 Gene Region Polymorphisms Associate with Antihypertensive Response to Hydrochlorothiazide in African Americans. 2010;87:46. (Abstract) [Google Scholar]
- 70.Psaty BM, Smith NL, Heckbert SR, Vos HL, Lemaitre RN, Reiner AP, Siscovick DS, Bis J, Lumley T, Longstreth WT, Jr, Rosendaal FR. Diuretic therapy, the alpha-adducin gene variant, and the risk of myocardial infarction or stroke in persons with treated hypertension. JAMA. 2002;287:1680–1689. doi: 10.1001/jama.287.13.1680. [DOI] [PubMed] [Google Scholar]
- 71.Davis BR, Arnett DK, Boerwinkle E, Ford CE, Leiendecker-Foster C, Miller MB, Black H, Eckfeldt JH. Antihypertensive therapy, the alpha-adducin polymorphism, and cardiovascular disease in high-risk hypertensive persons: the Genetics of Hypertension-Associated Treatment Study. 2007;7:112–122. doi: 10.1038/sj.tpj.6500395. [DOI] [PubMed] [Google Scholar]
- 72.Gerhard T, Gong Y, Beitelshees AL, Mao X, Lobmeyer MT, Cooper-DeHoff RM, Langaee TY, Schork NJ, Shriver MD, Pepine CJ, Johnson JA. Alpha-adducin polymorphism associated with increased risk of adverse cardiovascular outcomes: results from GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST-GENES) Am Heart J. 2008;156:397–404. doi: 10.1016/j.ahj.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beitelshees AL, Gong Y, Wang D, Schork NJ, Cooper-DeHoff RM, Langaee TY, Shriver MD, Sadee W, Knot HJ, Pepine CJ, Johnson JA. KCNMB1 genotype influences response to verapamil SR and adverse outcomes in the INternational VErapamil SR/Trandolapril STudy (INVEST) 2007;17:719–729. doi: 10.1097/FPC.0b013e32810f2e3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beitelshees AL, Navare H, Wang D, Gong Y, Wessel J, Moss JI, Langaee TY, Cooper-DeHoff RM, Sadee W, Pepine CJ, Schork NJ, Johnson JA. CACNA1C gene polymorphisms, cardiovascular disease outcomes, and treatment response. 2009:362–370. doi: 10.1161/CIRCGENETICS.109.857839. 2009/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niu Y, Gong Y, Langaee TY, Davis HM, Elewa H, Beitelshees AL, Moss JI, Cooper-DeHoff RM, Pepine CJ, Johnson JA. Genetic variation in the beta2 subunit of the voltage-gated calcium channel and pharmacogenetic association with adverse cardiovascular outcomes in the INternational VErapamil SR-Trandolapril STudy GENEtic Substudy (INVEST-GENES) 2010:548–555. doi: 10.1161/CIRCGENETICS.110.957654. 2010/12/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O'Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. 2009 doi: 10.1038/ng.384. 2009/05/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherva R, Ford CE, Eckfeldt JH, Davis BR, Boerwinkle E, Arnett DK. Pharmacogenetic Effect of the Stromelysin (MMP3) Polymorphism on Stroke Risk in Relation to Antihypertensive Treatment: The Genetics of Hypertension Associated Treatment Study. 2011;42:330–335. doi: 10.1161/STROKEAHA.110.593798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maitland-van der Zee A-H, van Wieren-de Wijer DB, de Boer A, Kroon AA, de Leeuw PW, Schiffers P, Janssen RG, Psaty BM, van Duijn CM, Stricker BH, Klungel OH. Genetic variation in the renin-angiotensin system, use of renin-angiotensin system inhibitors and the risk of myocardial infarction. J Renin Angiotensin Aldosterone Syst. 2010 doi: 10.1177/1470320310391834. 2010 Dec 16. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 79.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, Anderson JL, Carlquist JF, Krause-Steinrauf HJ, Lazzeroni LC, Port JD, Lavori PW, Bristow MR. A polymorphism within a conserved {beta}1-adrenergic receptor motif alters cardiac function and {beta}-blocker response in human heart failure. PNAS. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.ARCA Biopharma: ARCA announces special protocol assessment agreement with FDA for bucindolol development in genotype-defined heart failure population. 2011 [Google Scholar]
- 81.Biolo A, Clausell N, Santos KG, Salvaro R, shton-Prolla P, Borges A, Rohde LE. Impact of beta1-adrenergic receptor polymorphisms on susceptibility to heart failure, arrhythmogenesis, prognosis, and response to beta-blocker therapy. 2008:726–732. doi: 10.1016/j.amjcard.2008.04.070. 2008/09/09. [DOI] [PubMed] [Google Scholar]
- 82.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, Dorn GW. Clinical and genetic modifiers of long-term survival in heart failure. 2009:432–444. doi: 10.1016/j.jacc.2009.05.009. 2009/07/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sehnert AJ, Daniels SE, Elashoff M, Wingrove JA, Burrow CR, Horne B, Muhlestein JB, Donahue M, Liggett SB, Anderson JL, Kraus WE. Lack of association between adrenergic receptor genotypes and survival in heart failure patients treated with carvedilol or metoprolol. J Am Coll Cardiol. 2008;52:644–651. doi: 10.1016/j.jacc.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 84.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- 85.Lobmeyer MT, Gong Y, Terra SG, Beitelshees AL, Langaee TY, Pauly DF, Schofield RS, Hamilton KK, Herbert PJ, Adams KF, Jr, Hill JA, Aranda JM, Jr, Johnson JA. Synergistic polymorphisms of beta1 and alpha2C-adrenergic receptors and the influence on left ventricular ejection fraction response to beta-blocker therapy in heart failure. Pharmacogenet Genomics. 2007;17:277–282. doi: 10.1097/FPC.0b013e3280105245. [DOI] [PubMed] [Google Scholar]
- 86.Bristow MR, Murphy GA, Krause-Steinrauf H, Anderson JL, Carlquist JF, Thaneemit-Chen S, Krishnan V, Abraham WT, Lowes BD, Port JD, Davis GW, Lazzeroni LC, Robertson AD, Lavori PW, Liggett SB. An alpha2C-adrenergic receptor polymorphism alters the norepinephrine-lowering effects and therapeutic response of the beta-blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3:21–28. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 87.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, Spertus JA, Koch WJ, Kardia SL, Dorn GW. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 89.Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, Bentley D, McGinnis R, Deloukas P. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2006;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Limdi NA, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, Baird MF, Acton RT. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, Acton RT. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, Nicolae D, Cox NJ. The Missing Association: Sequencing-Based Discovery of Novel SNPs in VKORC1 and CYP2C9 That Affect Warfarin Dose in African Americans. Clin Pharmacol Ther. 2011;89:408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin Pharmacogenetics: CYP2C9 and VKORC1 Genotypes Predict Different Sensitivity and Resistance Frequencies in the Ashkenazi and Sephardi Jewish Populations. Am J Hum Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, King CR, Brower A, Schmelzer JR, Glurich I, Vidaillet HJ, Yale SH, Qi ZK, Berg RL, Burmester JK. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharm. 2009;75:1337–1346. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kringen MK, Haug KB, Grimholt RM, Stormo C, Narum S, Opdal MS, Fosen JT, Piehler AP, Johansen PW, Seljeflot I, Berg JP, Brors O. Genetic variation of VKORC1 and CYP4F2 genes related to warfarin maintenance dose in patients with myocardial infarction. J Biomed Biotechnol. 2011;2011 doi: 10.1155/2011/739751. 739751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.International Warfari Pharmacogenetics Consortium. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gage BF, Eby D, Johnson JA, Deych E, Rieder MJ, Ridker PM, Milligan PE, Grice G, Lenzini P, Rettie AE, Aquilante CL, Grosso L, Marsh S, Langaee T, Farnett LE, Voora D, Veenstra DL, Glynn RJ, Barrett A, McLeod HL. Use of Pharmacogenetic and Clinical Factors to Predict the Therapeutic Dose of Warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roper N, Storer B, Bona R, Fang M. Validation and comparison of pharmacogenetics-based warfarin dosing algorithms for application of pharmacogenetic testing. J Mol Diagn. 2010;12:283–291. doi: 10.2353/jmoldx.2010.090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schelleman H, Chen J, Chen Z, Christie J, Newcomb CW, Brensinger CM, Price M, Whitehead AS, Kealey C, Thorn CF, Samaha FF, Kimmel SE. Dosing Algorithms to predict Warfarin Maintenance Dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84:332–39. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langley MR, Booker JK, Evans JP, McLeod HL, Weck KE. Validation of clinical testing for warfarin sensitivity: comparison of CYP2C9-VKORC1 genotyping assays and warfarin-dosing algorithms. J Mol Diagn. 2009;11:216–225. doi: 10.2353/jmoldx.2009.080123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, Kahn SFS, May HT, Samuelson KM, Muhlestein JB, Carlquist JF, Couma-Gen I. Randomized Trial of Genotype-Guided Versus Standard Warfarin Dosing in Patients Initiating Oral Anticoagulation. Circulation. 2007 doi: 10.1161/CIRCULATIONAHA.107.737312. CIRCULATIONAHA. [DOI] [PubMed] [Google Scholar]
- 103.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 104.Epstein RS, Moyer TP, Aubert RE, DJ OK, Xia F, Verbrugge RR, Gage BF, Teagarden JR. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 105.Gurbel PA, Bliden KP, Dichiara J, Newcomer J, Weng W, Neerchal NK, Gesheff T, Chaganti SK, Etherington A, Tantry US. Evaluation of dose-related effects of aspirin on platelet function: results from the Aspirin-Induced Platelet Effect (ASPECT) study. Circulation. 2007;115:3156–3164. doi: 10.1161/CIRCULATIONAHA.106.675587. [DOI] [PubMed] [Google Scholar]
- 106.Faraday N, Yanek LR, Mathias R, Herrera-Galeano JE, Vaidya D, Moy TF, Fallin MD, Wilson AF, Bray PF, Becker LC, Becker DM. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007;115:2490–2496. doi: 10.1161/CIRCULATIONAHA.106.667584. [DOI] [PubMed] [Google Scholar]
- 107.Shen H, Herzog W, Drolet T, Cosentino R, Newcomer S, Sack P, Karon H, Ryan KA, Zhao Y, Shi X, Mitchell BD, Shuldiner AR. Aspirin resistance in healthy drug-naïve men versus women (from the Heredity and Phenotype Intervention [HAPI] Heart Study. Am J Cardiol. 2009 doi: 10.1016/j.amjcard.2009.04.027. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gum PA, Kottke-Marchant K, Welsh PA, White J, Topol EJ. A prospective, blinded determination of the natural history of aspirin resistance among stable patients with cardiovascular disease. J Am Coll Cardiol. 2003;41:961–965. doi: 10.1016/s0735-1097(02)03014-0. [DOI] [PubMed] [Google Scholar]
- 109.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Alfonso F, Macaya C, Bass TA, Costa MA. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 2007;49:1505–1516. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 110.Gurbel PA, Bliden KP, Hiatt BL, O'Connor CM. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107:2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 111.Gurbel PA, Tantry US. Drug insight: Clopidogrel nonresponsiveness. Nat Clin Pract Cardiovasc Med. 2006;3:387–395. doi: 10.1038/ncpcardio0602. [DOI] [PubMed] [Google Scholar]
- 112.O'Donoghue M, Wiviott SD. Clopidogrel response variability and future therapies: clopidogrel: does one size fit all? Circulation. 2006;114:e600–e606. doi: 10.1161/CIRCULATIONAHA.106.643171. [DOI] [PubMed] [Google Scholar]
- 113.Wang TH, Bhatt DL, Topol EJ. Aspirin and clopidogrel resistance: an emerging clinical entity. Eur Heart J. 2006;27:647–654. doi: 10.1093/eurheartj/ehi684. [DOI] [PubMed] [Google Scholar]
- 114.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, Pollin TI, Post W, Parsa A, Mitchell BD, Faraday N, Herzog W, Gurbel PA. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mitchell BD, McArdle PF, Shen H, Rampersaud E, Pollin TI, Bielak LF, Jaquish C, Douglas JA, Roy-Gagnon MH, Sack P, Naglieri R, Hines S, Horenstein RB, Chang YP, Post W, Ryan KA, Brereton NH, Pakyz RE, Sorkin J, Damcott CM, O'Connell JR, Mangano C, Corretti M, Vogel R, Herzog W, Weir MR, Peyser PA, Shuldiner AR. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am Heart J. 2008;155:823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goodman T, Ferro A, Sharma P. Pharmacogenetics of aspirin resistance: a comprehensive systematic review. Br J Clin Pharmacol. 2008;66:222–232. doi: 10.1111/j.1365-2125.2008.03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Savi P, Combalbert J, Gaich C, Rouchon MC, Maffrand JP, Berger Y, Herbert JM. The antiaggregating activity of clopidogrel is due to a metabolic activation by the hepatic cytochrome P450-1A. Thromb Haemost. 1994;72:313–317. [PubMed] [Google Scholar]
- 118.Richter T, Murdter TE, Heinkele G, Pleiss J, Tatzel S, Schwab M, Eichelbaum M, Zanger UM. Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Ther. 2004;308:189–197. doi: 10.1124/jpet.103.056127. [DOI] [PubMed] [Google Scholar]
- 119.Turpeinen M, Tolonen A, Uusitalo J, Jalonen J, Pelkonen O, Laine K. Effect of clopidogrel and ticlopidine on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Clin Pharmacol Ther. 2005;77:553–559. doi: 10.1016/j.clpt.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 120.Fontana P, Dupont A, Gandrille S, Bachelot-Loza C, Reny JL, Aiach M, Gaussem P. Adenosine diphosphate-induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation. 2003;108:989–995. doi: 10.1161/01.CIR.0000085073.69189.88. [DOI] [PubMed] [Google Scholar]
- 121.Fontana P, Hulot JS, De MP, Gaussem P. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. J Thromb Haemost. 2007;5:2153–2155. doi: 10.1111/j.1538-7836.2007.02722.x. [DOI] [PubMed] [Google Scholar]
- 122.Lau WC, Gurbel PA, Watkins PB, Neer CJ, Hopp AS, Carville DG, Guyer KE, Tait AR, Bates ER. Contribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistance. Circulation. 2004;109:166–171. doi: 10.1161/01.CIR.0000112378.09325.F9. [DOI] [PubMed] [Google Scholar]
- 123.Kim KA, Park PW, Hong SJ, Park JY. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther. 2008;84:236–242. doi: 10.1038/clpt.2008.20. [DOI] [PubMed] [Google Scholar]
- 124.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2008 doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 125.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost. 2008;6:1439–1441. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 126.Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, Aiach M, Lechat P, Gaussem P. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 127.Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, Lachno DR, Salazar D, Winters KJ. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 128.Fontana P, Hulot JS, De MP, Gaussem P. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. J Thromb Haemost. 2007;5:2153–2155. doi: 10.1111/j.1538-7836.2007.02722.x. [DOI] [PubMed] [Google Scholar]
- 129.Frere C, Cuisset T, Morange PE, Quilici J, Camoin-Jau L, Saut N, Faille D, Lambert M, Juhan-Vague I, Bonnet JL, Alessi MC. Effect of cytochrome p450 polymorphisms on platelet reactivity after treatment with clopidogrel in acute coronary syndrome. Am J Cardiol. 2008;101:1088–1093. doi: 10.1016/j.amjcard.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 130.Geisler T, Schaeffeler E, Dippon J, Winter S, Buse V, Bischofs C, Zuern C, Moerike K, Gawaz M, Schwab M. CYP2C19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics. 2008;9:1251–1259. doi: 10.2217/14622416.9.9.1251. [DOI] [PubMed] [Google Scholar]
- 131.Giusti B, Gori AM, Marcucci R, Saracini C, Sestini I, Paniccia R, Valente S, Antoniucci D, Abbate R, Gensini GF. Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4 IVS10 + 12G/A and P2Y12 T744C polymorphisms, is associated with response variability to dual antiplatelet treatment in high-risk vascular patients. Pharmacogenet Genomics. 2007;17:1057–1064. doi: 10.1097/FPC.0b013e3282f1b2be. [DOI] [PubMed] [Google Scholar]
- 132.Kim KA, Park PW, Hong SJ, Park JY. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther. 2008;84:236–242. doi: 10.1038/clpt.2008.20. [DOI] [PubMed] [Google Scholar]
- 133.Malek LA, Kisiel B, Spiewak M, Grabowski M, Filipiak KJ, Kostrzewa G, Huczek Z, Ploski R, Opolski G. Coexisting polymorphisms of P2Y12 and CYP2C19 genes as a risk factor for persistent platelet activation with clopidogrel. Circ J. 2008;72:1165–1169. doi: 10.1253/circj.72.1165. [DOI] [PubMed] [Google Scholar]
- 134.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost. 2008;6:1439–1441. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 135.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, Payot L, Brugier D, Cayla G, Beygui F, Bensimon G, Funck-Brentano C, Montalescot G. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2008 doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 136.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2008 doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 137.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, Steg PG, Ferrieres J, Danchin N, Becquemont L. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2008 doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 138.Trenk D, Hochholzer W, Fromm MF, Chialda LE, Pahl A, Valina CM, Stratz C, Schmiebusch P, Bestehorn HP, Buttner HJ, Neumann FJ. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51:1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 139.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot JS, Kastrati A, Montalescot G, Neumann FJ, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hulot JS, Collet JP, Silvain J, Pena A, Bellemain-Appaix A, Barthelemy O, Cayla G, Beygui F, Montalescot G. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol. 2010;56:134–143. doi: 10.1016/j.jacc.2009.12.071. [DOI] [PubMed] [Google Scholar]
- 141.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot JS, Kastrati A, Montalescot G, Neumann FJ, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pare G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, Simonsen K, Bhatt DL, Fox KA, Eikelboom JW. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 143.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, Morath T, Schomig A, von BN, Kastrati A. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 144.Gurbel PA, Tantry US, Shuldiner AR. Letter by Gurbel et al regarding article, "Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement". Circulation. 2010;122:478. doi: 10.1161/CIRCULATIONAHA.110.943548. [DOI] [PubMed] [Google Scholar]
- 145.Mega JL, Close SL, Wiviott SD, Shen L, Walker JR, Simon T, Antman EM, Braunwald E, Sabatine MS. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312–1319. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bouman HJ, Schomig E, van Werkum JW, Velder J, Hackeng CM, Hirschhauser C, Waldmann C, Schmalz HG, Ten Berg JM, Taubert D. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med. 2011;17:110–116. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 147.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias WL, Braunwald E, Sabatine MS. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009;119:2553–2560. doi: 10.1161/CIRCULATIONAHA.109.851949. [DOI] [PubMed] [Google Scholar]
- 148.Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J, Husted S, Katus H, Steg PG, Shah SH, Becker RC. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376:1320–1328. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 149.Gladding P, White H, Voss J, Ormiston J, Stewart J, Ruygrok P, Bvaldivia B, Baak R, White C, Webster M. Pharmacogenetic testing for clopidogrel using the rapid INFINITI analyzer: a dose-escalation study. JACC Cardiovasc Interv. 2009;2:1095–1101. doi: 10.1016/j.jcin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 150.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pao W, Iafrate AJ, Su Z. Genetically informed lung cancer medicine. J Pathol. 2010 doi: 10.1002/path.2788. n/a. [DOI] [PubMed] [Google Scholar]
- 152.Chen P, Lin JJ, Lu CS, Ong CT, Hsieh PF, Yang CC, Tai CT, Wu SL, Lu CH, Hsu YC, Yu HY, Ro LS, Lu CT, Chu CC, Tsai JJ, Su YH, Lan SH, Sung SF, Lin SY, Chuang HP, Huang LC, Chen YJ, Tsai PJ, Liao HT, Lin YH, Chen CH, Chung WH, Hung SI, Wu JY, Chang CF, Chen L, Chen YT, Shen CY. Carbamazepine-Induced Toxic Effects and HLA-B*1502 Screening in Taiwan. N Engl J Med. 2011;364:1126–1133. doi: 10.1056/NEJMoa1009717. [DOI] [PubMed] [Google Scholar]
- 153.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jagel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes A, Ryan S, Fitch N, Thorborn D, Benbow A, the PRED. HLA-B*5701 Screening for Hypersensitivity to Abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 154.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, Johnson JA, Hayes DF, Klein T, Krauss RM, Kroetz DL, McLeod HL, Nguyen AT, Ratain MJ, Relling MV, Reus V, Roden DM, Schaefer CA, Shuldiner AR, Skaar T, Tantisira K, Tyndale RF, Wang L, Weinshilboum RM, Weiss ST, Zineh I. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 156.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 157.Kurnik D, Loebstein R, Halkin H, Gak E, Almog S. 10 years of oral anticoagulant pharmacogenomics: what difference will it make? A critical appraisal. Pharmacogenomics. 2009;10:1955–1965. doi: 10.2217/pgs.09.149. [DOI] [PubMed] [Google Scholar]