Abstract
Pan-kinesin peptide antibodies were used to identify and isolate kinesin-related proteins (KRPs) from Drosophila melanogaster embryonic cytosol. These KRPs cosedimented with microtubules (MTs) polymerized from cytosol treated with AMP-PNP (adenyl-5′-yl imidodiphosphate), and one of them, KRP130, was further purified from ATP eluates of the embryonic MTs. Purified KRP130 behaves as a homotetrameric complex composed of four 130-kDa polypeptide subunits which displays a “slow” plus-end directed motor activity capable of moving single MTs at 0.04 ± 0.01 μm/s. The 130-kDa subunit of KRP130 was tested for reactivity with monoclonal and polyclonal antibodies that are specific for various members of the kinesin superfamily. Results indicate that the KRP130 subunit is related to Xenopus Eg5 (Sawin, K. E., Le Guellec, K. L., Philippe, M., Mitchinson, T. J. (1992) Nature 359, 540-543), a member of the BimC subfamily of kinesins. Therefore, KRP130 appears to be the first Drosophila KRP, and the first member of the BimC subfamily in any organism, to be purified from native a multimeric motor complex.
Kinesin and kinesin-related proteins (KRPs)1 comprise a family of motor proteins that play important and diverse roles in intracellular organelle transport and cell division (1-3). Kinesin was first purified from neural tissues (4, 5) and mitotic cells (6) using biochemical methods and was subsequently shown to be an asymmetric heterotetrameric complex consisting of two 110-130-kDa heavy chains (KHCs) and two 55-85-kDa light chains (7,8). At one end of the kinesin molecule, the KHCs form two globular NH2-terminal “motor domains” (9-11) capable of ATP-dependent MT gliding activity coupled to MT-activated ATP hydrolysis (12, 13). The heavy chains are dimerized via an α-helical coiled-coil region (14). The light chains and the COOH-terminal domains of the heavy chains are found at the other end of the molecule (9). Although the role of the light chains has not been demonstrated, the carboxyl-terminal domain of the KHC is believed to be responsible for the attachment of membranous cargo to kinesin (15).
Numerous KRPs have been identified at the nucleic acid level using genetic techniques or the polymerase chain reaction (2, 3). To complement these strategies, we prepared and used pan-kinesin antibodies against conserved kinesin motor domain peptides fort he purpose of identifying native kinesins in their natural tissues (16). A screen of sea urchin egg cytosol resulted in the purification of the first native KRP, sea urchin egg KRP85/95, a plus-end-directed, heterotrimeric motor complex composed of 85- and 95-kDa kinesin-related polypeptides plus an uncharacterized 115-kDa subunit (16, 17).
The fruit fly Drosophila melanogaster has proven to be a particularly rewarding system for studying kinesins. Drosophila kinesin was first isolated biochemically (18) leading to the cloning and molecular analysis of the KHC gene and its expressed product (10, 12, 19). Subsequently, severafly KRPs have been characterized using genetic approach(20-25), but no KRP has thus far been purified from native fruit fly tissue. In an effort to biochemically identify and purify native Drosophila KRP complexes, embryonic extracts were probed with the pan-kinesin peptide antibodies. We present here the purification and characterization of one of these kinesins, KRP130, a 490-kDa homotetrameric complex consisting of four 130-kDa subunits.
MATERIALS AND METHODS
Protein Preparation
Bovine phosphocellulose-chromatographed tubulin (PC tubulin) was prepared as described previously( 26) and stored in 1 mm MgGTP in PEM buffer (100 mM PIPES, pH 6.9, 2 mm EGTA, 1 mm MgSO4, and 2 mm dithiothreitol) in liquid N2 or at –80 °C until needed. Preparative MTs were formed by incubating PC tubulin in 1 mm MgGTP and 20 μm taxol (Sigma) at 37 °C for 30 min. Motility assay MTs were prepared by incubating 200 μg/ml PC tubulin in 1 mm MgGTP, 10 μm taxol at 37 °C for 60 min.
Drosophila cytosolic low speed supernatant was prepared from 0–24-h embryos as described previously (18) using PMEG buffer (100 mm PIPES, pH 6.9, 5 mm EGTA, 0.5 mm EDTA, 2.5 mm MgSO4, 0.9 m glycerol, and 1 mm dithiothreitol) with our standard protease inhibitor mixture (27) and stored at –80 °C. After thawing, fresh protease inhibitors were added to low speed supernatant prior to spinning at 175,000 × g for 45 min at 4 °C. The resulting high speed supernatant (100–150 ml) was supplemented with 1 mm GTP and 10 μm taxol, rocked for 15 min at 25 °C, then supplemented with 1 mm AMP-PNP and rocked for 20 min prior to spinning at 35,000 × g for 60 min at 10 °C. The MT pellet was washed with 10 ml of 10 mm EDTA in PEG (PMEG without MgSO4) buffer at 4 °C prior to repelleting at 100,000 × g for 25 min at 4 °C. The washed MT pellet was eluted with 6 ml of 10 mm MgATP, 200 mm KCl in PMEG for 6–14 h at °C prior to repelleting at 200,000 × g for 20 min at 4 °C. The eluate (ATP MAPs) was concentrated to 3 ml with a Centriprep 30 (Amicon) and fractionated on a Bio-Gel A-1.5m (1.6 × 90 cm) or Bio-Gel A-5m (1.0 × 90 cm) column equilibrated with 100 μm ATP, 150 mm KCl in PMEG buffer. The fractions containing kinesin and KRP130 were separately pooled and concentrated (Centriprep 30) to 1.5 ml prior to 20-min incubations with taxol MTs (PC tubulin) in 2 mm AMP-PNP, and either 50 mm KCl (kinesin) or 125-200 mm KCl (KRP130) in PMEG buffer at 25 °C. The MTs were respun (100,000 × g, 20 min), followed by release of kinesin and KRP130 from their respective pellets with 150 μl of 10 mm MgATP, 200 mm KCl in PMEG buffer for 30 min at 25 °C. The MTs were pelleted a final time (60,000 × g, 15 min), and the resulting kinesin and KRP130 supernatants were fractionated on linear 5-20% sucrose gradients in 100 μm ATP, 150 mm KCl in PMEG buffer formed by a piston-driven gradient former (Jule, Inc.) and spun at 300,000 × g for 8.5 h (4 °C).
SDS-polyacrylamide gel electrophoresis (28) and immunoblotting were done as described previously (29). The relative molecular mass of the KRP130 subunit was determined by using standard marker proteins; rabbit muscle myosin heavy chain (205 kDa), Escherichia coli β-galactosidase (116 kDa), rabbit muscle phosphorylase b (97.4 kDa), bovine serum albumin (66 kDa), chicken ovalbumin (45 kDa), bovine carbonic anhydrase (29 kDa), and soybean trypsin inhibitor (21.5 kDa).
Stoichiometry
The Stokes radius, RS, was determined as described previously (16) for two different preparations of Drosophila kinesin and KRP130 using the Bio-Gel A-1.5 m and Bio-Gel A-5m columns and running buffers described above. Plots of RS versus -log10 Kav1/2, where Kav = (elution volume – void volume)/(total volume – void volume) generated standard linear plots (r2 = 0.964; r2 = 0.981, where r is the correlation coefficient). The standard proteins and their Stokes radii included sea urchin egg kinesin(9.6 nm), sea urchin egg KRP85/95 (7.9 nm, Ref. 16), yeast alcohol dehydrogenase (4.6 nm), bovine serum albumin (3.5 nm), and bovine heart cytochrome c (1.7 nm).
The sedimentation coefficient S20,W was determined using two separate kinesin and two separate KRP130 preparations on 5-ml 5-20% linear sucrose gradients in 100 pr ATP, 150 μm KCl in PMEG. The gradients were overlaid with a solution containing kinesin or KRP130 and 100 μg of bovine liver catalase (11.3 S), 50 μg of bovine serum albumin (4.4 S), and 100 μg of ovalbumin (3.66 S). The gradients were centrifuged at 300,000 × g for 8.5 h. A graph of S20,W versus the distance traveled through the gradient for the two KRP130 preparations yielded standard linear plots (r2 = 0.997; r2 = 0.998). The native molecular weights of kinesin and KRP130 were calculated from the measured s values and Stokes radii as described previously (16, 30).
Motility Assays
MT gliding over a glass coverslip coated with sucrose gradient-purified kinesin and KRP130 was measured as described previously (31) with a few modifications. A bundling activity that copurified with KRP130 was purposely minimized by performing assays in 20-μl flow cells that produced widely dispersed MTs (32). In addition, MTs attached to the KRP130-coated coverslips displayed either intermittent or no motility unless the motility buffer (9 μm MgATP in PMEG) was supplemented with 50-75 μm KCl or NaCl. The video image was enhanced with an Argus-10 Image Processor (Hamamatsu). The polarity of MT gliding was determined with fluorescent “marked” MTs prepared using rhodamine-tubulin, N-ethylmaleimide-modified tubulin, GMP-CPP, and oxygen scavengers (all a generous gift from R. D. Vale) according to Howard and Hyman (33).
RESULTS AND DISCUSSION
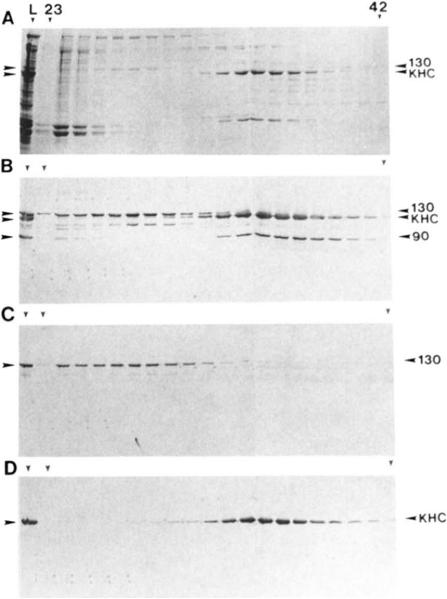
Pan-kinesin peptide antibodies (16, 34) were used to identify KRPs in fractions obtained during the purification protocol outlined under “Materials and Methods.” A number of polypeptides in the ATP MAPs fraction reacted with the peptide antibodies (Fig. 1B, lane L); the three with the greatest reactivity migrated at 130, 120, and 90 kDa (Fig. 1, A and B). Probing Western blots of the gel filtration column fractions with antibodies that are specific for various KRPs indicated that the 130-kDa polypeptide is related to Xenopus Eg5 (Fig. 1C) (35). The 120-kDa polypeptide was shown to be the kinesin heavy chain (Fig. 1D) based on its reactivity with the KHC-specific monoclonal antibody SUK4 (29). We believe that the 90-kDa polypeptide is the protein product of the ncd gene (20,21) based on its cross-reactivity with an Ncd-specific antibody raised against a peptide corresponding to the carboxyl-terminal 15 amino acids of the deduced Ncd protein (data not shown; antibody provided by Drs. McDonald and Goldstein).
FIG. 1. Bio-Gel A-1.5m gel filtration of ATP-eluted Drorophila MT-binding proteins.
A, Coomassie Blue-stained SDS gel. Lane L shows the ATP-eluted MAPs that were loaded onto the column. Duplicate immunoblots were probed with “LAGSE” antihody (similar results were obtained using other pankinesin antibodies 16, 34)) (B), anti-Eg5 antibody (C), and SUK4 antibody (D). The column (1.6 × 85 cm) was equilibrated with 100 μm ATP, 150 mm KCl in PMEG buffer. Fractions (3.0 ml) 23–42 are shown on the gels and blots here. The void volume corresponded to fraction 24, and the included volume corresponded to fraction 59.
KRP130 was purified further, based on its nucleotide-sensitive rebinding to and release from microtubules. The gel filtration fractions containing the peak of KRP130 were pooled and concentrated to 1.5 ml, then mixed with AMP-PNP and MTs formed from bovine brain PC tubulin. Critical to the purification of KRP130 was the finding that it bound to MTs in PMEG buffers containing AMP-PNP supplemented with significant amounts of KCI. In preliminary MT pelleting experiments, it was striking tha >90% and >60% of the KRP130 copelleted with MTs in AMP-PNP when the final concentration of KCl was 75 and 325 mm, respectively (data not shown), but negligible amounts of KRP130 cosedimented with MTs in ATP. By comparison, only 50% and <10% of Drosophila kinesin cosedimented with AMP-PNP MTs under the same conditions. Consequently, in the KRP130 purification protocol, we rebound KRP130 to MTs in the presence of AMP-PNP plus 12.5-200 mm KCl prior to elution with 100-200 μl of PMEG containing 10 mm MgATP and 200 mm KCl; these conditions optimized the removal of contaminating polypeptides and served to concentrate KRP130 in the eluates.
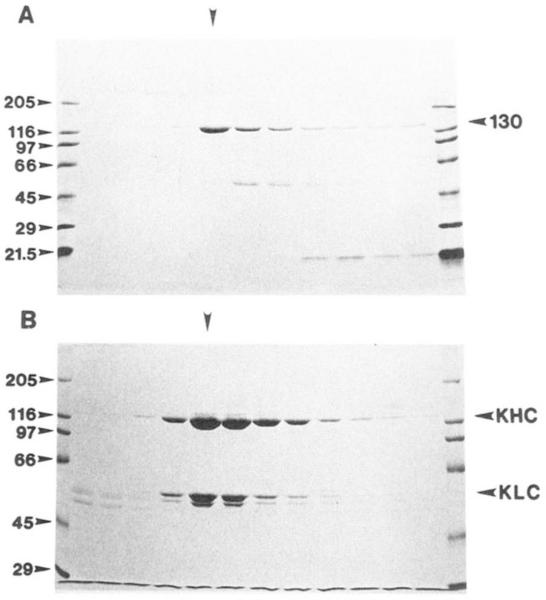
The final purification step involved sucrose density gradient centrifugation (Fig. 2). The 130-kDa polypeptide does not cosediment with significant amounts of any other polypeptides greater than 20 kDa (Fig. 2A). This leads us to conclude that the 130-kDa polypeptide is the only subunit present in the KRP130 complex. This is in contrast to Drosophila kinesin (Fig. 2B), which is believed to consist of two 120-kDa heavy chains and two 55-kDa light chains (the minor polypeptide hand just below the KLC in Fig. 2B may represent a modified light chain; Ref. 36). Unlike bovine and sea urchin egg kinesins, we detected no KLC-depleted fly kinesin (15, 37).
FIG. 2. Fractionation of Drosophila kinesin and KRP130 by sucrose density gradient centrifugation.
A, KRP130. Coomassie Blue-stained 5-20% acrylamide gradient SDS gel of a 5-20% sucrose gradient shows that KRP130 sediments as a single 7.6 S peak. No detectable polypeptides greater than 20 kDa coelute with the 130-kDa polypeptide. B, kinesin (KHC and KLC). Coomassie Blue-stained 7.5% acrylamide SDS gel of 5-20% sucrose gradient shows that kinesin sediments as a single 9.1 S peak. Vertical arrowheads indicate that peak KRP130 and kinesin fractions.
The properties of Drosophila KRP130 are compared with those of fly kinesin in Table I (the quaternary structure of fly kinesin has not been reported previously). We determined the Stokes radii of Drosophila kinesin and KRP130 to be approximately 9.0 and 16.2 nm, respectively. From sucrose density gradient centrifugation performed under the same buffer conditions as the gel filtration, we estimate the sedimentation coefficients of kinesin and KRP130 to be 9.1 and 7.6 S, respectively (data not shown). Using the method of Seigel and Monty (30), we estimate the relative molecular masses of kinesin and KRP130 to be 340 and 490 kDa, respectively. The calculated molecular mass of kinesin agrees closely with the value of 337 kDa predicted from cDNA cloning of fly KHC (19) and KLC (36), demonstrating the validity of our technique. Considering that each subunit of KRP130 is approximately 130 kDa, we calculate a subunit to complex ratio of 3.8 to 1, suggesting that KRP130 is composed of four 130-kDa kinesin-related motor subunits. Our attempts to visualize KRP130 by rotary shadowing and electron microscopy have thus far not revealed any consistent structure. Identical conditions produced high quality micrographs of Drosophila kinesin, suggesting that KRP130 may be too fragile for this imaging technique. However, the presence of four motor subunits in one complex is consistent with a bipolar structure consisting of two antiparallel 130-kDa dimers.
Table I.
Comparison of Drosophila KRP130 and Drosophila kinesin
| Kinesin | KRP130 | |
|---|---|---|
| Motility | Plus-end. 0.8 ± 0.06 μm/s | Plus-end, 0.04 ± 0.01 μm/s |
| MT binding | Strong: AMP-PNP | Strong: AMP-PNP |
| Weak: ATP | Weak: ATP | |
| Mr (subunits) | 120 kDa, 55 kDa | 130-kDa |
| RS (complex) | 9.0 nm | 16.2 nm |
| Sedimentation Coefficient | 9.1 S | 7.6 S |
| Molecular mass (complex) | 340 kDa | 490 kDa |
| Stoichiometry | 2 × 120 kDa | 4 × 130 kDa |
| 2 × 55 kDa |
To test the ability of KRP130 to act as an MT-based motor, standard MT gliding assays were used (see “Materials and Methods”). The purified KRP130 complex acts as a “slow” ATP dependent plus-end-directed motor (0.04 ± 0.01 μm/s). This motility and a bundling activity copurify with the KRP130 on the sucrose gradients. The motility was labile; the activity apparent in “fresh” KRP130 deteriorated during storage of KRP130 on ice over a 24-h period.
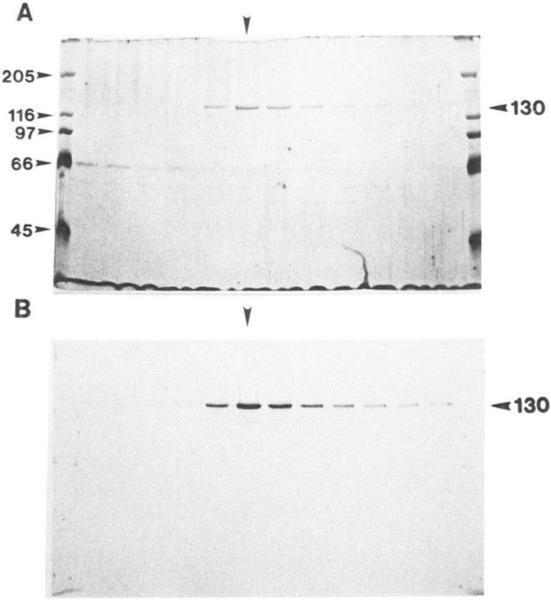
The 130-kDa motor subunit of KRP130 cross-reacts with anti-Xenopus Eg5 antibody (Figs. 1C and 3B), suggesting that KRP130 may represent a Drosophila homolog of Eg5 (35, 38) and may therefore be a member of the BimC subfamily of kinesins (3). This hypothesis is supported by the observations that slow plus-end-directed motility is a property of both purified KRP130 (0.04 ± 0.01 μm/s) and bacterially expressed Xenopus Eg5 (0.035 μm/s; Ref. 35).
FIG. 3. Immunoreactivity of KRP130 with anti-Eg5 antibody.
A silver-stained SDS gel of a 5-20% sucrose gradient shows that KRP130 sediments as a single peak. B, duplicate immunoblot probed with anti-Eg5 antibody. Vertical arrowheads indicate that peak KRP130 fraction.
Members of the BimC subfamily have been identified genetically across a wide range of organisms and appear to play important roles in the formation and maintenance of the mitotic spindle (25, 35, 38–45). It is possible that the 130-kDa polypeptide of KRP130 is the product of the Drosophila KLP61F gene (25), a member of the BimC subfamily also known as urchin.2 Disruption of the KLP61F gene results in failed spindle pole separation during mitotic prophase (25). Similar defects in spindle assembly result from fungal bimC (39) and cut7 (40) mutations.
Our observation that KRP130 is a homotetramer may be relevant to the mechanism of spindle pole separation mediated by members of the BimC subfamily. In interphase, duplication of the centrosome occurs, so that during prophase, cells contain two spindle poles lying side by side with arrays of MTs emanating from them (plus ends of MTs distal to the poles). We speculate that KRP130 (and other members of the BimC subfamily) could cross-link MTs emanating from one pole to parallel MTs emanating from the neighboring pole, and could cause the attached MTs to “slide” with their minus ends leading, thereby exerting “pushing” forces on the attached poles. If we assume that the heads of KRP130 can swivel to permit MT motility in any direction (as found for kinesin by Hunt and Howard (Ref. 46)), then such a protein assembly would be expected to “self-organize” into a metaphase-like array, consisting of separated spindle poles linked by overlapping arrays of antiparallel MTs cross-linked in the region of MT overlap by KRP130 homotetramers. In this way, KRP130 homotetramers could function as the microtubule cross-linking motors described in a recent model of spindle pole separation (see Fig. 3 of Ref. 47). Our hypothesis for KRP130 function is based on the notion that its four subunits are organized into a bipolar array capable of crosslinking adjacent microtubules. Additional studies of the structure and function of KRP130 are being initiated to test the hypothesis that it is indeed a bipolar assembly.
Acknowledgments
We thank Ken Sawin and Tim Mitchinson for their generous gifts of affinity-purified pan-kinesin peptide and anti-Eg5 antibodies. We thank Heather McDonald and Larry Goldstein for their generous gift of anti-ncd antibody. We thank Ron Vale for the generous gift of polarity assay components and Krsten Hall for assistance with motility assays. We thank all the members of the Scholey lab for helpful discussions.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: KRP, kinesin-related protein; KHC, kinesin heavy chain; KLC, kinesin light chain; MAP, microtubule-associated protein; MT, microtubule; PC tubulin, phosphocellulose-chromatographed tubulin; PIPES, 1,4-piperazine diethanesulfonic acid; AMP-PNP, adenyl-5′-yl imidodiphosphate; GMP-CPP, α,β-methylene guanosine 5′-triphosphate.
P. G. Wilson and M. T. Fuller, manuscript in preparation.
REFERENCES
- 1.Vale RD. Trends Biochrn. Sci. 1992;17:300–304. doi: 10.1016/0968-0004(92)90440-k. [DOI] [PubMed] [Google Scholar]
- 2.Skoufias DA, Scholey JM. Curr. Opin. Cell Biol. 1993;5:95–104. doi: 10.1016/s0955-0674(05)80014-6. [DOI] [PubMed] [Google Scholar]
- 3.Pereira A, Goldstein LSB. In: Microtubules. Hyams JS, Lloyd CW, editors. Wiley-Liss, Inc.; New York: 1994. pp. 269–284. [Google Scholar]
- 4.Vale RD, Reese TS, Sheetz MP. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady ST. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 6.Scholey JM, Porter ME, Grissom PM, McIntosh JR. Nature. 1985;318:483–486. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- 7.Bloom GS, Wagner MC, Pfister KK, Brady ST. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- 8.Kuznetsov SA, Vaisberg YA, Shanina NA, Magretova NN, Chernyak VY, Gelfand VI. EMBO J. 1988;7:353–356. doi: 10.1002/j.1460-2075.1988.tb02820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 10.Yang JT, Laymon RA, Goldstein LSB. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- 11.Scholey JM, Heuser J, Yang JT, Goldstein LSB. Nature. 1989;338:355–357. doi: 10.1038/338355a0. [DOI] [PubMed] [Google Scholar]
- 12.Yang JT, Saxton WM, Russell JS, Raff EC, Goldstein LSB. Science. 1990;249:42–47. doi: 10.1126/science.2142332. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert SP, Johnson KA. Biochemistry. 1993;32:4677–4684. doi: 10.1021/bi00068a028. [DOI] [PubMed] [Google Scholar]
- 14.de Cuevas M, Tao T, Goldstein LSB. J. Cell Biol. 1992;116:957–965. doi: 10.1083/jcb.116.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skoufias DA, Cole DG, Wedaman KP, Scholey JM. J. Bid. Chem. 1994;269:1477–1485. [PubMed] [Google Scholar]
- 16.Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM. J. Cell Sci. 1992;101:291–301. doi: 10.1242/jcs.101.2.291. [DOI] [PubMed] [Google Scholar]
- 17.Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 18.Saxton WM, Porter ME, Cohn SA, Scholey JM, Raff EC, McIntosh JR. Proc. Natl. Acad. Sci. U. S. A. 1988;86:1109–1113. doi: 10.1073/pnas.85.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JT, Saxton WM, Goldstein LSB. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1864–1868. doi: 10.1073/pnas.85.6.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endow SA, Henikoff S, Soler-Niedzeila L. Nature. 1990;345:81–83. doi: 10.1038/345081a0. [DOI] [PubMed] [Google Scholar]
- 21.McDonald HB, Goldstein LSB. Cell. 1990;61:991–1000. doi: 10.1016/0092-8674(90)90064-l. [DOI] [PubMed] [Google Scholar]
- 22.Zhang P, Knowles BA, Goldstein LSB, Hawley RS. Cell. 1990;62:1053–1062. doi: 10.1016/0092-8674(90)90383-p. [DOI] [PubMed] [Google Scholar]
- 23.Endow SA, Hatsumi M. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4424–4427. doi: 10.1073/pnas.88.10.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart RJ, Pesavento PA, Woerpel DN, Goldstein LSB. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8470–8474. doi: 10.1073/pnas.88.19.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heck MMS, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LSB. J. Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RC, Lee JC. Methods Enzymol. 1982;85:376–385. doi: 10.1016/0076-6879(82)85038-6. [DOI] [PubMed] [Google Scholar]
- 27.Buster D, Scholey JM. J. Cell Sci. Suppl. 1991;14:109–115. doi: 10.1242/jcs.1991.supplement_14.22. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Ingold AL, Cohn SA, Scholey JM. J. Cell Biol. 1988;107:2657–2667. doi: 10.1083/jcb.107.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel LM, Monty KJ. Biochin. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 31.Cohn SA, Ingold AL, Scholey JM. J. Biol. Chem. 1989;264:4290–4297. [PubMed] [Google Scholar]
- 32.Howard J, Hunt AJ, Baek S. Methods Cell Biol. 1993;39:137–147. doi: 10.1016/s0091-679x(08)60167-3. [DOI] [PubMed] [Google Scholar]
- 33.Howard J, Hyman AA. Methods Cell Biol. 1993;39:105–113. doi: 10.1016/s0091-679x(08)60164-8. [DOI] [PubMed] [Google Scholar]
- 34.Sawin KE, Mitchinson TJ, Wordeman LG. J. Cell Sci. 1992;101:303–313. doi: 10.1242/jcs.101.2.303. [DOI] [PubMed] [Google Scholar]
- 35.Sawin KE, Le Guellec KL, Philippe M, Mitchinson TJ. Nature. 1992;359:540–443. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 36.Gauger AK, Goldstein LSB. J. Biol. Chem. 1993;268:13657–13666. [PubMed] [Google Scholar]
- 37.Hackney DD, Levitt JD, Wagner DD. Biochem. Biophys. Res. Commun. 1991;174:810–815. doi: 10.1016/0006-291x(91)91490-4. [DOI] [PubMed] [Google Scholar]
- 38.Le Guellec R, Paris RB, Couturier A, Roghi C, Philippe M. Mol. Cell. Biol. 1991;11:3395–4398. doi: 10.1128/mcb.11.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enos AP, Morris NR. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- 40.Hagan I, Yanagida M. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- 41.Hagan I, Yanagida M. Nature. 1992;366:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- 42.Hoyt MA, He L, Loo KK, Saunders WS. J. Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connell MJ, Meluh PB, Rose MD, Moms NR. J. Cell Biol. 1993;120:153–162. doi: 10.1083/jcb.120.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roof DM, Meluh PB, Rose MD. J. Cell Bid. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saunders WS, Hoyt MA. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 46.Hunt AJ, Howard J. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11653–11657. doi: 10.1073/pnas.90.24.11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saunders WS. Trends Cell Biol. 1993;3:432–437. doi: 10.1016/0962-8924(93)90032-v. [DOI] [PubMed] [Google Scholar]