Abstract
Rhodopseudomonas palustris, a nonsulphur purple photosynthetic bacteria, has been extensively investigated for its metabolic versatility including ability to produce hydrogen gas from sunlight and biomass. The availability of the finished genome sequences of six R. palustris strains (BisA53, BisB18, BisB5, CGA009, HaA2 and TIE-1) combined with online bioinformatics software for integrated analysis presents new opportunities to determine the genomic basis of metabolic versatility and ecological lifestyles of the bacteria species. The purpose of this investigation was to compare the functional annotations available for multiple R. palustris genomes to identify annotations that can be further investigated for strain-specific or uniquely shared phenotypic characteristics. A total of 2,355 protein family Pfam domain annotations were clustered based on presence or absence in the six genomes. The clustering process identified groups of functional annotations including those that could be verified as strain-specific or uniquely shared phenotypes. For example, genes encoding water/glycerol transport were present in the genome sequences of strains CGA009 and BisB5, but absent in strains BisA53, BisB18, HaA2 and TIE-1. Protein structural homology modeling predicted that the two orthologous 240 aa R. palustris aquaporins have water-specific transport function. Based on observations in other microbes, the presence of aquaporin in R. palustris strains may improve freeze tolerance in natural conditions of rapid freezing such as nitrogen fixation at low temperatures where access to liquid water is a limiting factor for nitrogenase activation. In the case of adaptive loss of aquaporin genes, strains may be better adapted to survive in conditions of high-sugar content such as fermentation of biomass for biohydrogen production. Finally, web-based resources were developed to allow for interactive, user-defined selection of the relationship between protein family annotations and the R. palustris genomes.
Keywords: aquaporins, biohydrogen production, comparative genomics, functional annotation, fermentation, Pfam domains, Rhodopseudomonas palustris, strain-specific genes, uniquely shared genes, visual analytics
Introduction
Rhodopseudomonas are rod-shaped, gram-negative, purple nonsulfur, anoxygenic, phototrophic bacteria belonging to the alpha subclass of the Proteobacteria that inhabits diverse natural habitats including soil and wastewater systems.1,2 These ubiquitous organisms can grow in both anaerobic and aerobic conditions3,4 and are genetically tractable.5 Members of the genus are capable of growth using light, inorganic, or organic compounds as energy sources and carbon dioxide or organic compounds as carbon sources.4
Rhodopseudomas palustris are metabolically versatile species6,7 with strains that can convert atmospheric carbon dioxide into biomass,7 produce hydrogen gas,8–10 have multiple metal resistances11 and fix atmospheric nitrogen.12 Furthermore, R. palustris strains are also able to degrade a wide range of toxic organic compounds, and may be of use in bioremediation of polluted sites.4 The finished genome sequences and functional annotation of genes for six R. palustris strains (BisA53, BisB18, BisB5, CGA009, HaA2 and TIE-1) are publicly available,6,13 while the genome sequence of a 7th strain, DX-1, is in production.14 Strain DX-1 can produce high power densities that allow it to generate bioelectricity from the biodegration of organic and inorganic waste in low-internal-resistance microbial fuel cells. The ability of R. palustris strains to adapt and live under various environmental constraints as well as biodegrade pollutants to be used as biofuel, make them a model system for research on renewable energy from biological sources.
The assignment of functions to predicted genes from sequenced genomes is an approach to identify biological pathways that encode desirable phenotypes for diverse applications.13 A search of the Integrated Microbial Genomes (IMG) system (version 3.3)15 for genomes annotated with the hydrogen production phenotype revealed that six R. palustris strains (BisA53, BisB18, BisB5, CGA009, DX-1 and HaA2) were annotated with relevance for hydrogen production. Additionally, strain TIE-1 was annotated as an iron oxidizer. A strain of R. palustris is able to intracellularly synthesize cadmium sulfide nanoparticles and then secrete from cells.16 The availability of the finished genome sequences of six R. palustris strains combined with online bioinformatics software for integrated analysis presents new opportunities to elucidate the genomic basis of metabolic versatility and ecological lifestyles of the bacteria species. The purpose of this investigation was to compare the functional annotations available for multiple R. palustris genomes to identify annotations that could be further investigated as strain-specific or uniquely shared phenotypic characteristics.
The genome statistics, functional relatedness and functional annotations of the six R. palustris genomes were extracted or predicted using tools available on the IMG resource.15 Specifically, Pfam abundance data were extracted and encoded as a 6-digit binary accession to facilitate comparative analysis including strain-specific (annotation for only one genome) and uniquely shared annotations (annotation for only two genomes) for the genomes compared. We refer collectively to these bioinformatics analyses as functional annotation analytics since they can be accomplished within the IMG resource. The analytics process among others identified uniquely shared annotations for cell membrane water/glycerol transporter in strains BisB5 and CGA009. The observation orthologous aquaporins in R. palustris was of interest because of our ongoing and published research on aquaporins.17–19 Homology modeling predicted that the orthologous aquaporins in BisB5 and CGA009 are water-specific transporters.
Microbial aquaporins are known to function in freeze tolerance20 while loss of aquaporins is advantageous for utilization of high-sugar substrates.21 Investigation into the presence or absence of aquaporin in R. palustris strains could provide molecular basis for nitrogen fixation at low temperatures, a process affected by availability of liquid water, as well as the efficient utilization of high-sugar substrates in biohydrogen production.
Methods
Genome statistics
The complete genome sequences of six Rhodopseudomonas palustris strains (HaA2, NCBI Taxon ID 316058; BisA53, Taxon ID 316055; BisB18, NCBI Taxon ID 316056; BisB5, NCBI Taxon ID 316057; CGA009, NCBI Taxon ID 258594, TIE-1, NCBI Taxon ID 395960) are available in the public genome databases.6,22 The statistics of selected genome features were obtained for each of the R. palustris genomes and were retrieved from the Organism Details page on the Integrated Microbial Genomes website (version 3.3, February 2011). The Integrated Microbial Genomes (IMG) system is a data management, analysis and annotation platform for all publicly available genomes. 15 The statistics were then integrated to allow for comparative analysis of the DNA sequence (number of bases and guanine-cytosine content) and various functional classifications (the total genes predicted per genome and the proportion of the total genes annotated).
Functional relatedness of genomes based on Pfam domains
Functional relatedness of genomes is a measure of similarity between two genomes based on the similarity of the functional annotation of genes.15 The relationship between the six R. palustris genomes and Pfam domain annotation of genes were determined using the Genome Clustering Tool on the IMG system. This bioinformatics tool enables the use of the hierarchical clustering method to group genomes.
Genomes were also compared for the presence or absence of Pfam domain annotations to determine annotations that are specific to one or two of the six completely sequenced strains of R. palustris. The Abundance Profile Toolkit on the IMG system was used to generate and view the Pfam annotation abundance matrix for Pfam domains with at least one gene annotation. The resulting matrix was processed using customized PERL and UNIX scripts to generate a 6-digit binary accession for each Pfam domain. Digit 1 through 6 of the binary accession corresponds to BisA53, BisB18, BisB5, CGA009, HaA2 and TIE-1. Thus a Pfam domain with binary accession ‘100000’ indicated that the category was found only in genome of strain BisA53. To facilitate searching for user-defined combinations, we constructed a visual analytic view using Tableau Public (www.tableausoftware.com/public), a free data visualization software.
The availability of a matrix consisting of binary accessions for multiple Pfam domains allowed the clustering of R. palustris genomes based on total number of genomes annotated by a Pfam domain. Following similar approaches by Huang et al23 of hierarchical clustering analysis, the binary patterns were clustered using Cluster 3.024 with “Pfam domains” and “Genomes” as axes. The similarity matrix used was produced via the correlation (uncentered) method, and an average linkage clustering was performed. The figure generated by Cluster 3.0 was visualized in Java TreeView 1.1.5.r2.25
Gene orthology, sequence analysis and comparative protein structure modeling
Genes of interest with strain-specific or uniquely shared annotations were further analyzed for (i) gene orthology: genes in different genomes that evolved from a common ancestral gene by speciation (ii) sequence analysis: multiple sequence alignment of protein sequences of uniquely shared; and (iii) comparative protein structure modeling: inferring protein structure using a known template to understand structure-function relationship of strain-specific protein or uniquely shared proteins.
In the IMG system, orthologs are defined as bidirectional best hits from BLASTP comparisons and can be retrieved using the Gene Homolog Tool. Multiple sequence alignment was performed using ClustalW.26 Theoretical homology models of protein of interest were generated using MODELLER7V727 using a high resolution X-ray crystal structure of a homolog of the protein as template. The models were relaxed using a quick minimization routine with Amber force field and molecular surfaces were generated using the Molecular Operating Environment (MOE) (Chemical Computing Group, Montreal, Canada). Graphics were generated using University of California San Francisco (UCSF) Chimera Molecular Visualization package.28
Results
Genome statistics
The counts of DNA bases as well as selected annotations applied to assign functions to the six strains are presented in Table 1. The total number of bases sequenced for the R. palustris strains ranged from 4892717 (BisB5) to 5744041 (TIE-1) bases. The guanine-cytosine (GC) content of the genomes ranged from 64.44% (BisA53) to 66.04% (HaA2). The order of increasing genome size observed was BisB5, HaA2, CGA009, BisA53, BisB18 and TIE-1. The total number of genes also followed the order of genome size. Strain CGA009 had the highest coverage in four of the eight annotation schemes. Among the functional annotations methods applied to the protein coding genes, the Pfam had the highest coverage for all the genomes.
Table 1.
Genome features of Rhodopseudomonas palustris strains.
| Genome feature | BisA53 | BisB18 | BisB5 | CGA009 | HaA2 | TIE-1 |
|---|---|---|---|---|---|---|
| DNA, total number of bases | 5505494 | 5513844 | 4892717 | 5467640 | 5331656 | 5744041 |
| DNA coding, number of bases | 4766372 | 4765045 | 4276914 | 4810459 | 4677918 | 5024837 |
| DNA G+C, number of bases | 3547887 | 3581639 | 3170860 | 3555665 | 3520939 | 3725574 |
| Genes, total number | 4996 | 5028 | 4501 | 4920 | 4788 | 5377 |
| Protein coding genes | 4914 | 4943 | 4418 | 4838 | 4712 | 5318 |
| Pseudo genes | 36 | 57 | 21 | 18 | 29 | 72 |
| RNA genes | 82 | 85 | 83 | 82 | 76 | 59 |
| Enzymes | 1196 | 1233 | 1192 | 1253 | 1259 | 1317 |
| COGs | 3529 | 3688 | 3357 | 3791 | 3637 | 3897 |
| Pfam | 3594 | 3889 | 3505 | 3810 | 3834 | 4144 |
| TIGRfam | 1501 | 1528 | 1374 | 1520 | 1451 | 1536 |
| InterPro | 3720 | 3857 | 3522 | 1850 | 3823 | 4132 |
| IMG terms | 1266 | 1303 | 1232 | 1298 | 1331 | 1259 |
| IMG pathways | 355 | 386 | 378 | 382 | 385 | 370 |
| IMG parts List | 569 | 556 | 440 | 517 | 526 | 462 |
Pfam domain annotation statistics
In the abundance profile of the IMG system, a total of 2,355 Pfam domains were used to annotate at least one gene among the six finished R. palustris genomes analyzed. Further, 57 binary patterns of the possible 64 (26) patterns were used to label each Pfam domain with 1,641 domains present in all the genomes (ie, Pfam domains with binary pattern ‘111111’) (Table 2). The total Pfam annotations for CGA009, BisA53, TIE-1, BisB5, BisB18 and HaA2 were 1955, 1961, 2005, 1886, 1986, and 1944 respectively. A total of 245 Pfam domains were strain-specific annotations for the genomes compared (Table 2). Strain BisB18 had a total of 65 unique Pfam domain annotations; the highest among the strains analyzed. A total of 132 Pfam domains were uniquely shared by two strains. Further, 31 uniquely shared annotations that included CGA009 when the six genomes were compared. We prioritized Pfam domains shared by CGA009 and BisB5, Bis18 and HaA2 by verifying in the IMG system if they were used to annotate genes in the draft genome of strain DX-1.
Table 2.
Binary accessions generated for Pfam Categories in six Rhodopseudomonas palustris genomes.
| Six-digit binary accession* | Pfam category count |
|---|---|
| 000110 001011 100011 100110 | 1 |
| 101001 101010 101011 101110 | |
| 011011 011110 | 2 |
| 010001 010011 010100 011010 | 3 |
| 111110 | |
| 000011 100010 111100 | 4 |
| 001100 011000 100001 110001 | 5 |
| 110011 111001 | |
| 011101 101000 110110 | 6 |
| 001001 101101 111000 111011 | 7 |
| 000100 010101 100111 | 9 |
| 010111 111010 | 10 |
| 001101 100101 110010 | 11 |
| 001010 010010 110100 110101 | 14 |
| 000111 001000 | 19 |
| 000101 | 22 |
| 001111 | 25 |
| 111101 | 26 |
| 110111 | 30 |
| 101111 | 32 |
| 011111 | 34 |
| 110000 | 39 |
| 000001 000010 | 49 |
| 100000 | 54 |
| 010000 | 65 |
| 111111 | 1641 |
Note:
Digit 1 to 6 represent BisA53, BisB18, BisB5, CGA009, HaA2 and TIE-1 respectively.
Pfam domains shared exclusively with CGA009 are presented in Table 4. The numbers of Pfam domains shared by 3, 4 and 5 R. palustris genomes were 98, 107, and 132 respectively. The seven binary patterns of Pfam domains that were not observed in the matrix are 000000; 001110 (shared by only BisB5, CGA009 and HaA2), 010110 (shared by only BisB18, CGA009 and HaA2); 011001 (shared by only BisB18, BisB5 and TIE-1); 011100 (shared by only BisB18, BisB5 and CGA009); 100100 (shared by only BisA53 and CGA009); and 101100 (shared by only BisA53, BisB18 and CGA009).
Table 4.
Functional categories of genome-unique Pfam domain annotations for genomes of six Rhodopseudomonas palustris strains.*
| Functional category | BisA53 | BisB18 | BisB5 | CGA009 | HaA2 | TIE-1 |
|---|---|---|---|---|---|---|
| Information processing and storage | ||||||
| Translation, ribosomal structure and biogenesis [J] | 0 | 0 | 0 | 0 | 0 | 1 |
| RNA processing and modification [A] | 0 | 0 | 0 | 0 | 0 | 0 |
| Transcription [K] | 1 | 0 | 0 | 2 | 2 | 1 |
| Replication, recombination and repair [L] | 1 | 1 | 1 | 0 | 0 | 2 |
| Chromatin structure and dynamics [B] | 0 | 0 | 0 | 0 | 0 | 0 |
| Cellular processes and signaling | ||||||
| Cell cycle control, cell division, chromosome partitioning [D] | 2 | 1 | 0 | 0 | 0 | 0 |
| Nuclear structure [Y] | 0 | 0 | 0 | 0 | 0 | 0 |
| Defense mechanisms [V] | 0 | 1 | 1 | 0 | 0 | 0 |
| Signal transduction mechanisms [T] | 2 | 2 | 1 | 0 | 0 | 1 |
| Cell wall/membrane/envelope biogenesis [M] | 0 | 2 | 0 | 0 | 0 | 0 |
| Cell motility [N] | 1 | 0 | 0 | 0 | 0 | 0 |
| Cytoskeleton [Z] | 0 | 0 | 0 | 0 | 0 | 0 |
| Extracellular structures [W] | 0 | 0 | 0 | 0 | 1 | 0 |
| Intracellular trafficking, secretion, and vesicular transport [U] | 0 | 0 | 0 | 0 | 1 | 0 |
| Posttranslational modification, protein turnover, chaperones [O] | 0 | 0 | 2 | 0 | 0 | 0 |
| Metabolism | ||||||
| Energy production and conversion [C] | 3 | 6 | 0 | 0 | 0 | 0 |
| Carbohydrate transport and metabolism [G] | 5 | 3 | 0 | 0 | 1 | 0 |
| Amino acid transport and metabolism [E] | 0 | 3 | 0 | 0 | 4 | 0 |
| Nucleotide transport and metabolism [F] | 1 | 1 | 0 | 0 | 0 | 0 |
| Coenzyme transport and metabolism [H] | 2 | 1 | 0 | 0 | 1 | 0 |
| Lipid transport and metabolism [I] | 2 | 1 | 0 | 0 | 0 | 0 |
| Inorganic ion transport and metabolism [P] | 2 | 1 | 0 | 0 | 1 | 0 |
| Secondary metabolites biosynthesis, transport and catabolism [Q] | 0 | 4 | 0 | 1 | 4 | 0 |
| Poorly characterized | ||||||
| General function prediction only [R] | 2 | 3 | 2 | 0 | 4 | 12 |
| Function unknown [S] | 6 | 4 | 1 | 1 | 8 | 6 |
| Unmapped | 29 | 33 | 11 | 5 | 25 | 26 |
| Total Genome-Unique Pfam Domain Annotations | 59 | 67 | 19 | 9 | 52 | 49 |
Notes:
Pfam domains are genome-unique based on comparison of the six R. palustris genomes. Inclusion of additional genomes may change the count of genome-unique Pfam domains. To facilitate comparison of unique annotations for biological insights, a visual representation of the data in Table 3 is presented in Figure 4.
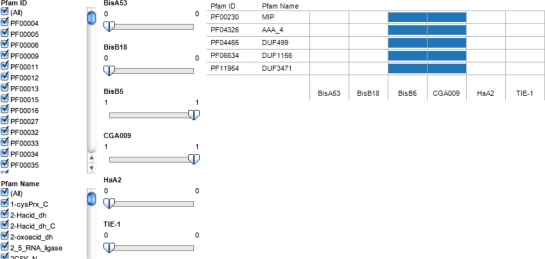
A visual analytics interactive view of binary patterns encoding the availability of the Pfam annotation for six R. palutris strains was also developed (Fig. 1). This interactive visualization resource enables user to specify the binary patterns (Table 2) to retrieve the Pfam domains clusters with the pattern. Figure 1 is an example of output of search for uniquely shared Pfam annotations for CGA009 and BisB5. The website for the resource is http://public.tableausoftware.com/views/pfam2rpalustris/pfamviz.
Figure 1.
Visual analytics resource for functional annotation analytics of six Rhodopseudomonas palustris genomes. The view illustrates selection of options to display on Pfam categories present in only strains BisB5 and CGA009. Five Pfam categories were identified including PF00230 (MIP—Major Intrinsic Protein family) the domain for water and/or glycerol transport. To achieve this view, the filters for only strains BisB5 and CGA009 were set to 1 equivalent to the 6-digit binary accession 001100. The position of the digit corresponds to the column number for each strain. Thus, the annotation of BisB5 and CGA009 are represented by digits 3 and 4 in the binary accession. Web page for interactive view: http://public.tableausoftware.com/views/pfam2rpalustris/pfamviz.
Functional relatedness of genomes based on Pfam domains
The overall functional relatedness of the six R. palustris genomes using hierarchical clustering based on the Pfam domains is presented in Figure 2. Two major groups were observed: genomes BisA53 and BisB18 clustered together while genomes BisB5, CGA009, TIE-1 and HaA2 clustered together with BisB5 on a distinct branch. CGA009 and TIE-1 clustered on the same node.
Figure 2.
Clustering of Rhodopseudomonas palustris genomes based on Pfam domain annotation of genes. Proximity of grouping indicates the relative degree of similarity of genomes to each other. The genome tree illustrating the relationship between genomes and Pfam domains was generated using Genome Clustering Tool on the Integrated Microbial Genomes (IMG) system (http://img.jgi.doe.gov/).
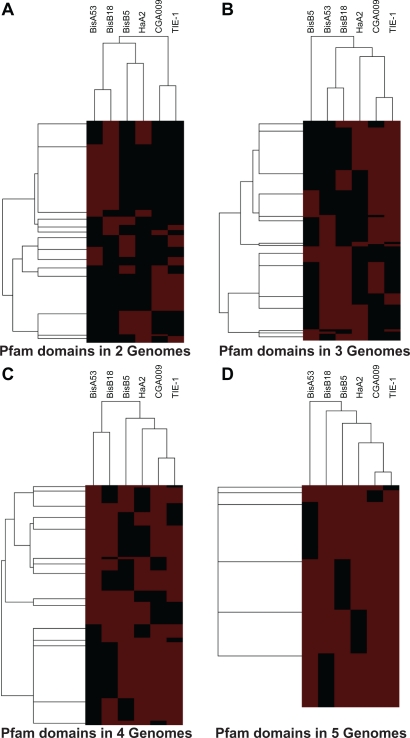
Pfam domains were grouped into six groups based on the number of genomes with the annotation. Clusters of Pfam domains by binary patterns for each of the group were determined using hierarchical clustering (Fig. 3). Again, in all the clustering CGA009 and TIE-1 clustered together. The number of clusters observed for Pfam in 2, 3, 4 and 5 genomes were 14 (Fig. 3A), 15 (Fig. 3B), 15 (Fig. 3C) and 6 (Fig. 3D) respectively.
Figure 3.
Relationship between six Rhodopseudomonas palustris genomes for Pfam annotations defined by presence in genomes. The hierarchical clustering of genomes (horizontal axis) and Pfam domains (vertical axis) are shown for Pfam domains present in 2, 3, 4 and 5 genomes. Data for clustering was obtained from matrix of binary patterns representing presence or absence of 2,255 Pfam domains in Rhodopseudomonas palustris genomes. The number of outermost branches is equivalent to the number of clusters. Red and black indicate presence and absence respectively of Pfam annotation in a genome.
Functional categories of strain-specific Pfam domain annotations
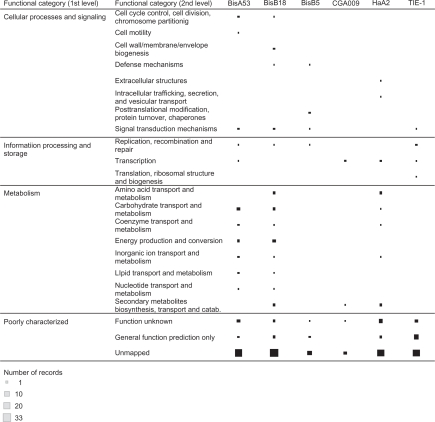
The annotations in the Cluster of Orthologous Groups (COGs) of Proteins system are classified into functional categories that allow for inferences on biological processes. The IMG system has 25 functional categories for Pfam domains based on the COG categories (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi?section=FindFunctions&page=pfamCategories). Therefore, we decided to extract deeper functional information on strain-specific Pfam domains for the six genomes. In this investigation, Pfam domains that have not been mapped to functional categories were categorized as “Unmapped”.
The 245 strain-specific Pfam domains among the genomes compared (Table 3) were mapped to four first level and 25 second level functional categories (Table 4 and Fig. 4). The first level categories were: Information Processing and Storage; Cellular Processes and Signaling; Metabolism and Poorly Characterized. A list of mappings of Pfam domains to functional categories is found in the Supplementary File. A Pfam domain can map to multiple categories. At least 40% of the Pfam domains for each of the strain were unmapped (Table 3). Strain CGA009 had the least number of unique Pfam domain annotations (9) and their second-level functional categories were as follows: BsuBI_PstI_RE (BsuBI/PstI restriction endonuclease C-terminus, PF06616, Unmapped); DUF2081 (Uncharacterized conserved protein, PF09854, Unmapped); DUF2806 (Domain of unknown function, PF10987, Unmapped); DUF364 (Domain of unknown function, PF04016, Function unknown), HTH_7 (Helix-turn-helix domain of resolvase, PF02796, Unmapped), LCM (Leucine carboxyl methyltransferase, PF04072, Secondary metabolites biosynthesis, transport and catabolism) Nudix_N (Hydrolase of X-linked nucleoside diphosphate N terminal, PF12535, Unmapped), RepB (RepB plasmid partitioning protein, PF07506, Transcription) and RNA_pol (DNA-dependent RNA polymerase, PF00940, Transcription).
Table 3.
Listing of unique Pfam domain annotations for finished genomes of six Rhodopseudomonas palustris strains.
| Strain | Pfam domains | Pfam domain count |
|---|---|---|
| CGA009 | RNA_pol HTH_7 DUF364 LCM BsuBI_PstI_RE RepB DUF2081 DUF2806 Nudix_N | 9 |
| BisB5 | Y_phosphatase Histone_HNS HIPIP Peptidase_M13 Transposase_12 TIG DUF389 DmsC TniB Peptidase_M13_N Endostatin Curlin_rpt GRDB CPT PHA_synth_III_E DUF2239 DUF2304 AlcB DUF3302 | 19 |
| TIE-1 | Phage_lysozyme Transposase_14 Cytochrom_CIII Transposase_Tn5 Mu_DNA_bind Bro-N Terminase_1 ANT BTAD DUF411 Phage_Mu_F DUF421 ERF Terminase_3 Baseplate_J DUF646 Phage_sheath_1 Phage_tube Phage_tail_S Terminase_4 Phage_tail_X NinB DUF847 Phage_GPD DUF935 Lambda_tail_I Phage_CP76 Phage_P2_GpU DUF1320 Phage_Mu_Gam Glyco_hydro_88 DUF1622 PglZ DUF1788 DUF1799 DUF1847 DALR_2 DUF1983 PG_binding_3 ATPase_gene1 YqaJ Potass_KdpF Tail_P2_I DUF2134 Mu-like_gpT DUF3164 DUF2933 DUF3486 DUF3732 | 49 |
| HaA2 | Tyrosinase ROK Ring_hydroxyl_A Ring_hydroxyl_B Cu_amine_oxid Fe_dep_repress TIR UPF0052 DUF108 CofC F420_ligase Gal_Lectin AT_hook Laminin_G_2 Cu_amine_oxidN2 Cu_amine_oxidN3 Fe_dep_repr_C HEAT DUF288 Lipoprotein_15 DUF304 GSP_synth YadA DUF350 NAPRTase SoxD SoxG Hep_Hag HIM DUF897 DUF971 5-nucleotidase DUF1185 Gp37_Gp68 Lipoprotein_Ltp PepX_C PNK3P TnsA_N YqcI_YcgG DUF1933 DUF2219 DUF2220 DUF2314 T5orf172 MraY_sig1 DUF2786 EcoR124_C DUF3604 DUF3696 | 49 |
| BisA53 | Peptidase_C1 UDPGT PAX PLDc Peptidase_C2 Peptidase_S7 Grp1_Fun34_YaaH Endonuclease_NS DUF82 PYC_OADA LacAB_rpiB DUF155 DUF258 Peptidase_C39 C4dic_mal_tran MinC_C MinE CitX CheD PspA_IM30 DUF399 LuxE AstE_AspA Mn_catalase DUF692 LuxC Glyco_transf_36 CBM_X GT36_AF DUF1234 DUF1542 VPEP DUF1624 Citrate_ly_lig Exonuc_X-T_C Cytotoxic ChAPs DUF1998 Exosortase_EpsH DUF2063 DUF2075 DUF2200 DUF2235 DUF2282 DUF2329 Muc_lac_enz Vir_act_alpha_C P63C Z1 DUF2581 DUF2809 DUF2971 DUF3280 DUF3485 | 54 |
| BisB18 | BMC A_deaminase IRK 3Beta_HSD Aldose_epim Avidin DeoC Glyco_transf_15 Glyco_hydro_3_C Peptidase_M29 OCD_Mu_crystall DUF161 Nitrate_red_del SCFA_trans Prismane EutN_CcmL Sulfotransfer_2 MbtH KdgT NapB NapD ALO ST7 PQ-loop NA37 LytTR RgpF Phage_portal_2 DUF763 Zot NACHT DUF889 DUF930 PduL EutQ PrpR_N His_kinase MipA MreB_Mbl Plasmid_Txe NapE HycH 5TM-5TMR_LYT Abi_2 DOT1 NRPS KR TrwC Acetone_carb_G DUF1993 Peptidase_M75 CbtA DHC CRISPR_Cas2 DUF2190 DUF2252 DUF2335 Hist_Kin_Sens RNA_bind_2 TrwB_AAD_bind DUF2817 DUF3072 DUF3387 DUF3494 DUF3644 | 65 |
Figure 4.
Visual analytic view of functional categories for strain-specific Pfam domain annotation of six Rhodopseudomonas palustris genomes. The view allows for user-interaction to select search fields including functional categories defined Cluster of Orthologous Groups (COGs) of Proteins. Interactive version of the figure can be found at http://public.tableausoftware.com/views/rhodo_palustris/uniquepfam2strain.
These mappings also helped to identify (i) functional categories that are unique to a genome in the comparison genome set; and (ii) identify strains in which Pfam domains were mapped to multiple functional categories (Table 3 and Fig. 4). Strain TIE-1 had the only entry “Translation, ribosomal structure and biogenesis [J]” with unique Pfam domain (PF05746, DALR_1) being an all alpha helical domain is the anticodon binding domain in Arginyl and glycyl tRNA synthetase. Strain BisB18 is unique for the “Cell wall/membrane/envelope biogenesis” category with two Pfam domains: PF06629 (MltA-interacting protein MipA) and PF05045 (Rhamnan synthesis protein F RgpF). Strain BisA53 is unique for “Cell motility [N]” category with one Pfam domain: PF03975 (Chemotactic sensory transduction CheD). Strain HaA2 is unique for the “Extracellular structures [W]” and “Intracellular trafficking, secretion, and vesicular transport [U]” categories. One Pfam domain: PF03895 (YadA-like C-terminal region YadA) was mapped to both categories. Strain BisB5 is unique for “Posttranslational modification, protein turnover, chaperones [O]” with two domains: PF01431 (Peptidase family M13 Peptidase_M13) and PF05649 (Peptidase family M13 Peptidase_M13_N).
A web resource that enables selection of functional annotation categories for the strain-specific Pfam domains is available at http://public.tableausoftware.com/views/rhodo_palustris/uniquepfam2strain.
We were particularly interested in gene products annotated as containing protein domain for water and/or glycerol transport (PF00230) that was observed only in CGA009 and BisB5. Therefore, additional bioinformatics analyses were performed in the IMG system to verify strain-specific or uniquely shared annotations. A search using the IMG Function Tool in the six completely sequenced R. palustris genomes for genes annotated with the Pfam domain PF00230 (water/glycerol transport) retrieved 3 genes from genomes of 2 strains (RPA2485 from CGA009 and RPD_2467 and RPD_2519 from BisB5).
Gene orthology, sequence analysis and homology modeling of Rhodopseudomonas water/glycerol transporters
Orthologous proteins RPA2485 and RPD_2467 from strains CGA009 and BisB5 had a sequence length of 240 aa, The alignment of their sequences with the sequence of aquaporin of Agrobacterium tumerfaciens str. C58 (Protein Data Bank (PDB) with accession 3LLQ) is presented in Figure 5. Both R. palustris aquaporin (AQP) sequences have two conserved Asparagine-Proline-Alanine (NPA) motifs that is characteristic motif of aquaporin sequences. These motifs align with those found in the 3LLQ. In the two R. palustris aquaporin sequences, prediction of membrane protein topology using Topcons29 confirmed six transmembrane domains in the following residue positions: 10–30, 35–55, 83–103, 131–151, 162–182, 206–226 (Fig. 6) connected by 5 loops (Loop A to Loop E according to the nomenclature in Kruse et al).30 Furthermore, the first NPA motif (residues 64–66) is inside loop (Loop B) while the second NPA motif (residues 186–188) is located outside loop (Loop E).
Figure 5.
Multiple sequence alignment of orthologous aquaporins from Agrobacterium tumerfaciens str. C58 (PDB ID: 3LLQ) as well as strains CGA009 and BisB5 of Rhodopseudomonas palustris. RPA2485 from CGA009 and RPD_2467 from BisB5 are 240 aa long. Information on the match of the residues is placed below each block of residues: “*”, “:” and “.” indicate that residues in column are identical, conserved substitutions and semi-conserved substitutions respectively. Below the symbols for sequence match, “=” indicates the two Asparagine-Proline-Alanine (NPA) motifs (residues 64 to 66; residues 186 to 188) and “^” indicates the residues for the aromatic/Arginine (ar/R) region (F44, H174, T183 and R189). The percent sequence identities between the 3LLQ and RPD_2467 and RPA2485 are 67.6% and 66.7% respectively.
Figure 6.
Sequence and predicted topologies for aquaporin of Rhodopseudomonas palustris CGA009. Graphic was generated with TOPCONS (http://topcons.net/), which provides a consensus prediction of membrane protein topology from 5 topologies.
Notes: Alphabets represent the predicted location of the residue in the protein.
Abbreviations: M, membrane; i, on the inside of the membrane; or o, on the outside of the membrane.
RPD_2519 from strain BisB5 had a 95 aa predicted protein that lacked ortholog in any other genomes according to predictions in Integrated Microbial Genome system. Further, RPD_2519 had only one NPA motif and thus does not fit the typical definition of aquaporins, which have two NPA or NPA-like motifs to form the water/solute channel. Therefore, we did not continue to investigate the sequence.
Theoretical homology models of aquaporins of strains BisB5 and CGA009 of R. palustris were generated using MODELLER7V7 with the high resolution X-ray crystal structure of aquaporin from the plant pathogen Agrobacterium tumerfaciens str. C58 (PDB ID: 3LLQ) as the template. The percent identities of the modeled AQP from BisB5 and CGA009 with the template were 67.6% and 66.7% respectively. The final homology model was aligned with the widely studied human AQP1 crystal structure (1J4N) and to compare residue interactions, pore dynamics and the overall structure-function relationship with the reported structures of 34 AQP channels (PDB ID: 1H6I, 1IH5, 1LDA, 1LDF, 1LDI, 1RC2, 1YMG, 1Z98, 2ABM, 2B5F, 2B6O, 2B6P, 2C32, 2D57, 2EVU, 2F2B, 2O9D, 2O9E, 2O9F, 2O9G, 2W1P, 2W2E, 2ZZ9, 3CLL, 3CN5, 3CN6, 3D9S, 3GD8, 3IYZ, 3LLQ, 3NE2, 3NK5, 3NKA, 3NKC). In addition to the presence of the characteristic NPA motifs, a narrow constriction region called aromatic/arginine (ar/R) approximately 8 Amstrongs above the NPA site. The shape of ar/R constriction region determines channel transport selectivity.31,32
Discussion
Rhodospeudomonas palustris, a nonsulphur purple photosynthetic bacteria, has been extensively investigated for its metabolic versatility including ability to produce hydrogen gas from sunlight and biomass as well as production of nanoparticles.8,16,33 Therefore, the discovery of new knowledge on strain-specific adaptation or phenotypes can advance their use in industrial processes. The identification of unique and shared annotations from closely related bacteria species is a useful step to unraveling unique and shared biological processes that define their ability to survive. Further, functional annotation analytics relying on bioinformatics tools integrated in a microbial genome informatics resource can provide insights into the origin of novel functions encoded in microbial genomes.4,34–36
We have compared the genomes of six strains of R. palustris based on the Pfam domain functional annotations. These strains have been described as ecotypes or genomospecies, which indicates their heterogeneous genetic structure.2 Our analysis revealed strain-specific and uniquely shared protein family annotations of genes among the six strains that could be further investigated. Gene loss or gain can explain the presence of strain-specific or uniquely shared genes.13,37 In addition, we identified a set of 1,641 Pfam domain annotations common to all genomes. The classification of Pfam domains into strain-specific, uniquely shared or common to all genomes is dependent upon the number of strains compared. Thus, the inclusion of strain DX-1 in the analysis will generate a new set of profiles. We have not included DX-1 in the analysis since only a draft genome sequence is available and not yet published. Nonetheless, in the case of Pfam domain annotation for water/glycerol transport, inclusion of DX-1 confirmed that the annotation is present in only genomes for strains BisB5 and CGA009. Our bioinformatics algorithm can be adapted to include additional genomes as needed for comparative analysis of Pfam domain annotations.
The integrative bioinformatics tools on Integrated Microbial Genome (IMG) system allowed for a comparison of the functional annotations of encoded proteins in R. palustris genomes based on COG clusters,38 Pfam,39 TIGRfam,40 and InterPro.41 We choose to further explore Pfam functional annotations for the selected annotation groups because the annotation method had the highest annotation coverage for the six genomes when compared to TIGRFAM and COG annotation schema (Table 1). In addition, the Pfam database is a large collection of 12,273 families (as of March 2011, Release 25) and commonly used for functional annotation of genomic data.39 An innovation of our investigation is the inclusion of an interactive visualization of the binary accessions associated with 2,355 Pfam domain annotations for six R. palustris genomes.
The use of visual analytics software to allow human interaction with dataset is increasingly recognized as relevant to gaining novel insights into biological datasets beyond purely biostatistical approaches.42–46 The binary-based integration provides rapid snapshots of the dataset that can facilitate deeper biological insights or relationships between the datasets to direct further analysis.19,47 The visual analytics web-based resources accompanying this report allows for user-defined queries beyond those reported here. The data visualizations could also yield novel insights on the functional annotations associated with the six strains. In this investigation, we have illustrated the use of these visual analytics resources to identify annotations shared by BisB5 and CGA009 (Fig. 3). In addition, a static visualization of the functional categories of the 245 strain-specific Pfam domains is presented in Figure 4. An interactive version of Figure 4 is available as a web resource.
Previous studies conducted by Oda et al13 revealed that Rhodopseudomonas populations isolated from sediment microenvironments contain unique genes that promote distinct physiological characteristics conducive for environmental adaptation. In addition, strain-specific adaptations that allow anaerobic fermentation, expanded biodegradation, or expanded light-harvesting capabilities are also potentially useful in applications for biohydrogen production by Rhodopseudomonas. We also used hierarchical clustering to define Pfam domain clusters using the number of annotated genomes (Fig. 2). Strains CGA009 and TIE-1 always clustered together in line with previous observations that the genomes of TIE-1 and CGA009 are 97.9% identical at the nucleotide level over 5.28 Mb of shared DNA.13 Further, strains BisA53 and BisB18 clustered together in our analysis consistent with them having similar genome architecture. The genome clusters observed in this investigation is consistent with phylogenetic trees constructed using 3 molecular marker sequences from 33 Rhodopseudomonas strains.2
Comparison of the Pfam domain annotations revealed that proteins annotated with PF00230 (major intrinsic proteins) were restricted to strain CGA009 and BisB5. Protein sequences annotated PF00230 belong to a universal family of cellular water/solute channels. In terms of function, members are classified into orthodox aquaporins (AQP) (water-specific channels) and aquaglyceroporins (permeated by mainly glycerol and some other solutes, whereas water transport is strongly limited).48,49 Generally, permeation is strictly passive according to the osmotic or solute gradient. Orthodox aquaporins function in water homeostasis while aquaglyceroporins function in metabolism.
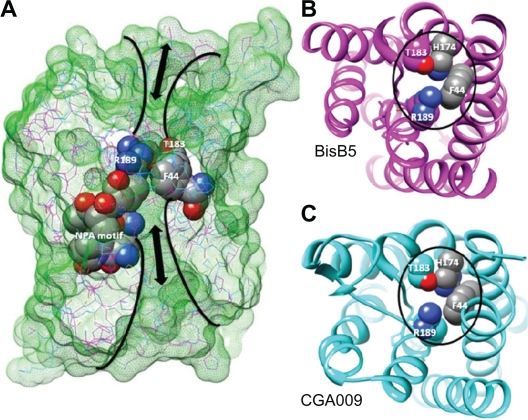
Our homology modeling and sequence analysis of the two 240 aa R. palustris proteins from strains BisB5 and CGA009 that were annotated with PF00230 annotation indicate they may function as water-specific channel (Fig. 7). The transport specificity in water-specific AQP channels have been clearly demonstrated by using mutational studies of three aromatic/Arginine (ar/R) constriction region residues F56, H180 and R195 rat AQP1.32 Single or double mutants of ar/R residues to amino acids with small amino acid residues alanine or valine did not alter water permeability. However, the double mutants H180A/R195V allowed transport of larger molecules including glycerol and urea indicating a clear ar/R pore constriction versus transport relationship. 32 The corresponding ar/R region in the aquaporins from R. palustris is occupied by F44, H174, T183 and R189 (Fig. 7) indicating the similar selectivity towards water molecules.31,32,50
Figure 7.
Homology model of aquaporins encoded in two strains of Rhodopseudomonas palustris. (A) Superposition of theoretical models of R. palustris water channel proteins from BisB5 (magenta) and CGA009 (cyan) strains. Molecular surfaces (green mesh) clearly illustrate the role of residues F44, H174, T183 and R189 in conferring selectivity towards water molecules in BisB5 (B) and CGA009 (C).
Aquaporins have function beyond water/glycerol transport including cell adhesion,51 cell migration52 and transport of molecules such as arsenic and boron.53 The lack of genuine aquaporins in most microorganisms has led to the conclusion that aquaporins are not essential for basic cellular function in microorganisms.54 However, they could be advantageous for improving freeze tolerance in natural conditions of rapid freezing of microbes20 and insect larvae.55 Strains or genes of R. palustris have been isolated or cloned from cold soil environments including the high arctic56 and the sub-Antarctic57 in the context of nitrogen fixation, a process in which access to liquid water is a more limiting factor for continued activation of nitrogenase in low temperature.58 CGA009, a strain widely distributed in temperate soil and water, is well equipped for nitrogen fixation as it encodes three nitrogenases.22 The absence of aquaporins in strains BisA53, BisB18, HaA2 and TIE-1 may also have functional relevance. In natural Saccharomyces cerevisiae populations, the loss of aquaporins provides a major fitness advantage on high-sugar substrates such as fruits or fermentations common to many S. cerevisiae strains’ natural niche.21 Strains P4, PBUM001, M23, WP3-5, and W004 of R. palustris have been employed to produce hydrogen gas directly by fermenting sugars or improving the hydrogen gas production yield.59–63 Specifically, strain WP3-5 improved hydrogen gas production from cassava starch by using soluble metabolite products (eg, acetic acid, butyric acid) from dark fermentation.63 Research to determine the presence or absence of aquaporin in R. palustris strains of known phenotype could provide molecular basis for nitrogen fixation at low temperatures as well as efficient utilization of substrates with high sugar content.
Conclusions
Functional annotation analytics of six genomes of Rhodopseudomonas palustris revealed sets of annotations that could be verified as strain-specific or uniquely shared phenotypes. Genes encoding water/glycerol transport were present in genome sequences of strains CGA009 and BisB5 but absent in strains BisA53, BisB18, HaA2 and TIE-1. Based on observations in other microbes, the presence of aquaporin in R. palustris strains may improve freeze tolerance in natural conditions of rapid freezing such as nitrogen fixation at low temperatures where access to liquid water is a limiting factor for nitrogenase activation. In the case of adaptive loss of aquaporin genes, strains may be better adapted to survive in conditions of high sugar content such as fermentation of biomass for biohydrogen production. Finally, web-based resources were developed to allow for interactive, user-defined selection of the relationship between protein family annotation and the R. palustris genomes.
Supplementary Materials
Table 5.
Selected uniquely shared Pfam annotations in Rhodopseudomonas palustris strains.*
| Pfam accession | Pfam identifier | Strains with Pfam annotation | Function classification |
|---|---|---|---|
| PF06897 | DUF1269 | CGA009 BisB18 | Function unknown |
| PF04326 | AAA_4 | CGA009 BisB5 | Transcription |
| PF04465 | DUF499 | CGA009 BisB5 | Unmapped |
| PF00230 | MIP | CGA009 BisB5 | Carbohydrate transport and metabolism |
| PF06634 | DUF1156 | CGA009 BisB5 | Unmapped |
| PF09250 | Prim-Pol | CGA009 HaA2 | Unmapped |
Notes:
Based on comparison of strains BisA53, BisB18, BisB5, CGA009, HaA2 and TIE-1.
Acknowledgments
Mississippi NSF-EPSCoR Award (EPS-0903787); NSF-Undergraduate Research and Mentoring Program (DBI-0958179); Visual Analytics in Biology Curriculum Network (DBI-1062057); US Department of Homeland Security Science & Technology Directorate (2007-ST-104-000007; 2009-ST-062-000014; 2009-ST-104-000021); Research Centers in Minority Institutions (RCMI)—Center for Environmental Health at Jackson State University (NIH-NCRR G12RR013459); Pittsburgh Supercomputing Centre’s National Resource for Biomedical Supercomputing (T36GM095335); National Center for Integrative Biomedical Informatics, University of Michigan (NIH-U54DA021519); Mississippi IDeA Network for Biomedical Excellence (NIH-NCRR-P20RR016476); Arkansas IDeA Network for Biomedical Excellence (NIH-NCRR-P20RR016460); NIH RIMI Grant 1P20MD002725-01 to Tougaloo College. SSS was a Louis Stokes Mississippi Alliance for Minority Participation (LSMAMP) Fellow in 2005 and is currently a PhD Candidate in the Environmental Science PhD Program at Jackson State University. We thank Dr. Michael Allen and Dr. Carrie S. Harwood for their helpful suggestions and comments during the preparation of the manuscript. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the funding agencies.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Bent SJ, Gucker CL, Oda Y, Forney LJ. Spatial distribution of Rhodopseudomonas palustris ecotypes on a local scale. Appl Environ Microbiol. 2003;69:5192–7. doi: 10.1128/AEM.69.9.5192-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okamura K, Takata K, Hiraishi A. Intrageneric relationships of members of the genus Rhodopseudomonas. J Gen Appl Microbiol. 2009;55:469–78. doi: 10.2323/jgam.55.469. [DOI] [PubMed] [Google Scholar]
- 3.Harwood CS, Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol. 1988;54:712–7. doi: 10.1128/aem.54.3.712-717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karpinets TV, Pelletier DA, Pan C, et al. Phenotype fingerprinting suggests the involvement of single-genotype consortia in degradation of aromatic compounds by Rhodopseudomonas palustris. PLoS One. 2009;4:e4615. doi: 10.1371/journal.pone.0004615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Kappler A, Croal LR, Newman DK. Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl Environ Microbiol. 2005;71:4487–96. doi: 10.1128/AEM.71.8.4487-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larimer FW, Chain P, Hauser L, et al. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol. 2004;22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 7.VerBerkmoes NC, Shah MB, Lankford PK, et al. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. J Proteome Res. 2006;5:287–98. doi: 10.1021/pr0503230. [DOI] [PubMed] [Google Scholar]
- 8.Ren NQ, Liu BF, Ding J, Xie GJ. Hydrogen production with R. faecalis RLD-53 isolated from freshwater pond sludge. Bioresour Technol. 2009;100:484–7. doi: 10.1016/j.biortech.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Rey FE, Oda Y, Harwood CS. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J Bacteriol. 2006;188:6143–52. doi: 10.1128/JB.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rey FE, Heiniger EK, Harwood CS. Redirection of metabolism for biological hydrogen production. Appl Environ Microbiol. 2007;73:1665–71. doi: 10.1128/AEM.02565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehrabi S, Ekanemesang UM, Aikhionbare FO, Kimbro KS, Bender J. Identification and characterization of Rhodopseudomonas spp., a purple, non-sulfur bacterium from microbial mats. Biomol Eng. 2001;18:49–56. doi: 10.1016/s1389-0344(01)00086-7. [DOI] [PubMed] [Google Scholar]
- 12.Cantera JJ, Kawasaki H, Seki T. The nitrogen-fixing gene (nifH) of Rhodopseudomonas palustris: a case of lateral gene transfer? Microbiology. 2004;150:2237–46. doi: 10.1099/mic.0.26940-0. [DOI] [PubMed] [Google Scholar]
- 13.Oda Y, Larimer FW, Chain PS, et al. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc Natl Acad Sci USA. 2008;105:18543–8. doi: 10.1073/pnas.0809160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing D, Zuo Y, Cheng S, Regan JM, Logan BE. Electricity generation by Rhodopseudomonas palustris DX-1. Environ Sci Technol. 2008;42:4146–51. doi: 10.1021/es800312v. [DOI] [PubMed] [Google Scholar]
- 15.Markowitz VM, Chen IM, Palaniappan K, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–90. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai HJ, Zhang ZM, Guo Y, Yang GE. Biosynthesis of cadmium sulfide nanoparticles by photosynthetic bacteria Rhodopseudomonas palustris. Colloids Surf B Biointerfaces. 2009;70:142–6. doi: 10.1016/j.colsurfb.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Cohly HH, Isokpehi R, Rajnarayanan RV. Compartmentalization of aquaporins in the human intestine. Int J Environ Res Public Health. 2008;5:115–9. doi: 10.3390/ijerph5020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fadiel A, Isokpehi RD, Stambouli N, et al. Protozoan parasite aquaporins. Expert Rev Proteomics. 2009;6:199–211. doi: 10.1586/epr.09.10. [DOI] [PubMed] [Google Scholar]
- 19.Isokpehi RD, Rajnarayanan RV, Jeffries CD, Oyeleye TO, Cohly HH. Integrative sequence and tissue expression profiling of chicken and mammalian aquaporins. BMC Genomics. 2009;10(Suppl 2):S7. doi: 10.1186/1471-2164-10-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanghe A, Van DP, Dumortier F, et al. Aquaporin expression correlates with freeze tolerance in baker’s yeast, and overexpression improves freeze tolerance in industrial strains. Appl Environ Microbiol. 2002;68:5981–9. doi: 10.1128/AEM.68.12.5981-5989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Will JL, Kim HS, Clarke J, et al. Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 2010;6:e1000893. doi: 10.1371/journal.pgen.1000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oda Y, Samanta SK, Rey FE, et al. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J Bacteriol. 2005;187:7784–94. doi: 10.1128/JB.187.22.7784-7794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang XP, Setola V, Yadav PN, et al. Parallel functional activity profiling reveals valvulopathogens are potent 5-hydroxytryptamine(2B) receptor agonists: implications for drug safety assessment. Mol Pharmacol. 2009;76:710–22. doi: 10.1124/mol.109.058057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 25.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Marti-Renom MA, Stuart AC, Fiser A, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–8. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruse E, Uehlein N, Kaldenhoff R. The aquaporins. Genome Biol. 2006;7:206. doi: 10.1186/gb-2006-7-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliva R, Calamita G, Thornton JM, Pellegrini-Calace M. Electrostatics of aquaporin and aquaglyceroporin channels correlates with their transport selectivity. Proc Natl Acad Sci U S A. 2010;107:4135–40. doi: 10.1073/pnas.0910632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc Natl Acad Sci U S A. 2006;103:269–74. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gosse JL, Engel BJ, Rey FE, et al. Hydrogen production by photoreactive nanoporous latex coatings of nongrowing Rhodopseudomonas palustris CGA009. Biotechnol Prog. 2007;23:124–30. doi: 10.1021/bp060254+. [DOI] [PubMed] [Google Scholar]
- 34.Davidsen T, Beck E, Ganapathy A, et al. The comprehensive microbial resource. Nucleic Acids Res. 2010;38:D340–5. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeil LK, Reich C, Aziz RK, et al. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 2007;35:D347–53. doi: 10.1093/nar/gkl947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder EE, Kampanya N, Lu J, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–6. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marri PR, Hao W, Golding GB. Gene gain and gene loss in streptococcus: is it driven by habitat. Mol Biol Evol. 2006;23:2379–91. doi: 10.1093/molbev/msl115. [DOI] [PubMed] [Google Scholar]
- 38.Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selengut JD, Haft DH, Davidsen T, et al. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–4. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter S, Apweiler R, Attwood TK, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naumova EN. Visual analytics for immunologists: Data compression and fractal distributions. Self Nonself. 2010;1:241–9. doi: 10.4161/self.1.3.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shih DC, Ho KC, Melnick KM, et al. Facilitating the analysis of immunological data with visual analytic techniques. J Vis Exp. 2011 doi: 10.3791/2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445–55. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamel Boulos MN, Viangteeravat T, Anyanwu MN, Ra NV, Kuscu E. Web GIS in practice IX: a demonstration of geospatial visual analytics using Microsoft Live Labs Pivot technology and WHO mortality data. Int J Health Geogr. 2011;10:19. doi: 10.1186/1476-072X-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson MO, Cohly HH, Isokpehi RD, Awofolu OR. The case for visual analytics of arsenic concentrations in foods. Int J Environ Res Public Health. 2010;7:1970–83. doi: 10.3390/ijerph7051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isokpehi RD, Simmons SS, Cohly HH, et al. Identification of drought-responsive universal stress proteins in viridiplantae. Bioinform Biol Insights. 2011;5:41–58. doi: 10.4137/BBI.S6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agre P. The aquaporin water channels. Proc Am Thorac Soc. 2006;3:5–13. doi: 10.1513/pats.200510-109JH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozono D, Yasui M, King LS, Agre P. Aquaporin water channels: atomic structure molecular dynamics meet clinical medicine. J Clin Invest. 2002;109:1395–9. doi: 10.1172/JCI15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–8. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 51.Kumari SS, Varadaraj K. Intact AQP0 performs cell-to-cell adhesion. Biochem Biophys Res Commun. 2009;390:1034–9. doi: 10.1016/j.bbrc.2009.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saadoun S, Papadopoulos MC, Hara-Chikuma M, Verkman AS. Impairment of angiogenesis and cell migration by targeted aquaporin-1 gene disruption. Nature. 2005;434:786–92. doi: 10.1038/nature03460. [DOI] [PubMed] [Google Scholar]
- 53.Hove RM, Bhave M. Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol Biol. 2011;75:413–30. doi: 10.1007/s11103-011-9737-5. [DOI] [PubMed] [Google Scholar]
- 54.Tanghe A, Van DP, Thevelein JM. Why do microorganisms have aquaporins. Trends Microbiol. 2006;14:78–85. doi: 10.1016/j.tim.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Philip BN, Yi SX, Elnitsky MA, Lee RE., Jr Aquaporins play a role in desiccation and freeze tolerance in larvae of the goldenrod gall fly Eurosta solidaginis. J Exp Biol. 2008;211:1114–9. doi: 10.1242/jeb.016758. [DOI] [PubMed] [Google Scholar]
- 56.Deslippe JR, Egger KN. Molecular diversity of nifH genes from bacteria associated with high arctic dwarf shrubs. Microb Ecol. 2006;51:516–25. doi: 10.1007/s00248-006-9070-8. [DOI] [PubMed] [Google Scholar]
- 57.Rapley J. Phylogenetic diversity of nifH genes in marion island soil. Master of Science Thesis, University of the Western Cape, South Africa. 2006. http://etd.uwc.ac.za/usrfiles/modules/etd/docs/etd_gen8-Srv25Nme4_6147_1223533256.pdf.
- 58.Davey A. Effects of abiotic factors on nitrogen fixation by blue-green algae in Antarctica. Polar Biology. 1983;2:95–100. [Google Scholar]
- 59.Chen CY, Lu WB, Liu CH, Chang JS. Improved phototrophic H2 production with Rhodopseudomonas palustris WP3-5 using acetate and butyrate as dual carbon substrates. Bioresour Technol. 2008;99:3609–16. doi: 10.1016/j.biortech.2007.07.037. [DOI] [PubMed] [Google Scholar]
- 60.Oh YK, Seol EH, Lee EY, Park S. Fermentative hydrogen production by a new chemoheterotrophic bacterium Rhodopseudomonas palustris P4. Int J Hydrogen Energy. 2011;27:1373–9. [Google Scholar]
- 61.Jamil Z, Mohamad Annuar MS, Ibrahim S, Vikineswary S. Optimization of phototrophic hydrogen production by Rhodopseudomonas palustris PBUM001 via statistical experimental design. Int J Hydrogen Energy. 2009;34:7502–12. [Google Scholar]
- 62.Yang CF, Lee CM. Enhancement of photohydrogen production using phbC deficient mutant Rhodopseudomonas palustris strain M23. Bioresour Technol. 2011;102:5418–24. doi: 10.1016/j.biortech.2010.09.078. [DOI] [PubMed] [Google Scholar]
- 63.Su H, Cheng J, Zhou J, Song W, Cen K. Improving hydrogen production from cassava starch by combination of dark and photo fermentation. International Journal of Hydrogen Energy. 2009;34:1780–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.