Abstract
The translation factor eIF5 is an important partner of eIF2, directly modulating its function in several critical steps. First, eIF5 binds eIF2/GTP/Met-tRNAiMet ternary complex (TC), promoting its recruitment to 40S ribosomal subunits. Secondly, its GTPase activating function promotes eIF2 dissociation for ribosomal subunit joining. Finally, eIF5 GDP dissociation inhibition (GDI) activity can antagonize eIF2 reactivation by competing with the eIF2 guanine exchange factor (GEF), eIF2B. The C-terminal domain (CTD) of eIF5, a W2-type HEAT domain, mediates its interaction with eIF2. Here, we characterize a related human protein containing MA3- and W2-type HEAT domains, previously termed BZW2 and renamed here as eIF5-mimic protein 1 (5MP1). Human 5MP1 interacts with eIF2 and eIF3 and inhibits general and gene-specific translation in mammalian systems. We further test whether 5MP1 is a mimic or competitor of the GEF catalytic subunit eIF2Bε or eIF5, using yeast as a model. Our results suggest that 5MP1 interacts with yeast eIF2 and promotes TC formation, but inhibits TC binding to the ribosome. Moreover, 5MP1 is not a GEF but a weak GDI for yeast eIF2. We propose that 5MP1 is a partial mimic and competitor of eIF5, interfering with the key steps by which eIF5 regulates eIF2 function.
INTRODUCTION
During translation initiation, eukaryotic initiation factors (eIFs) assemble initiator methionyl tRNAiMet (Met-tRNAiMet) and m7G-capped mRNA with the 40S ribosome subunit, precisely matching the tRNAiMet anticodon to the mRNA start codon at the 40S ribosomal P-site (1,2). This translation process occurs in multiple steps. First, the heterotrimeric factor eIF2 (composed of α, β and γ subunits) binds Met-tRNAiMet, a process that is dependent on GTP being bound to the γ subunit of eIF2. The resulting eIF2/GTP/Met-tRNAiMet ternary complex (TC) is incorporated into the 43S pre-initiation complex (PIC), which also contains eIF1A, eIF1, eIF3 and eIF5. The m7G-capped mRNA is then activated and is recruited to the 43S PIC by eIF4F, composed of the cap binding protein eIF4E, adaptor eIF4G and mRNA helicase eIF4A, forming the 48S PIC that includes the 40S subunit located at the 5′-end of the mRNA. The PIC then scans along the 5′-leader of the transcript until the start codon of the mRNA base-pairs to the tRNAiMet anticodon in the P-site of the ribosome. The concerted actions of the assembled eIFs coupled to the eIF2 GTPase activation (GAP) function catalyzed by the N-terminal domain (NTD) of eIF5 are thought to provoke conformational changes within the PIC, promoting the release of most eIFs, including eIF1, eIF2 and eIF5. The second GTP binding factor eIF5B then mediates the joining of the 60S subunit to form an 80S initiation complex, which is then ready to accept the second aminoacyl tRNA into the ribosomal A-site during the elongation phase of translation. eIF2 is released from the PIC as the GDP-bound form, which is reactivated to the GTP-form by eIF2B-catalyzed guanine nucleotide exchange, allowing for subsequent rounds of translation initiation. eIF5 appears to be ejected from the PIC in complex with eIF2-GDP, and antagonize eIF2B-catalyzed eIF2 reactivation (3) via its GDP dissociation inhibition (GDI) activity (4).
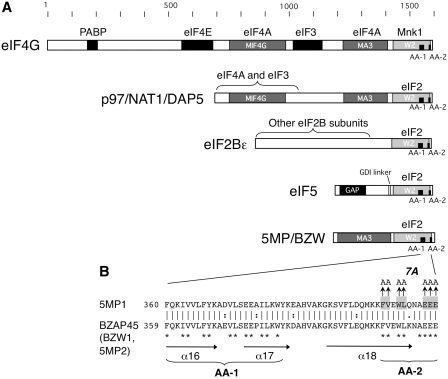
Many critical protein–protein interactions mediating the eukaryotic translation initiation process involve the HEAT domain, a globular domain comprising several repeats of conserved anti-parallel α-helical structures (5). For example, mammalian eIF4G molecules possess three distinct HEAT domains termed MIF4G, MA3 and W2 (Figure 1A). The MIF4G and MA3 domains of eIF4G together form a binding pocket for a single molecule of eIF4A, thereby regulating mRNA recruitment and scanning (6), whereas the last W2 domain is the binding site for Mnk eIF4E kinase (7) (Figure 1A). Mammalian eIF4G also possesses an unstructured N-terminal segment capable of binding poly(A) binding protein (PABP) and the m7G-cap binding subunit, eIF4E. W2-type HEAT domains (W2-CTDs) are also found in the C-terminal segments of eIF5 and eIF2Bε but these are substrate binding sites for the lysine-rich N-terminal half of the eIF2β subunit (8,9). The role of W2-CTDs of eIF5 and eIF2Bε in the regulation of eIF2 activity has been well-established. The W2-CTD of eIF2Bε functions in guanine nucleotide exchange on eIF2 (10), whereas the W2 domain in eIF5 mediates the formation of a multifactor complex (MFC) with eIF1, eIF2-GTP, eIF3 and Met-tRNAiMet (9,11). Finally the eIF5 W2-CTD together with the adjacent N-terminal linker region is responsible for the GDI activity against eIF2-GDP (3,4).
Figure 1.
HEAT domain-containing translation initiation factors and regulators. (A) Primary structures of human eIF4G1, p97/NAT1/DAP5, 5MP1/BZW2 and yeast eIF2Bε and eIF5 are drawn to scale with filled boxes indicating segments known to interact with their partners listed across the top. Bracket indicates an approximate area of interaction with indicted partners. Light gray boxes indicate the W2 domains, with short thick lines representing the location of AA-boxes 1 and 2 (AA-1, AA-2, respectively). Gray boxes indicate other HEAT domains. (B) Alignment of C-terminal parts of amino acid sequences (starting with residue numbered to the left) from human 5MP1/BZW2 and 5MP2/BZW1/BZAP45. Asterisks indicate residues making up AA-boxes, while vertical arrows indicate mutations in 7A. Horizontal arrows indicate the location of α-helices predicted from the homology with eIF4G.
Two distinct copies of eIF4G are found in most eukaryotes examined so far. However, vertebrates encode a third eIF4G-like species termed p97/NAT1/DAP5 (Figure 1A). This protein lacks the N-terminal segment but contains the three HEAT domains analogous to mammalian eIF4G. Thus, p97/NAT1/DAP5 is a macromolecular mimic of the eIF4G C-terminal half (12,13), and it has been implicated in the modulation of translation for specific mRNAs (14). Interestingly the DAP5-CTD 3D structure is very similar to that of the W2-CTDs of eIF5 and eIF2Bε, and in common with these factors, but unlike eIF4G, DAP5 also binds to eIF2β (15).
In this study, we characterize another human HEAT domain-containing protein that is related to eIF5, known as BZW or MA3+W2 protein (Figure 1A) (16). Human BZW1 and BZW2 share 72% sequence identity and contain two HEAT domains, MA3 and W2 (hence the family name), comprising 10 and 8 α-helices that correspond to five and four HEAT repeats, respectively (http://famshelp.gsc.riken.jp/famsbase). While BZW1 was characterized as a transcription factor termed BZAP45 (17), its GFP-fusion form is localized in the cytoplasm (18). In addition, a single MA3+W2 protein is found in Drosophila melanogaster and is termed Krasavietz (Kra) (also known as eIF5C, Exba, or Ecp). Lee et al. (19) presented evidence that Kra co-migrates with 40S ribosomes and interacts with eIF2β more weakly than eIF5, and that Kra AA-box 1 and 2 mutations weaken this interaction with eIF2β. Furthermore, Kra reduces luciferase expression in rabbit reticulocyte lysate (RRL) (19). Interestingly, the Drosophila Kra mutants are defective in long-term memory (20) and consistent with this, Kra is implicated in neuronal development and interacts with Shot, the cytoskeletal element that links F-actin and microtubules required for axon formation (19).
The above-mentioned study of Kra protein suggests that all W2-containing proteins (eIF5, eIF2Bε, p97/NAT1/DAP5 and BZW/Kra), except eIF4G, bind eIF2. Because the eIF5-NTD that activates eIF2 GTPase in response to start codon selection is not present in the BZW proteins (Figure 1A), we hypothesized that BZW/Kra may serve as a mimic or competitor of eIF2Bε or an antagonist of eIF5 functions in translation. Here we tested these predictions and provide evidence from multiple systems (RRL, mammalian cells and yeast) that BZW2 does indeed antagonize eIF5 functions with eIF2 to modulate both general translation and the translation of specific genes at multiple steps. BZW stands for basic leucine-zipper with a W2 domain, although the conserved lysine/arginine and leucine residues that were characterized as basic leucine-zipper (17) are in fact a part of HEAT repeats that make up a MA3 domain (16). Based on this study emphasizing its role as a competitive inhibitor of eIF5 function by macro-molecular mimicry, we thus designate the BZW proteins as the eIF5-mimic proteins (5MPs), and rename human BZW2 and BZW1 as 5MP1 and 5MP2, respectively.
MATERIALS AND METHODS
Plasmids and yeast strains
Plasmids used in this study and oligodeoxyribonucleotides used for their constructions are listed in Supplementary Tables S1 and S2, respectively. Details of their construction and yeas strains used are described in Supplementary Data.
Yeast methods
We used the standard genetic/ molecular biology techniques and prepared yeast growth media. Immuno-affinity purification of FLAG-tagged protein complexes was performed as described (3,11,21).
In vitro protein–protein interaction assay
eIF3 and eIF2 were purified from HeLa cells (kindly provided by J. Hershey and M. Sokabe). FL-5MP1 and its mutants were purified from H2557 transformants carrying pEMBL-FL-5MP1 and its derivatives, that had been grown in synthetic complete galactose medium lacking uracil (SCGal-ura), as described (22). Recombinant eIF5 was purified by G50 column fractionation after thrombin treatment of GST-human eIF5 (23).
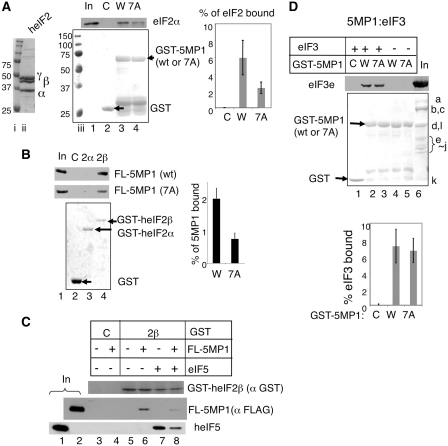
To perform GST pull-down assays, GST-fusion proteins were expressed in BL21(DE3) transformants carrying the relevant pGEX- series of plasmids (Table S1), purified in one-step with a glutathione resin and used immediately for the interaction with their purified partners, essentially as described (24). In each experiment and its repeats, we used a fixed amount of GST-fusion proteins (5 µg each of GST, GST-5MP1 and GST-5MP1-7A in Figure 2A and D; 5 µg GST, 3 µg each of GST-eIF2α and -eIF2β in Figure 2B) and 10 times less (in molarity) purified partners. For the latter, we used 1 µg human eIF2 (Figure 2A) or 8 µg human eIF3 (Figure 2D), or 300 ng of FL-5MP1 or FL-5MP1-7A (Figure 1E). After incubation, protein complexes adsorbed to the resin were washed, dissolved into a SDS–PAGE buffer and analyzed by immunoblotting.
Figure 2.
Human 5MP1/BZW2 interacts with eIF2 and eIF3. (A) Pulldown assays with GST-5MP1 (W, wild-type), its 7A derivative (7A) or GST alone (C). The complexes with human eIF2 were analyzed with 10% in-put (In) control by immunoblotting using anti-eIF2α (Santa Cruz Biotech.) (top gel) followed by Ponceau S staining (bottom gel). Arrows in the bottom gel indicate the location of GST-fusions used. Lanes i and iii, size standards. Lane ii, human eIF2 used visualized by Coomassie staining. Graph summarizes percentage of eIF bound to GST-fusions used, with bars indicating SD. (B) Interactions of 5MP1 with individual eIF2 subunits fused to GST. GST-eIF2α (2α), and -eIF2β (2β) bound to FL-5MP1 (wt) or its 7A version (7A) were analyzed with 2.5% in-put (lanes 1) by immunoblotting (top gels) with anti-FLAG M2 antibodies (Sigma), followed by Ponceau S staining shown on bottom and presented as in (A). C, control with GST alone. Graph indicates the percentage of FL-5MP1 or FL-5MP1-7A bound to GST-eIF2β, with bars denoting SD. (C) Competition assay. FL-5MP1 (200 ng) and/or human eIF5 (1.6 µg) were incubated with GST-heIF2β (2β) or GST alone (C) (1 µg) pre-adsorbed to glutathione resin, and the complex was precipitated and analyzed by immunoblotting with anti-GST (top), anti-FLAG (middle) and anti-heIF5 (bottom) antibodies, as in (A). Lane 1, 5% in-put of heIF5. Lane 2, 25% in-put of FL-5MP1. (D) Pulldown assays with GST-5MP1 and its 7A derivative (7A) were performed with human eIF3, similar to (A), and analyzed with 50% in-put (In) control by immunoblotting using anti-eIF3e (41) antibodies (top gel), followed by Ponceau S staining (bottom gel). The results were presented as in (A). In the ‘in-put’ lane, eIF3 subunit names are identified to the right. Graphs summarize the percentage of eIF3 bound to GST-fusions used, with bars indicating SD.
Analysis of 43S/48S PIC in RRL
m7G-capped luciferase mRNA was synthesized and purified using the mMegascript mMessage kit (Ambion) and SmaI-linearized plasmid pRG166 as template (25). Translation reaction (20 µl) was performed with RRL in the presence of cold amino acids using 2.5 ng cap-LUC mRNA, as recommended by the manufacturer (Promega). After incubation at 30°C for 20 min, the sample was supplemented with 160 ng BSA or FL-5MP1 and incubated for an additional hour. The samples were fixed with 1% HCHO on ice for 30 min, quenched with 0.1 M glycine and layered on 5–40% sucrose gradient containing 20 mM Tris–HCl (pH 7.5), 100 mM NaCl and 1 mM MgCl2. After ultracentrifugation at 39 000 rpm for 4.5 h, the gradient sample was fractionated by ISCO gradient fractionator. The top nine fractions were precipitated and analyzed by immunoblotting, together with an in-put control and purified recombinant eIF5 as references.
Polysome profiling of the HeLa cells
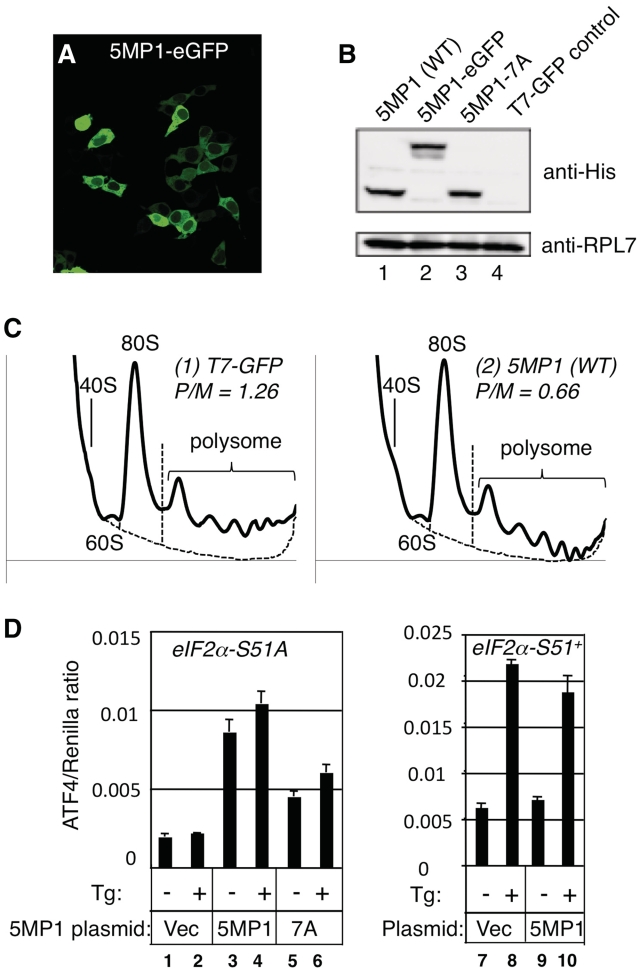
HeLa cells grown in 6-cm petri dish containing 2 ml complete Dulbecco’s modified Eagle’s medium (D-MEM, GIBCO) supplemented with 10% fetal bovine serum (GIBCO) were transfected with indicated plasmids using PolyFect Transfection Reagent (QIAGEN), and after 24 h, the medium was replaced with 2 ml D-MEM (no serum) and the cells were incubated for additional 24 h. Then FBS (GIBCO) was added to 10% and the cells were collected for polysome profiling after 20 min. The collected cells (4 × 106 cells) were lysed in 500 µl lyse buffer (20 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.25% NP-40, 100 µg/ml cycloheximide, 100 U/ml RNase inhibitor, protease inhibitor cocktail) by passing through a sterile syringe with a 26 or 27 gauge needle several times. Then the lysate was cleared by a brief centrifugation (10 000g, 5 min) to remove non-ribosomal nuclear components, which otherwise increase the base-line of the profile towards the bottom of the gradient. Subsequently, 400 µl of the cleared lysate was layered on 5 ml 10–45% sucrose gradient (20 mM HEPES, 100 mM KCl, 10 mM MgCl2) in 151–513 tubes and fractionated by centrifugation at 40 000 rpm for 1 h at 4°C in a Beckman MLS-50 rotor. The gradient was analyzed by the Biocomp Piston Gradient FractionatorTM. The reminder of the lysate was used for checking the total protein yield by the Lowry method and immunoblotting with anti-RPL7 antibodies. As shown by anti-RPL7 immunoblot in Figure 4B, the total ribosome content of the lysate was not altered significantly by expression of 5MP1 under the conditions examined. We also confirmed by immunoblotting with these antibodies that the above-mentioned treatment of clearing the lysate does not affect the amount of ribosomes in the lysate and that the area shown by bracket in Figure 4C covers the entire polysome. Thus, the lysate clearing is essential in evaluating polysome abundance in mammalian cultured cells by densitometry under our experimental conditions.
Figure 4.
5MP1/BZW2 controls general and ATF4-specific translation in mammalian cells (A) Fluorescence emitted from 293 T cells transfected with pCDNA-5MP1-GFP. (B) HeLa cells were transfected with plasmids indicated across the top and subjected for immunoblotting with antibodies indicated to the right. Anti-RPL7 was used as loading control. (C) Polysome profiles of serum-activated HeLa cells transfected with pCDNA-5MP1 (panel 2) or pT7-GFP control (panel 1) were monitored as described (29) with the P/M ratio presented on top right. Dotted line indicates an arbitrary base-line used to calculate P/M ratio. Identical base-line and scale in y axis were set for each set of experiments for accurate comparison. (D) Effect of 5MP1/BZW2 on ATF4 expression. Wild-type (eIF2α-S51+) MEF (Columns 7–10) or MEF homozygous for eIF2α-S51A (Columns 1–6) were triple-transfected with ATF4-luc, Renilla luciferase plasmind, and an empty vector (Vec), pCDNA-5MP1 (WT) or −7A (7A), treated with (+) or without (−) 0.1 µM Tg, and assayed for luciferase expression. Graphs in (D) show the ratio of expression from ATF4-luc to that from the Renilla luc with bars indicating SD (n = 3). These are a typical result of reproduced experiments.
ATF4 expression assay in MEF cell lines
Details of ATF4 expression assay were reported previously (26). Briefly, MEF cells homozygous for eIF2α-S51A mutation obtained from Randall Kaufman (University of Michigan, Ann Arbor) were immortalized by infection of a recombinant retrovirus expressing simian virus 40 large T antigen. MEF cells were grown in 24-well plates in D-MEM (BioWhittaker) supplemented with 10% FBS, 2 mM glutamine, 1 mM nonessential amino acids, 100 units of penicillin per ml and 100 µg of streptomycin per milliliter. Plasmid transfections were performed by using the MEF cells at 40% confluency and the FuGENE tranfection reagent (Roche Applied Science). Cotransfections were carried out in triplicate by using the pCDNA plasmids, the TK-ATF4-Luc fusion plasmid (p759) and a Renilla luciferase plasmid (p851) serving as an internal control (Promega). After transfection (40 h), MEF cells were treated with thapisgargin (Tg) at 0.1 µM, for 6 h or with no ER stress. Dual luciferase assays were carried out as described by the Promega instruction manual (cat #E1960). Values shown in Figure 4D are a measure of a ratio of firefly versus Renilla luciferase units (relative light units, RLU), and represent the mean values of three independent transfections. We reproducibly obtained similar levels of Renilla LUC activity in wild-type and eIF2α-S51A MEFs, unless translation is strongly perturbed e.g. by high dose of Tg. Thus, the transfection efficiency is stable under our experimental conditions. At least two sets of independent experiments were performed to confirm reproducibility.
Fluorescent microscopy
Live yeast cells were mounted onto 0.5% poly-l-lysine–coated slides and visualized on a Leica SP5 confocal microscope with a 63 × 0.6–1.40 NA Plan Apochromat oil objective (Leica). Images were acquired using Application suite 1.6.3 (Leica). For densitometric analysis, a merged ~80 Z-series projection image was taken and analyzed using NIH ImageJ software. For data analysis, we took Z-projection images of cells from at least three independent mid-log cultures that had been grown in SCGal-ura.
RESULTS
5MP1 interacts with human eIF2 and eIF3 in vitro
We first wished to test if the human 5MP1 protein binds to eIF2. For this purpose, we incubated eIF2 purified from HeLa cells (Figure 2A, lane ii) with glutathione resin pre-adsorbed with GST-5MP1 fusion protein, or with GST alone. Associated proteins were then analyzed by immunoblotting with anti-eIF2α antibodies. eIF2α was detected in the complex with GST-5MP1 (Figure 2A, lane 3, top gel); thus, GST-5MP1 interacts with human eIF2. To test if the 5MP1 interaction with eIF2 depends on AA-boxes that characterize the W2 domain (Figure 1B), we introduced 7 alanine-substitutions to the AA-box 2 of GST-5MP1. This mutation, here called 5MP1-7 A, reduced the interaction between GST-5MP1 and eIF2 by >2-fold (P = 0.027, n = 4) (Figure 2A, lane 4; quantified in the graph on top). This shows that AA-box 2 of 5MP1 contributes to the eIF2 binding site similar to the AA-box motifs in eIF5, eIF2Bε and Kra.
Because eIF5, eIF2Bε and Kra interact with eIF2 via the eIF2β subunit (8,19,27), we also examined if 5MP1 binds directly to eIF2β. As shown in Figure 2B (top gel) purified FLAG-tagged 5MP1 (FL-5MP1) bound specifically to GST-fused human eIF2β (GST-eIF2β), but not to GST-fused human eIF2α (lanes 2–4). Again, the 7A mutation in 5MP1 reduced its interaction with GST-eIF2β by 2-fold (P = 0.002, n = 6) (Figure 2B, lower gel and graph). Thus, 5MP1 interacts with eIF2β at least partially via its AA-box 2 motif, suggesting eIF2β represents a major site of interaction between 5MP1 and eIF2.
Since the observed interaction between 5MP1 and eIF2β proceeds via the same AA-box 2 motif as eIF5 uses to bind eIF2β, we hypothesize that 5MP1 and eIF5 may compete with each other for binding to eIF2β. To test this idea directly in vitro, we incubated GST-eIF2β with 5MP1 and eIF5, alone or in combination. As shown in Figure 2C (middle gel), human eIF5 outcompeted the binding of FL-5MP1 to GST-eIF2β (compare lanes 6 and 8). Interestingly, the amount of human eIF5 bound to GST-eIF2β was also reduced by the presence of FL-5MP1 (Figure 2C, bottom gel, lane 8), even though eight times more eIF5 than FL-5MP1 was present. This suggests that interactions of eIF2β with eIF5 and 5MP1 are mutually exclusive, and 5MP1 may act as a competitive inhibitor of eIF5.
The eIF5 W2 domain also interacts with eIF3. Therefore, we next examined whether 5MP1 could also bind to purified eIF3. The eIF3 preparation used here contains all 13 subunits including eIF3e subunit, which was used for immunodetection of eIF3 in this study. As shown by immunoblotting with anti-eIF3e, eIF3 specifically co-precipitated with GST-5MP1 but not with GST alone (Figure 2D, lanes 1 and 2). The absence of anti-eIF3e signals with GST-5MP1 or -5MP1-7A alone confirms the authenticity of the eIF3 subunit detected here (lanes 4 and 5). Importantly, this interaction was not compromised by the AA-box 2 mutation in the 5MP1-7A protein (lanes 2 and 3, see graph to the right for quantification), indicating that this mutation specifically affects the interaction with eIF2. These results together indicate that 5MP1 can interact with both eIF2 and eIF3, similar to the eIF5 W2 domain.
5MP1 inhibits translation in RRL
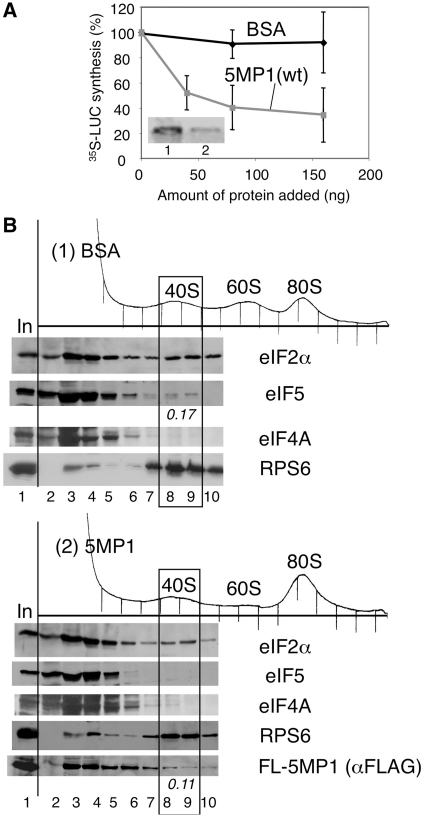
Next, we examined the effect of 5MP1 on protein synthesis using a cell-free RRL translation assay. As shown in Figure 3A (inset), purified FL-5MP1 repressed 35S-luciferase synthesis in RRL programmed with capped luciferase (LUC) mRNA (lane 2), while the addition of equivalent amount of BSA did not (lane 1). The titration experiments in Figure 3A indicated that ~40 ng or 0.8 pmol of FL-5MP1 is required for nearly full inhibition: Because the RRL used here contains 7 and 3 pmol of eIF2 and eIF5, respectively (data not shown), translational inhibition by 5MP1 is quite efficient.
Figure 3.
5MP1/BZW2 inhibits cell-free translation in RRL. (A) Cell-free translation of 35S-luciferase (LUC) was performed in the presence of different amounts of indicated proteins in 10 µl RRL programmed with 1.25 ng capped LUC mRNA. The reaction was run at 30°C for 1 h in the presence of [35S]-methionine. Inset shows autoradiography indicating the amount of LUC production. Graph shows the plot of relative 35S-LUC synthesis against the amount of proteins added from at least three independent experiments, with bars indicating SD. (B) RRL programmed with cap-LUC mRNA was supplemented with 160 ng BSA (panel 1) or FL-5MP1 (panel 2) and incubated for 1 h. The samples were fixed with HCHO and resolved by sucrose gradient-velocity sedimentation. Fractions were analyzed by immunoblotting with antibodies indicated to the right. In, 10% in-put RRL used. The amounts of eIF5 or FL-5MP1 (in picomoles) associated with the 40 S subunit are listed to the bottom of the gel.
To address the mechanisms by which 5MP1 can inhibit translation in RRL, we fractionated RRL programmed for luciferase mRNA translation with or without FL-5MP1 by sucrose gradient–velocity sedimentation. As shown in Figure 3B, anti-eIF2α immunoblot of gradient fractions of control RRL revealed that eIF2 was across the gradient; however, there was a specific peak indicating eIF2 comigration with the 40S subunit (panel 1). The amount of eIF2 comigrating with the 40S was not significantly affected by 5MP1 addition to RRL (Figure 3B, panels 1 and 2), a trend that was confirmed among many repeated experiments. Importantly, the addition of FL-5MP1 reduced eIF5 binding to the ribosome to levels that were not detectable (Figure 3B, panels 1 and 2). The eIF5 comigration with the 40S subunit in the control RRL appears to reflect specific interaction, as the 40S fractions do not contain mRNA helicase eIF4A, which does not bind stably to the 40S subunit (Figure 3B, panels 1 and 2). Instead, we found that FL-5MP1 appeared to comigrate with the 40S subunit (Figure 3B, panel 2, bottom panel) at a level equivalent to that of eIF5 in the control RRL (panel 1). These results, combined with the competition experiments shown in Figure 2D, suggest that 5MP1 may competitively inhibit eIF5 recruitment to the 40S subunit.
Human 5MP1 is capable of repressing translation and modulating ATF4 expression in mammalian cells
To express 5MP1 in mammalian cells, we cloned 5MP1 and its GFP-fusion and 7A mutant derivatives under a CMV promoter (pCDNA-5MP1 and derivatives in Supplementary Table S1). Transfection of 5MP1-GFP led to cytoplasmic GFP localization as judged by fluorescent microscopy (Figure 4A), in agreement with the cytoplasmic location of 5MP2/BZW1 reported previously (18). pCDNA-5MP1 and its derivatives express a similar level of 5MP1 in transfected HeLa cells (Figure 4B), and immunoblotting indicated that the expressed 5MP1 proteins were found specifically in the cytoplasmic, but not in the nuclear fractions of HeLa cell extracts (data not shown). The transfection of pCDNA-5MP1 expressing 5MP1 alone led to a small reduction in the polysome content of the cells, especially the highly sedimenting polysomes (Figure 4C, compare panels 1 and 2). Given that 5MP1 is cytoplasmic and its expression impacts upon polysome profiles, the data are consistent with the idea that 5MP1 modulates translation in mammalian cells.
The relatively weak effect of enhanced 5MP1 expression upon general protein synthesis (Figure 4C) led us to address whether elevated 5MP1 changes the translation of specific mRNAs. ATF4 mRNA is translationally enhanced by phosphorylation of eIF2 in response endoplasmic reticulum (ER) stress, a process that can be triggered by addition of the well-characterized ER stress agent thapsigargin (Tg) (28). The regulation of ATF4 translation depends on its upstream open reading frames (uORFs). In short, ATF4 translation requires prior translation of uORF1, resumed 40S scanning, bypassing of uORF2 and re-initiation at ATF4 AUG, whereas re-initiation at uORF2 prevents subsequent translation of ATF4 (28). This choice is modulated by eIF2 phosphorylation, which transforms eIF2 into a competitive inhibitor of eIF2B, thereby reducing the cellular eIF2-GTP/Met-tRNAiMet (TC) level and delaying TC binding to the re-initiating 40S subunit. This allows uORF2 to be bypassed during times of stress and instead, promotes re-initiation at ATF4 located downstream. Thus, if 5MP1 modulates TC recruitment or abundance, this may alter re-initiation and affect ATF4 expression in the absence of eIF2 phosphorylation.
ATF4 expression was measured by firefly luciferase activity from a transiently transfected ATF-LUC plasmid and was normalized by Renilla luciferase activity expressed from a second co-transfected plasmid. In mouse embryonic fibroblasts (MEF) containing an alanine mutation in the serine-51 phosphorylation site of eIF2α (eIF2α-S51A) there is no induction of ATF4-regulated luciferase activity in response to ER stress, emphasizing the specificity of this translational control (Figure 4D, Columns 1 and 2). As a control, the same Tg treatment induced ATF4 expression >3-fold in MEF with wild-type eIF2α (eIF2α-S51+) (Figure 4D, Columns 7 and 8). Expression of 5MP1 significantly increased the level of ATF4 translation in the eIF2α-S51A cells independent of stress treatment (Figure 4D, Columns 3 and 4). However, 5MP1 expression did not increase ATF4 expression in the eIF2α-S51+ cells (Figure 4D, Columns 9 and 10). Interestingly, the increase in ATF4 expression observed here with enhanced 5MP1 expression was alleviated partially by the 7A mutation, and was observed to be independent of Tg treatment (Figure 4D, lanes 5 and 6). These results suggest that the alteration in 5MP1 interaction via the AA-box 2, perhaps with eIF2, can modulate ATF4 translation by affecting TC activity or recruitment in mammalian cells.
Human 5MP1 can inhibit GDP dissociation from yeast eIF2
The data described thus far are consistent with the idea that 5MP1 can modulate translation initiation by binding to eIF2 and competitively excluding eIF5 through its W2 HEAT domain. This is consistent with a major function of the W2-CTD, which is to bridge the interaction between eIF2 and eIF3, hence mediating the formation of the MFC also containing eIF1 (11). Because the eIF5 GAP function requires the NTD that is not conserved in 5MP/BZW (30) it appeared highly unlikely that 5MP1 was a functional GAP, so this activity was not assessed. However, the W2-CTD of yeast eIF5, together with its juxtaposed linker region (LR), has also been described to inhibit GDP dissociation from eIF2: GDI activity (4). This activity opposes the guanine nucleotide exchange activity of eIF2B by stabilizing GDP binding. While our data so far suggest that 5MP is a competitive inhibitor of eIF5 function in promoting translation, because theW2-type HEAT domains (W2-CTD) of eIF2Bε, p97/NAT1/DAP5 and eIF5 are structurally similar to that of 5MP (Figure 1) and all interact with eIF2, it was also possible that 5MP1 is a mimic of eIF2Bε or acts to functionally replace this factor. Indeed, the W2-CTD of eIF2Bε is the catalytic domain that can function alone in a direct GEF assay (10). Therefore by assessing whether 5MP1 affects GDP dissociation, we were able to directly assess if it mimics either GEF (accelerates GDP release) or GDI (retards GDP release) functions of eIF2Bε and eIF5, respectively. For this purpose, we purified GST-5MP1 and its 7A mutant derivative and examined their effect on GDP retention by yeast eIF2 in a standard assay examining the dissociation of [3H]GDP from purified eIF2. As shown in Figure 5A, GDP dissociation was modestly, but significantly, reduced by wild-type 5MP1, but not by 5MP1-7A. Since GDP dissociation was not increased, as has been observed by adding eIF2Bε (10), this result indicates that 5MP1 is not a functional mimic of eIF2Bε. The retardation of GDP dissociation by 5MP1 was weaker than by GST-yeast eIF5 (yeIF5) (Figure 5A). Thus, 5MP1 appears to act as a mimic/competitor of the GDI activity, in addition to its macromolecular mimicry and competitive binding to eIF2 and antagonizing pre-initiation factor complex formation. These activities are potentially opposed to each other, as weak GDI function with eIF2-GDP could promote nucleotide exchange and enhance eIF2 recycling. However binding to eIF2 TC may prevent its productive recruitment to 40S ribosomes and reduce translation initiation. We therefore decided to examine effects of 5MP1 expression in yeast, as this has been a sensitive in vivo system for examining translation and its control.
Figure 5.
The GDI activity test of 5MP1/BZW2 and its interaction with eIFs in yeast S. cerevisiae. (A) GDI assay. Off-rates for eIF2-GDP was measured in the presence of indicated proteins fused to GST (4). C, Buffer control. Bars and line indicate mean and SD, respectively. Gray filled bars, not significantly different from buffer alone; unfilled bars are significant with P-value and the number of experiments (n) shown. (B) Immunoprecipitation. An amount of 200 µg of WCE prepared from H2557 transformants carrying an empty vector (Vec) or pEMBL-FL-5MP1 (FL-5MP1) which had been grown in SCGal-ura was used for anti-FLAG IP, and entire IP fractions (P), 20% in-put (In) and 10% supernatant (S) fractions were analyzed by immunoblotting with anti-FLAG (top) and anti-yeIF2α (SUI2) (bottom) antibodies. (C and D) Affinity-purification of FL-5MP1 complexes in yeast. Complexes containing the FLAG-tagged proteins listed across the top were purified from H2557 transformants carrying pEMBL-FL-5MP1 (lane 1) or a vector control (lane 2) and KAY50 (TIF5-FL) (lane 3), as described (11). The former two strains were grown in SCGal-ura medium and the latter was grown in YPD. Portions of the eluates were analyzed by Coomassie Blue staining (C) and immunoblotting with antibodies raised against yeast eIFs listed to the right (D). In (C), arrowheads indicate the positions of eIF subunit identified. Gray labels, eIF2 subunits; Iron labels, eIF3 subunits; Black labels, yeIF5-FL (5-FL) or FL-5MP1. (E) Table summarizes the relative molar ratio of co-purifying factors, compared to the molarity of eIF2 (Rows 2 and 3) as defined as 1. These ratios were determined in reference to purified external standards as described previously (3,21). (F) Models depicting the interaction of FLAG-yeIF5 (panel 1, oval labeled eIF5) and FLAG-human 5MP1 (panel 2, gray oval) with yeast factors (open ovals). Direct contact, strong interaction; arrow, intermediate interaction; dotted arrow, weak interaction.
5MP1 interacts with eIF2 and weakly with eIF3 in Saccharomyces cerevisiae
We expressed 5MP1 in yeast to take advantage of various assays established for analyzing eIF2 regulation in translational control. We first examined its interaction with the yeast translation initiation factor (yeIF) components in vivo. When overexpressed from the GAL promoter, FLAG-tagged 5MP1 (FL-5MP1) expressed ~25 times more abundantly than endogenous eIF5 or eIF2 in galactose medium. Anti-FLAG immunoprecipitation (IP) indicated that ~25% of total eIF2 is specifically associated with the galactose-induced FL-5MP1 (Figure 5B). We previously reported that ~60% of yeIF5, expressed in single-copy, is associated with total eIF2 (21). Because the level of 5MP1/yeIF2 complex was lower than that of yeIF5/yeIF2 complex even when 5MP1 was overexpressed, the interaction of 5MP1 with yeast eIF2 is likely to be weaker than that between yeast eIF5 and eIF2 in vivo.
To investigate the interaction of other MFC components with FL-5MP1, we analyzed the eIF constituents of FL-5MP1 complexes that were purified by anti-FLAG affinity chromatography (Figure 5C, lane 1). As shown in Figure 5C, Coomassie blue staining of the gel indicated that the FL-yeIF5 immunopurified sample contains yeast eIF3, eIF5 and eIF2 (lane 3), representing part of the MFC, as reported previously (3,11,21). The FL-5MP1 immunopurified sample contained three proteins, which appeared to comigrate with the three eIF2 subunits (? in lane 1) in quantities roughly equivalent to those found in the FL-yeIF5 sample (gray arrowheads in lane 3). The identity of six of the MFC polypeptides was confirmed by immunoblotting (Figure 5D), and the relative molar abundances of eIF1, eIF2, eIF3 and eIF5 (compared to eIF2 subunits) were determined using purified recombinant proteins as external standards and presented in Figure 5E. These data showed that the FL-5MP1 (lane 1) and FL-yeIF5 (lane 3) immunopurified fractions contained similar amounts of co-purifying yeIF2 (Rows 2 and 3). Because much more FL-5MP1 was present in lane 1 than FL-yeIF5 found in lane 3 (Figure 5D and E, Row 1), this also highlighted that the FL-5MP1 interaction with yeIF2 is likely to be weaker than yeIF5/yeIF2 interaction. Importantly, yeIF5 was not detected at all in the FL-5MP1 complex (Figure 5E, Row 5) providing in vivo evidence that 5MP1 and eIF5 form mutually exclusive complexes with eIF2 (Figure 5F). This is consistent with each factor competing for the same interaction site on eIF2. In addition, a small but significant amount of yeIF3 was found associated with immunopurified FL-5MP1 (Figure 5D and E, Row 4), consistent with the observed 5MP1 interaction with human eIF3 (Figure 2D). However, no yeIF1 was associated with FL-5MP1, indicating that the 5MP1 cannot mimic the role of eIF5 in MFC formation (Figure 5D and E, Row 6).
5MP1 decreases eIF2 localization to eIF2B-associated bodies in yeast
Despite the substantial interaction observed between 5MP1 and yeIF2, the 5MP1 overexpression did not retard the yeast growth rate. To detect any perturbation in translation initiation in vivo, especially one related to eIF2 activity, we took an independent cell biology approach. A significant fraction of yeast mother cells usually harbor a single cytoplasmic body containing eIF2B (termed the eIF2B body) (34). These are distinct from other cytoplasmic foci, e.g. the processing bodies, EGP bodies or stress granules (31–33). Photobleaching experiments have shown that eIF2B is a fixed component of this body, while eIF2 is more dynamic, shuttling through the body. Under conditions that reduce GEF activity (stress or eIF2B mutations); the rate of eIF2 cycling through the body was significantly reduced, suggesting that the eIF2B bodies represent a concentrated site of nucleotide exchange (34). We predicted that if 5MP1 interacts with yeIF2 and inhibits translation, this would decrease the shuttling of yeIF2 through the eIF2B bodies. To test this idea, we introduced the GAL-5MP1 plasmid, or hc yeIF5 plasmid as a control, to strains bearing GFP-tagged yeIF2Bγ or GFP-tagged yeIF2α. The introduction of neither plasmid affected the frequency or size of the eIF2B body in the eIF2B-GFP strain (data not shown). In contrast, when introduced to YMK883 expressing GFP-eIF2α (Figure 6A inset), we observed that both the GAL-5MP1 and hc yeIF5 plasmids reduced the proportion of cells eliciting medium to strong GFP-yeIF2 signals present in the bodies (Figure 6A, graph, Rows 3 and 4; Figure 6B, Row 3), suggesting that eIF2 activity is reduced by the overexpression of 5MP1 or yeIF5. This thesis is supported by additional statistical analyses of the microscopic data; we measured the proportion of the GFP-yeIF2 signal intensity localized in the eIF2B body, compared to total GFP-yeIF2 signal intensity in each cell across an entire population of cells. This value decreased from 9.2% (vector control) to 2.7 and 4.0% by overexpression of yeIF5 and 5MP1, respectively (Figure 6B, Row 4). Importantly, the reduced localization was restored to normal by introduction of the AA-box 2 mutation to the GAL-5MP1 construct (Figure 6A, column d; Figure 6B, Rows 3 and 4). This result is the opposite of the impact of conditions that lead to induced or constitutive eIF2α phosphorylation, under which increased association of GFP-yeIF2 with eIF2B bodies was observed (34). Thus, the result shown in Figure 6B suggests that overproduced 5MP1 reduces the rate of shuttling of yeIF2 through the eIF2B body, by inhibiting yeIF2 recruitment to eIF2B bodies, directly or indirectly, in a manner that is dependent upon the 5MP1 AA-box 2. This observation is consistent with excess yeIF5 or 5MP1 exerting an overall negative effect on translation initiation rates in cells. We explored this idea next using a genetic strategy.
Figure 6.
Effect of 5MP1 on GFP-eIF2 association with eIF2B bodies. (A) Inset shows a typical image of yeast YMK883 cell expressing GFP-eIF2, with its localization in large cytoplasmic eIF2B bodies (34). YMK883 transformants carrying an empty vector (Vec, column a), YEpL-TIF5-FL (hc yeIF5, column b), pEMBL-FL-5MP1 (5MP1, column c) and pEMBL-FL-5MP1-7A (5MP1-7A, column d) (Supplementary Table S1) were grown in SCGal-ura medium to a mid-log phase. Then, fluorescent images of the cells were taken under a confocal microscope, and quantified for the amount of GFP-eIF2 signal localized in the bodies compared to one in the whole cell. Graph indicates the proportions of the groups of cells (in each column) with no, weak, medium and strong GFP-eIF2 signals in the bodies, as defined in each row of the graph. (B) Table summarizing the plasmid used (Row 1), the number of transformed cells measured (Row 2), percentage of cells with med to strong GFP-eIF2 signals in the body (the sum of percentage values in Rows 3 and 4 of graph in A) (Row 3), and percentage of GFP-eIF2 signals localized in the bodies, averaged for the entire population of the cells carrying the same plasmid (Row 4), together with its standard error of the mean (SD/n1/2; Row 5).
5MP1 increases the yeast general control response independently of Gcn2p activation
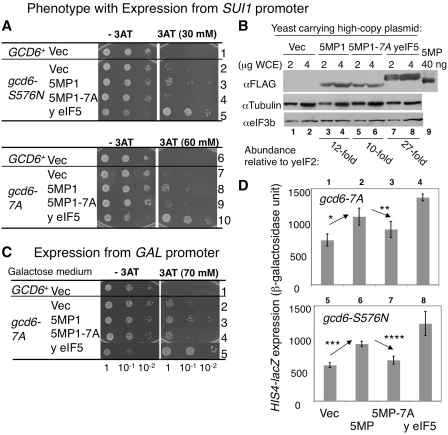
We reasoned that if 5MP1 directly binds yeIF2, then it might influence GCN4 translation, which is activated by alterations in TC levels and recruitment in a mechanism similar to that described for ATF4 above. For example, 5MP1 might trigger preferential translation of GCN4, due to the depletion of eIF2 from the TC (35). This would be entirely consistent with the effect of 5MP1 on ATF4 translation in the absence of eIF2 phosphorylation (Figure 4D). Phosphorylation of eIF2 in response to amino acid starvation in yeast, via the Gcn2p eIF2α kinase, typically lowers TC abundance and delays the re-initiation process that otherwise occurs on an uORF in the GCN4 leader, and instead increases re-initiation at the GCN4 ORF. We overexpressed 5MP1 to investigate whether the increased eIF2 binding could deplete the TC leading to enhance GCN4 translation in a Gcn2p-independent manner. In practice, this entailed the measurement of both cell growth and the expression of a Gcn4p transcriptional target (HIS4) during histidine starvation conditions induced by the inhibitor 3-aminotriazole (3AT). As shown in Figure 7A, Row 1, the growth of a gcn2Δ strain is sensitive to 3AT because the absence of eIF2 phosphorylation prevents the preferential translation of GCN4 and the subsequent transcriptional induction of those genes required to alleviate the nutrient starvation.
Figure 7.
5MP1 overexpression increases GCN4 translation in gcn2Δ gcd6 backgrounds. (A) Yeast dilution assays. Transformants of KAY16 (Row 1), GP3578 (Rows 2–5), KAY33 (Row 6) and KAY34 (Rows 7–10) carrying an empty vector (Vec), YEpL-FL-5MP1 (5MP1 in Row 3), YEpL-FL-5MP1-7A (5MP1-7A in Row 4), YEpTIF5-FL (yeIF5 in Row 5), YEpU-FL-5MP1 (5MP1 in Row 8), YEpU-FL-5MP1-7A (5MP1-7A in Row 9), and YEpU-TIF5 (yeIF5 in Row 10) were grown in SC-His-Leu or SC-His-Ura medium. Fixed amounts (A600 = 0.15) of the culture and its 10-fold serial dilutions were spotted onto the agar plates of the same medium without or with indicated amounts of 3AT and incubated at 30°C for 2 and 6 days, respectively. (B) Expression check. Indicated amounts of WCE prepared from KAY16 transformants carrying the plasmids used in (A) Rows 1–5 were subjected for immunoblotting with antibodies listed to the left. Bottom; molar amounts of FLAG-tagged proteins present in WCE were determined using 40 ng purified FL-5MP1 (lane 9) as standard, and compared to known amount of yeIF2 present in WCE (21). (C) Cultures of KAY33 (Row 1) or KAY34 (Rows 2–5) transformants carrying an empty vector (Vec), pEMBL-FL-5MP1 (5MP1), pEMGL-FL-5MP1-7A (5MP1-7A) and YEpU-TIF5 (yeIF5) were spotted and incubated as in (A), except that they were grown in SCGal-His-Ura medium. (D) Expression from chromosomally integrated HIS4-lacZ in transformants used in Panel A, Rows 10–13 and 5–8 were presented by β-galactosidase units. P-values for the differences indicated by arrows are: *, 0.004; **, 0.035; *** and ****, <0.00001.
We envisaged that 5MP1 overexpression might mimic the effect of eIF2 phosphorylation to confer 3AT resistance to the gcn2Δ strain. However, 5MP1 expression from the constitutive SUI1 promoter or GAL promoter did not confer 3AT resistance to gcn2Δ strains (data not shown), although the abundance of 5MP1 expressed from SUI1 and GAL promoters was ~12 (Figure 7B) and ~25-fold (see above) higher than that of yeIF2, respectively. We reasoned that eIF2B mutations known to limit TC levels could sensitize gcn2Δ strains to a reduction in eIF2 activities, which might be caused by 5MP1 expression. Indeed, alteration of either S576 or the AA-box 2 of the eIF2Bε catalytic domain (in the gcd6-S576N gcn2Δ and gcd6-7A gcn2Δ mutant strains, respectively) sensitizes the strains such that 5MP1 expression now increases the 3AT resistance (Figure 7A, Rows 3 and 8; Figure 7C, Row 3). It should be noted that these gcd6 mutant strains are slightly resistant to high doses of 3AT (Figure 7A, Rows 2 and 7; Figure 7C, Row 2) (8,36) and that expression of 5MP1 provides further increase in this 3AT resistance. This growth of 5MP1-expressing strains suggests a modulatory role for 5MP1 in the regulation of translation initiation. However, its effect on yeast translation is only kinetic, as examined below, but may not be clearly physiological.
The overexpression of yeIF5 conferred much stronger growth in the presence of 3AT in the gcn2Δ gcd6 strains than did 5MP1 (Figure 7A, Rows 5 and 10; Figure 7C, Row 5). This result may reflect either the fact that yeIF5 is expressed at a higher level than 5MP1 (Figure 7B), or that the yeIF5/yeIF2 interaction is stronger than 5MP1/yeIF2 interaction (Figure 2), or both. The enhanced resistance to histidine starvation shown by both excess eIF5 and 5MP1 was accompanied by corresponding increase in HIS4-lacZ expression, whose transcription is activated by Gcn4p (Figure 7D). Furthermore, both growth and HIS4-lacZ effects of 5MP1 are alleviated by the 5MP1 AA-box 2 mutation 7A (Figure 7A, C and D) without decreasing 5MP1 abundance (Figure 7B). Overall, these results suggest that 5MP1 has the capacity to impact upon GCN4 translation in a manner dependent upon its AA-box 2. Given the exquisite sensitivity of the GCN4 system to changes in TC recruitment to the ribosome, these results further suggest that 5MP1 impacts upon the level or recruitment of TC to the pre-initiation complex.
5MP1 promotes eIF2 TC formation and inhibits MFC formation in yeast
A key question arising from the experiments described above is whether 5MP1 impacts upon the levels of complexes such as the TC or the multi-factor complex. To assess this, we quantified the abundance of yeIF2-containing complexes. For this purpose, the HA-tagged 5MP1 (HA-5MP1) was overexpressed in a strain encoding yeIF2β-FL (encoded by SUI3-FL), and the initiation components which co-immunoprecipitated with FLAG-eIF2 were quantitatively analyzed (21). Immunoblotting indicated that HA-5MP1 was expressed at approximately four to five times higher levels than yeIF5 (data not shown). As shown in Figure 8A, the expressed HA-5MP1 significantly increased TC levels (P = 0.019, n = 7), as determined by the amount of tRNAiMet immunoprecipitated (see graph 1 to the right). The reason for this increase will be discussed below. Importantly, this TC increase was accompanied by a decrease in yeIF5 and yeIF3b that are co-associated with FL-yeIF2 (Figure 8A, graph 2 and 3, respectively, P < 0.05, n = 3). This decrease in the interaction of both eIF5 and eIF3 with FL-yeIF2 implicates HA-5MP1 in the inhibition of MFC formation.
Figure 8.
Effect of 5MP1 on the abundance of eIF complexes in yeast. (A) Quantitative anti-FLAG IP. An amount of 1 mg of WCE prepared from KAY107 (TIF34-3HA) transformant carrying an empty vector (C), KAY128 (SUI3-FL) transformants carrying an empty vector (V) or pEMBL-HA-5MP1 (5MP) was used for anti-FLAG IP and 80% (top gel) and 20% (bottom gels) of the precipitated fractions (lanes under αFLAG IP) were analyzed by northern and western blotting, respectively, with 2% in-put amount (lanes under In-put), as described (21). HA or FLAG-tagged alleles present in the transformants are indicated across the top. Graphs to the right summarize the relative amount of indicated components associated with FLAG-eIF2, after correction by the amount of eIF2α precipitated. (B) Co-IP with HA-eIF3. An amount of 200 µg of WCE prepared from KAY127 (SUI3-FL) transformant carrying an empty vector (C), KAY113 (TIF34-3HA) transformants carrying an empty vector (V), pEMBL-FL-5MP1 (HP), or pEMBL-FL-5MP-7A (7A) was used for anti-HA IP, and entire IP fractions and 10% in-put amounts were analyzed by immunoblotting with antibodies raised against HA-epitope (top) and yeast eIF2α (bottom). (C) Possible models to explain increased TC abundance by 5MP1 in yeast. Numbers in circles refer to eIFs (e.g. 1, eIF1). Gray squares, 5MP1. Model 1. 5MP1/TC interaction slows down MFC formation (dotted arrow). The accumulation of 5MP1/TC complex contributes to increase in overall TC abundance (both free TC and TC/5MP1). Model 2. 5MP1/eIF2-GDP interaction antagonizes the GDI function (stopped bar to GDP) of eIF5. The steps of GDI and GEF are highlighted with light gray round squares.
To confirm and extend these observations, we next examined the impact of 5MP1 expression on the yeIF3-containing complexes. Hence, we overproduced FL-5MP1 in a strain encoding the HA-tagged yeIF3i subunit. As shown in Figure 8B, less yeIF2α co-precipitated with HA-yeIF3 in the presence of FL-5MP1 (lane 7) than in its absence (lane 6). Because eIF2/eIF3 association depends on bridging by eIF5, this is the hallmark of MFC formation. Thus, these results confirm that FL-5MP1 indeed inhibits MFC formation. The more dramatic effect on MFC relative to Figure 8A is likely due to a higher expression level of this FL-5MP1 construct than that of HA-5MP1. The 5MP1-mediated inhibition of MFC formation was alleviated by the AA-box 2 mutation 7A introduced to FL-5MP1 (Figure 8B, lane 8). Since 5MP1 expression did not reduce the interaction of HA-yeIF3 with yeIF5 (Figure 8B), it seems that 5MP1 does not compete with yeIF3 binding to yeIF5 in yeast, consistent with the weak interaction observed between FL-5MP1 and yeIF3 (Figure 5D and E). These observations suggest that 5MP1 inhibits TC binding to the 40S subunit, primarily by sequestering TC away from yeIF5 and yeIF3, which otherwise promote TC binding to the 40S subunit (Figure 8C, Model 1). This would mean that an inhibition of TC recruitment would explain why 5MP1 enhanced GCN4 translation in the gcn2Δ gcd6 mutant strains (Figure 7).
DISCUSSION
In this article, we have assessed the function of the human MA3-W2 HEAT repeat protein previously called BZW2 and here renamed as 5MP1. We have shown that the human 5MP1 directly interacts with eIF2 via the β subunit in vitro (Figure 2). Together with the finding that the 5MP1 Drosophila homolog, Kra, interacts with eIF2β, this result strengthens the idea that translation factors and regulators with the W2-type HEAT domains (i.e. eIF5, eIF2Bε and p97/NAT1/DAP5) can generally interact with eIF2. We also showed that 5MP can modulate overall translation (Figure 4C) and translation of a luciferase reporter gene in RRL (Figure 3A). These observations suggest that 5MP binding to eIF2 (and perhaps eIF3) can modulate the efficiency of protein synthesis.
How does 5MP regulate protein synthesis?
5MP interacts directly with eIF2 or eIF3 (Figure 2), and therefore could competitively inhibit or even replace the functional interactions of any of the W2-domain containing proteins such as eIF5 or eIF2Bε. Our in vitro studies directly examining the effect of 5MP on GDP binding to eIF2 showed that the human 5MP1 is not a GEF, but instead has weak GDI activity for eIF2 (Figure 5A). GDI activity was recently ascribed to yeast eIF5 as a second function in addition to its GAP activity (4). Therefore eIF5 GAP activity switches eIF2 into an inactive GDP-bound state, and its GDI activity prevents reactivation of eIF2 by the GEF eIF2B. Similarly the observed inhibition of GDP release by 5MP1 (GDI activity) could contribute to locking eIF2 in an inactive state; however, because 5MP1 GDI activity is weak, 5MP1/eIF2-GDP binding may antagonize eIF5 GDI and therefore promote GDP release. Consistent with this idea, we made an interesting observation that 5MP1 overexpression increases TC (eIF2-GTP-Met-tRNAiMet) abundance (Figure 8A). These observations led us to propose one mechanism for 5MP function (Figure 8C, Model 2). In this model, 5MP1 expression would compete directly with eIF5 GDI function, resulting in an increase in TC abundance by preventing eIF5 antagonizing the GEF activity of eIF2B. However an alternative explanation is also possible (see below).
The ability of 5MP1 to interact with both eIF2 and eIF3 (Figure 2A, B and D) strengthened the idea that 5MP1 could function as a competitive inhibitor and structural mimic of eIF5 functions in pre-initiation multifactor complex (MFC) assembly necessary for efficient translation initiation by the standard scanning model. This model is supported by a number of experimental observations. First, 5MP1 can bind eIF2β in competition with eIF5 (Figure 2C). Second, 5MP1 can compromise eIF5 co-migration with 40S ribosomes in the RRL (Figure 3B). Third, 5MP1 complex with yeast eIF2 does not include eIF5 (Figure 5D) and finally it can antagonize the binding of eIF2 TC to eIF3, which is necessary for MFC formation in yeast (Figure 8B). This latter observation provides an alternative explanation for the observed increase in TC levels following 5MP1 expression (Figure 8A). The observations are consistent with a fraction of TC becoming sequestered into a TC/5MP1 complex and being unavailable for MFC assembly. This idea is also shown diagrammatically in Figure 8C (Model 1). Thus, 5MP can act as a competitive inhibitor of eIF5 functions in promoting the assembly of the ribosomal pre-initiation complex, or act to antagonize eIF5 GDI function and thereby act positively to promote GEF function. Either or both of the models together could account for the observed increase in TC. We also acknowledge that the effect of the human 5MP1 on yeast translation is overall weak and not clearly physiological, perhaps owing to the evolutionary distance from the model organism. To take better advantage of the yeast system, it would be attractive to study the effect of 5MP from lower eukaryotes (16), those from fungi (Basidiomycota), in particular.
Is 5MP a part of inhibitory mRNP complexes?
The models proposed in Figure 8C consider the inhibitory effect of 5MP1 on eIF2, as observed in yeast. Because 5MP1 can also interact with eIF3 (Figure 2D), its effect on translation in mammalian cells could be more complex. For example, with a link to eIF3, 5MP1 that lacks the GAP domain might replace eIF5 recruitment to the 40S subunit (Figure 3B) and thereby prevent 60S subunit joining. An alternative, but not mutually exclusive, idea consistent with its partial effect on translation in mammalian cells (Figure 4C) is that effective translational control by this protein requires additional binding proteins in complex with 5MP1, eIF2 and eIF3. In agreement with this idea, evidence was provided previously that the Drosophila homolog Kra binds simultaneously to eIF2 and Shot, a cytoskeletal element. Furthermore, Kra co-expresses with the mRNA-specific binding factors Pumilio and Staufen in certain areas of the central nervous system, suggesting that Kra is a component of multiple inhibitory mRNA/protein (mRNP) complexes (20). Specific expression of Kra in cholinergic local neurons of antennal lobe has been established and a krasavietz enhancer trap line is frequently used to study Drosophila CNS function (37). Together these findings suggest an intriguing possibility that 5MP1 and eIF2 form part of a larger translationally repressed mRNA complex that are undergoing cytoskeleton-mediated intracellular transport. The observed GDI activity of 5MP1 (Figure 5A), if it occurs in vivo, would inhibit GDP dissociation during the transport process in favor of translational inhibition. A 5MP interaction with cytoskeletal elements in humans was suggested previously by the physical interaction of human BZW1/5MP2 with PSTPIP1 (38), which in turn interacts with Wiskott-Aldrich Syndrome Protein (39). The latter two proteins, which regulate F-actin formation, are expressed predominantly in hematopoietic cells, where higher levels of BZW1/5MP2 transcripts are also observed (40). Although work to address these ideas is beyond the scope of the present manuscript, it is one avenue to pursue in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health GM64781, NCRR K-INBRE Pilot Grant P20 RR016475; K-state Terry Johnson Cancer Center (to K.A.); NIDCR intramural grant (to J.A.C.); GM49164 (to R.C.W.); BBSRC grants BB/E002005/1 and BB/H010599/1 (to G.D.P.); and short-term fellowship for his sabbatical to Manchester, UK in Feb-Mar, 2008 (to K.A.). Funding for open access charge: K-state Terry Johnson Cancer Center.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks are due to John Hershey and Masaaki Sokabe for purified human eIF2 and eIF3 and Hans Gross for critical reading of the manuscript. The authors also thank Assen Marinchev and Gota Kawai for advice on 5MP structure, Futoshi Shibasaki for anti-eIF3e, Alan Hinnebusch, Tom Donahue, John McCarthy and Tom Dever for antibodies against yeast eIFs, Miwako Homma for human eIF5 plasmids and members of Ashe and Pavitt labs for frank discussion.
REFERENCES
- 1.Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–128. [Google Scholar]
- 2.Hinnebusch AG, Dever TE, Asano K. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 225–268. [Google Scholar]
- 3.Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006;25:4537–4546. doi: 10.1038/sj.emboj.7601339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jennings MD, Pavitt GD. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature. 2010;465:378–381. doi: 10.1038/nature09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 6.Marintchev A, Edmonds KA, Maritcheva B, Hendrickson E, Oberer M, Suzuki C, Herby B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raught B, Gigras A-C. Signaling to Translation Initiation. In: Mathews MB, Sonenberg N, and Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 369–400. [Google Scholar]
- 8.Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3 and eIF4G. Proc. Natl Acad. Sci. USA. 2005;102:16164–16169. doi: 10.1073/pnas.0507960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez E, Mohammad SS, Pavitt GP. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 2002;21:5292–5301. doi: 10.1093/emboj/cdf515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imataka H, Olsen S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV, Jr, Iwao H, Innerarity TL. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 2000;19:5533–5541. doi: 10.1093/emboj/19.20.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SH, McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cylce proteins. EMBO J. 2006;25:4008–4019. doi: 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberman N, Dym O, Unger T, Albeck S, Peleg Y, Jacobovitch Y, Branzburg A, Eisenstein M, Marash L, Kimchi A. The crystal structure of the C-terminal DAP5/p97 domain sheds light on the molecular basis for its processing by caspase cleavage. J. Mol. Biol. 2008;383:539–548. doi: 10.1016/j.jmb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: Implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- 17.Mitra P, Vaughan PS, Stein JL, Stein GS, van Wijnen AJ. Purification and functional analysis of a novel leucine-zipper/nucleotide-fold protein, BZAP45, stimulating cell cycle regulated histone H4 gene transcription. Biochemistry. 2001;40:10693–10699. doi: 10.1021/bi010529o. [DOI] [PubMed] [Google Scholar]
- 18.Hoya MR, Wahlestedt C, Höög C. A visual intracellular classification strategy for uncharacterized human proteins. Exp. Cell Res. 2000;259:239–246. doi: 10.1006/excr.2000.4948. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Nahm M, Lee M, Kwon M, Kim E, Zadeh AD, Cao H, Kim H-J, Lee ZE, Oh SB, et al. The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development. 2007;134:1767–1777. doi: 10.1242/dev.02842. [DOI] [PubMed] [Google Scholar]
- 20.Dubnau J, Chiang A-S, Grady L, Barditch J, Gossveiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 21.Singh CR, Udagawa T, Lee B, Wassink S, He H, Yamamoto Y, Anderson JT, Pavitt GD, Asano K. Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J. Mol. Biol. 2007;370:315–330. doi: 10.1016/j.jmb.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammad-Qureshi SS, Haddad R, Palmer KS, Richardson JP, Pavitt GD. Purification of FLAG-tagged eukaryotic initiation factor 2B complexes, subcomplexes, and fragments from Saccharomyces cerevisiae. Methods Enzymol. 2007;431:1–13. doi: 10.1016/S0076-6879(07)31001-X. [DOI] [PubMed] [Google Scholar]
- 23.Homma MK, Wada I, Suzuki T, Yamaki J, Krebs EG, Homma Y. CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cylce progression. Proc. Natl Acad. Sci. USA. 2005;102:15688–15693. doi: 10.1073/pnas.0506791102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh CR, Asano K. Localization and characterization of protein-protein interaction sites. Methods Enzymol. 2007;429:139–161. doi: 10.1016/S0076-6879(07)29007-X. [DOI] [PubMed] [Google Scholar]
- 25.Asano K, Lon P, Krishnamoorthy T, Pavitt GD, Gomez E, Hannig EM, Nika J, Donahue TF, Huang H-K, Hinnebusch AG. Analysis and reconstitution of translation initiation in vitro. Methods Enzymol. 2002;351:221–247. doi: 10.1016/s0076-6879(02)51850-4. [DOI] [PubMed] [Google Scholar]
- 26.Teske BF, Baird TD, Wek RC. Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 2011;493:333–356. doi: 10.1016/B978-0-12-385114-7.00019-2. [DOI] [PubMed] [Google Scholar]
- 27.Das S, Maitra U. Mutational analysis of mammalian translation initiation factor 5 (eIF5): role of interaction between the beta subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol. Cell. Biol. 2000;20:3942–3950. doi: 10.1128/mcb.20.11.3942-3950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a poly(A)- and eIF4A-dependent manner. Mol. Cell. 2009;36:1007–1017. doi: 10.1016/j.molcel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Conte MR, Kelly G, Babon J, Sanfelice D, youell J, Smerdon SJ, Proud CG. Structure of the eukaryotic initiation factor 5 reveals a fold common to several translation factors. Biochemistry. 2006;45:4550–4558. doi: 10.1021/bi052387u. [DOI] [PubMed] [Google Scholar]
- 31.Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grousl T, Ivanov P, Frýdlová I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcová I, Nováková L, Janosková D, et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- 34.Campbell SG, Hoyle NP, Ashe MP. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translational control. J. Cell Biol. 2005;170:925–934. doi: 10.1083/jcb.200503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinnebusch AG. Translational regulation of gcn4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 36.Gomez E, Pavitt GD. Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2β) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell Biol. 2000;20:3965–3976. doi: 10.1128/mcb.20.11.3965-3976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenbock GM. Excitatory local circuits and their implications for alfactory processing in the fly antennal lobe. Cell. 2007;128:601–612. doi: 10.1016/j.cell.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Spencer SD, Lasky LA. Trysine phosphorylation regulates the SH3-mediated binding of the Wiskott-Aldrich Syndome protein to PSTPIP, a cytoskeletal-associated protein. J. Biol. Chem. 1999;273:5765–5770. doi: 10.1074/jbc.273.10.5765. [DOI] [PubMed] [Google Scholar]
- 40.Chunlei W, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Uchida K, Endler A, Shibasaki F. Mammalian tumor suppressor Int6 specifically targets hypoxia inducible factor 2alpha for degradation by hopoxia- and pVHL-independent regulation. J. Biol. Chem. 2007;282:12707–12716. doi: 10.1074/jbc.M700423200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.