Abstract
The design of synthetic gene networks requires an extensive genetic toolbox to control the activities and levels of protein components to achieve desired cellular functions. Recently, a novel class of RNA-based control modules, which act through post-transcriptional processing of transcripts by directed RNase III (Rnt1p) cleavage, were shown to provide predictable control over gene expression and unique properties for manipulating biological networks. Here, we increase the regulatory range of the Rnt1p control elements, by modifying a critical region for enzyme binding to its hairpin substrates, the binding stability box (BSB). We used a high throughput, cell-based selection strategy to screen a BSB library for sequences that exhibit low fluorescence and thus high Rnt1p processing efficiencies. Sixteen unique BSBs were identified that cover a range of protein expression levels, due to the ability of the sequences to affect the hairpin cleavage rate and to form active cleavable complexes with Rnt1p. We further demonstrated that the activity of synthetic Rnt1p hairpins can be rationally programmed by combining the synthetic BSBs with a set of sequences located within a different region of the hairpin that directly modulate cleavage rates, providing a modular assembly strategy for this class of RNA-based control elements.
INTRODUCTION
The field of synthetic biology encompasses the engineering of new cellular functions through the design of synthetic gene networks. The tuning of protein levels is critical for proper functioning of integrated genetic networks. For example, the optimization of metabolic networks often requires the precise tuning and regulation of enzyme levels and activities to avoid undesired consequences associated with metabolic burden due to gene overexpression (1,2), the accumulation of toxic intermediates (3–5) and the redirection of metabolic flux from pathways critical to cell growth and viability (6–8). Altered levels of protein components can be achieved by controlling transcription (9–14), post-transcriptional stability and translation (15–20) and protein stability (21,22). In addition, libraries of genetic control elements have been generated to increase the precision with which protein levels can be modulated (3,10,11,14,17,18). However, the majority of gene regulatory tools developed to date function in bacterial hosts, such as Escherichia coli. Therefore, extending toolsets of genetic control elements to other cellular chassis is essential to supporting the design of more complex, integrated genetic networks in those organisms.
The budding yeast, Saccharomyces cerevisiae, is a relevant cellular chassis in industrial bioprocessing (23–28). The current genetic toolbox for S. cerevisiae gene regulation relies primarily on transcriptional control mechanisms such as inducible and constitutive promoter systems. Many inducible promoters depend on accurately controlling the level of the exogenously-applied inducer molecule, where intermediate expression levels are determined through the partitioning of cells in the population between either being fully repressed or expressing the desired protein (29). While engineered variants have been constructed that offer more tunable responses to varying inducer concentrations (11,12,14), these systems can exhibit other undesirable properties, such as pleiotropic effects of the inducer molecules, undesired effects of altering the natural regulatory networks associated with the native promoter system and the cost associated with the inducing molecule in scale-up processes. RNA-based control modules based on post-transcriptional mechanisms may offer an advantage since their activities are independent of the choice of promoter. Moreover, RNA-based controllers can be combined with transcriptional controllers to expand the design of integrated regulatory networks and thus provide more sophisticated control strategies. While post-transcriptional regulatory elements have been described in yeast (30–33), synthetic libraries of post-transcriptional control elements that exhibit tuned activities across a broad regulatory range had not been developed.
We previously developed a novel class of RNA control modules that act through post-transcriptional cleavage by the S. cerevisiae Rnt1p enzyme (20). Rnt1p recognizes RNA hairpins that contain a consensus AGNN tetraloop, which forms a predetermined fold that is recognized by the dsRNA binding domain (dsRBD) of Rnt1p (34–36). RNA hairpins cleaved by Rnt1p have three critical regions: the initial binding and position box (IBPB), comprising the tetraloop; the binding stability box (BSB), comprising the base-paired region immediately adjacent to the tetraloop and the cleavage efficiency box (CEB), comprising the region containing and surrounding the cleavage site (36). Rnt1p hairpins were inserted as genetic control elements within the 3′-untranslated region (UTR) of a transcript in order to direct cleavage to that region, thereby inactivating the transcript and lowering target protein levels. We designed an initial library based on randomization of the Rnt1p substrate CEB and screened this library to identify a set of Rnt1p control modules that tune expression levels through differential Rnt1p processing rates (20). In contrast to other post-transcriptional control elements, which exhibit activities that vary substantially with genetic context (37), the Rnt1p control elements exhibited conserved gene regulatory activities across varied genetic contexts. The utility of the Rnt1p control modules was demonstrated for achieving predictable control over protein levels and manipulating biological networks.
Here, we examined the role of a different critical region of the Rnt1p substrate, the BSB, on Rnt1p processing efficiencies and thus gene regulatory activities. We generated a library of hairpins based on randomization of the BSB to identify sequences that modulated Rnt1p binding affinity. Rigid structural constraints imposed by the BSB resulted in a low percentage (~0.6%) of sequence variants in the library that exhibit Rnt1p binding activity, and thus required the development of a selection strategy based upon fluorescence-activated cell sorting (FACS) by enriching for cells exhibiting low fluorescence. In total, 16 unique BSBs were identified that span an intermediate range of protein expression levels. In vitro characterization assays indicated that altered expression levels are due to the ability of BSBs to determine the hairpin cleavage rate and to form active cleavable complexes with Rnt1p. The integration of the synthetic BSB sequences with different synthetic CEB modules demonstrated that the BSB sequences function as modules that retain their relative activities under the context of different CEBs. Further characterization indicated that proportional deviation from the ‘parent’ BSB was inversely related to the strength of the coupled CEB. Our work establishes a set of BSB sequences and a previously developed set of CEB sequences as modular units that can be implemented combinatorially to build synthetic Rnt1p hairpins exhibiting finely-tuned processing properties and an extended range of gene regulatory activities. The development of such well-characterized and distinct activity modules is critical for synthetic biology applications, where they can be combined with other components to build more sophisticated control devices (38).
MATERIALS AND METHODS
Plasmid construction
Standard molecular biology techniques were utilized to construct all plasmids (39). DNA synthesis was performed by Integrated DNA Technologies (Coralville, IA, USA) or the Protein and Nucleic Acid Facility (Stanford, CA, USA). All enzymes, including restriction enzymes and ligases, were obtained through New England Biolabs (Ipswich, MA, USA) unless otherwise noted. Pfu polymerases were obtained through Stratagene. Ligation products were electroporated with a GenePulser XCell (Bio-Rad, Hercules, CA, USA) into E. coli DH10B (Invitrogen, Carlsbad, CA, USA), where cells harboring cloned plasmids were maintained in Luria-Bertani media containing 50 mg/ml ampicillin (EMD Chemicals). Clones were initially verified through colony PCR and restriction mapping. All cloned constructs were sequence verified by Elim Biopharmaceuticals (Hayward, CA, USA) or the Protein and Nucleic Acid Facility (Stanford, CA, USA). Plasmid maps are available in Supplementary Figure S1.
The construction of the Rnt1p characterization plasmid, pCS321, and the Rnt1p expression plasmid, pRNT1, have been previously described (20). A screening plasmid (pCS1585) was constructed from pCS321 by replacing the GAL1-10 promoter with the endogenous TEF1 promoter (J.C. Liang, unpublished data). A second screening plasmid (pCS1748) was constructed from pCS1585 by inserting an additional open reading frame (ORF) containing the yeast enhanced mCherry gene, ymCherry, flanked by a TEF1 promoter and a CYC1 terminator (J.C. Liang et al., manuscript in preparation).
Insertion of engineered Rnt1p substrates and appropriate controls into the 3′-UTR of yEGFP3 in pCS321 and pCS1585 was performed through either digestion with appropriate restriction endonucleases and ligation-mediated cloning or homologous recombination-mediated gap-repair during transformation into S. cerevisiae strain W303 (MATa, his3-11,15 trp1-1 leu2-3 ura3-1 ade2-1) through standard lithium acetate procedures (40). The Rnt1p substrates were amplified for insertion with both techniques using the forward and reverse primers RntGap321_fwd (5′-ACCCATGGTATGGATGAATTGTACAAATAAAGCCTAGGTCTAGAGGCG) and RntGap321_rev2 (5′-TAAGAAATTCGCTTATTTAGAAGTGGCGCGCCCTCTCGAGGGCG), respectively. The Rnt1p switch (RS) and the BSB variants (RS-B03, RS-B12) were amplified for insertion with both techniques using the forward and reverse primers RS_fwd (5′-ATGGTATGGATGAATTGTACAAATAAAGAGCCTAGGAAACAAACAAACTTGATGCCCTTGG) and RS_rev (5′-AAATTCGCTTATTTAGAAGTGGCGCGCCCTCTCGAGTTTTTATTTTTCTTTTTACGATGCTGGTATC), respectively. The oligonucleotide templates of RS, RS-B03 and RS-B12 for amplification are RS_temp (5′-AAACAAACTTGATGCCCTTGGCAGCCGGATGTCATGAGTCCATGGCATCTGGATACCAGCATCGTAAAAAGAAAAATAAA), RS-B03_temp (5′-AAACAAACTTGATGCCCTTGGCAGCCGGATGTTGAAAGTCTTCAGCATCTGGATACCAGCATCGTAAAAAGAAAAATAAA) and RS-B12_temp (5′-AAACAAACTTGATGCCCTTGGCAGCCGGATGTAGTGAGTCCACTGCATCTGGATACCAGCATCGTAAAAAGAAAAATAAA), respectively. In the case of digestion and ligation, the PCR products were digested with the unique restriction sites AvrII and XhoI, which are located 3 nt downstream of the yEGFP3 stop codon and upstream of the ADH1 terminator. Following construction and sequence verification of the desired vectors, 100–500 ng of each plasmid was transformed into strain W303. In the case of gap-repair, 250–500 ng of the PCR product and 100 ng of plasmid digested with AvrII and XhoI were transformed into the yeast strain. All yeast strains harboring cloned plasmids were maintained on synthetic complete media with an uracil dropout solution and 2% dextrose at 30°C.
Library-scale yeast transformation
Yeast transformations with the binding library were performed on Rnt1p substrates as previously described (41). The C13-based binding library was amplified with template BndLib_C13 (5′-AGCCTAGGTCTAGAGGCGCTATCGTGTCATGTNNNNAGTCNNNNGCATGGCATGATAGCGCCCTCGAGAGGG) and forward and reverse primers C13BindLibgap_fwd_prmr (5′-GTATTACCCATGGTATGGATGAATTGTACAAATAAAGCCTAGGTCTAGAGGCGCTATC) and C13BindLibgap_rev_prmr (5′-AATCATAAGAAATTCGCTTATTTAGAAGTGGCGCGCCCTCTCGAGGGCGCTATCA), respectively. The reaction was scaled-up to 800 µl to obtain roughly 40–50 µg of PCR product. Eight micrograms of plasmid (either pCS1585 or pCS1748) was digested overnight with AvrII and XhoI in 400 µl total reaction volume. Two tubes of DNA were made with 375 µl of PCR product (~20 µg) and 150 µl of digested plasmid (~3 µg). A third tube acting as a negative control contained 450 µl of water and 75 µl of digested plasmid (~1.5 µg). Each tube was extracted with phenol–chloroform (1:1) and ethanol-precipitated into fresh tubes.
Five hundred microliters of Tris–DTT [2.5 M DTT, 1 M Tris (pH 8.0)] was added to a 50-ml culture of yeast strain W303 that was grown in YPD to an OD600 of 1.3–1.5 at 30°C. Following 10–15 min of additional incubation, the cells were collected and washed in 25 ml of ice-cold Buffer E [10 mM Tris (pH 7.5), 2 mM MgCl2, 270 mM sucrose] and washed again in 1 ml of Buffer E before being resuspended to a final volume of 300 µl in Buffer E. Sixty microliters of this cell mixture was added to the negative control tube and 120 µl was added to the two tubes containing digested plasmid and the library. After allowing the precipitated DNA to resuspend, 50 µl of the negative control or the library suspension was transferred to a chilled 2-mm gap cuvette and electroporated (540 V, 25 µF, infinite resistance, 2 mm gap). Each 120-µl tube of library suspension contained enough material for two electroporations. Following electroporation, the cells were resuspended in 1 ml of pre-warmed YPD and added to a fresh 15-ml Falcon tube. The cuvette was washed a second time with a fresh 1-ml aliquot of YPD, which was added to the same Falcon tube. Library electroporations were collected in the same Falcon tube (8 ml total). The Falcon tubes were incubated with shaking for 1 h at 30°C. After incubation, the cells were collected and resuspended in 1 ml of synthetic complete media with an uracil dropout solution and 2% dextrose. The resuspension was added to 6 ml of fresh media to prepare for FACS.
FACS and sorted library retransformation
The transformed binding library was grown for 2–3 days in liquid culture. Following this growth period, the library and appropriate control cultures were collected and suspended in 1× PBS with 1% BSA and either 7-amino-actinomycin D (7-AAD; Invitrogen) or 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) was added as a viability stain. The cell suspension was passed through a 40-µm cell strainer (BD Falcon) prior to analysis on a FACSAria or FACSAria II flow cytometry cell sorter (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA). On the FACSAria, GFP was excited at 488 nm and measured with a bandpass filter of 530/30 nm. 7-AAD was excited at 488 nm and measured with a bandpass filter of 695/40 nm. On the FACS Aria II, GFP was excited at 488 nm and measured with a splitter of 505 nm and bandpass filter of 525/50 nm. mCherry was excited at 532 nm and measured with a splitter of 600 nm and a bandpass filter of 610/20 nm. DAPI was excited at 355 nm and measured with a bandpass filter of 450/50 nm. Detailed sorting procedures are presented in Supplementary Figures S2 and S3. The collected fractions were diluted to 100 ml in synthetic complete media with an uracil dropout solution and 2% dextrose and grown until an OD600 of ~1.5. The culture was continually back-diluted and grown in successively decreasing culture volume for 2 days at which time freezer stocks were made of the fractions. From the culture, 100 µl was collected and the library hairpins amplified by colony PCR with forward and reverse primers RntGap321_fwd and RntGap321_rev2, respectively. The PCR products representing the sorted binding library were recloned through a gap-repair method by transforming the DNA with the pCS321 plasmid in yeast strain W303.
Rnt1p substrate characterization assays
S. cerevisiae cells harboring pCS321-based plasmids were grown on synthetic complete media with an uracil dropout solution and the appropriate sugars (2% raffinose, 1% sucrose) overnight at 30°C. The cells were back-diluted the following morning into fresh media (4.5 ml total volume in test tubes and 450 µl in deep-well plates) to an optical density at 600 nm (OD600) of 0.1 and grown again at 30°C. After 1 h, 0.5 ml (test tubes) or 50 µl (plates) of 20% galactose (2% final concentration) or water (non-induced control) was added to the cell cultures. The cells were grown for another 4.5 h before measuring the fluorescence levels or collecting cells for RNA extraction. Cells harboring pCS1585-based and pCS1748-based plasmids followed the same procedure as pCS321-based plasmids, except 2% dextrose was the only sugar in the media and no induction was required.
Fluorescence quantification
On the Quanta flow cytometer (Beckman Coulter, Fullerton, CA, USA), the distribution of GFP fluorescence was measured with the following settings: 488-nm laser line, 525-nm bandpass filter and photomultiplier tube setting of 5.83 (pCS321-based) and 4.50 (pCS1585-based). Data were collected under low flow rates until 10 000 viable cell counts were collected. For pCS321-based plasmids, a non-induced cell population was used to set a gate to represent GFP-negative and GFP-positive populations. For pCS1585-based plasmids, a plasmid harboring the same backbone as pCS1585 but with no fluorescence gene was used to set the GFP-negative and GFP-positive gates.
The LSRII flow cytometer (Becton Dickinson Immunocytometry Systems) was used to measure mCherry and GFP fluorescence from pmCh-Y-based plasmids. GFP was excited at 488 nm and measured with a splitter of 505 nm and a bandpass filter of 525/50 nm. mCherry was excited at 532 nm and measured with a splitter of 600 nm LP and a bandpass filter of 610/20 nm. DAPI was excited at 405 nm and measured with a bandpass filter of 450/50 nm.
Quantification of cellular transcript levels
Total RNA from S. cerevisiae was collected by a standard hot acid phenol extraction method (42) and followed by DNase I (New England Biolabs) treatment to remove residual plasmid DNA according to manufacturer's instructions. cDNA was synthesized from 5 µg of total RNA with gene-specific primers for yEGFP3 and ACT1 (43) (rnt1p_rtpcr_rev2 and ACT1_rtpcr_rev, respectively) and SuperScript III Reverse Transcriptase (Invitrogen) according to manufacturer's instructions. The forward and reverse primers for yEGFP3 quantification are rnt1p_rtpcr_fwd2 (5′-CGGTGAAGGTGAAGGTGATGCTACT) and rnt1p_rtpcr_rev2 (5′-GCTCTGGTCTTGTAGTTACCGTCATCTTTG), respectively. The forward and reverse primers for ACT1 quantification are ACT1_rtpcr_fwd (5′-GGCATCATACCTTCTACAACGAAT) and ACT1_rtpcr_rev (5′-GGAATCCAAAACAATACCAGTAGTTCTA), respectively. Relative transcript levels were quantified in triplicate from three identical reactions from the cDNA samples by using an appropriate primer set and iQ SYBR Green Supermix (Bio-Rad) on an iCycler iQ qRT–PCR machine (Bio-Rad) according to the manufacturer's instructions. For each run, a standard curve was generated for yEGFP3 and a house-keeping gene, ACT1, using a dilution series for a control representing no insertion of an Rnt1p substrate. Relative yEGFP3 and ACT1 levels were first individually determined for each sample and then the yEGFP3 values were normalized by their corresponding ACT1 values.
In vitro transcription of Rnt1p substrates
All Rnt1p substrates were PCR-amplified to include an upstream T7 promoter site using forward and reverse primers Rnt1p-T7-PCR_fwd_prmr (5′-TTCTAATACGACTCACTATAGGGACCTAGGAAACAAACAAAGTTGGGC) and Rnt1p-T7-PCR_rev_prmr (5′-CTCGAGTTTTTATTTTTCTTTTTGCCGGGCG), respectively. From the PCR product, 1–2 µg was transcribed with T7 RNA Polymerase (New England Biolabs) in the presence and absence of α-P32-GTP. The 25-µl reaction consisted of the following components: 1× RNA Pol Reaction Buffer (New England Biolabs), 3 mM rATP, 3 mM rCTP, 3 mM rUTP, 0.3 mM rGFP, 1 µl RNaseOUT (Invitrogen), 10 mM MgCl2, 2 mM DTT, 1 µl T7 Polymerase and 0.5 µCi α-P32-GTP. Unincorporated nucleotides were removed from the reactions by running the samples through NucAway Spin Columns (Ambion, Austin, TX, USA) according to the manufacturer's instructions.
Rnt1p expression and purification
The pRNT1 plasmid was transformed into E. coli strain BL21 using the Z-competent E. coli Transformation Kit and Buffer Set (Zymo Research, Orange, CA, USA) according to manufacturer's instructions. Rnt1p was collected as a protein extract as previously described (44). Briefly, an overnight culture of BL21 cells harboring pRNT1 was back-diluted to an OD600 of 0.5. Once the culture reached an OD600 of 1.1–1.4, it was induced with 1 mM IPTG and grown for an additional 3 h. The cells were centrifuged at 2500 g for 12 min at 4°C and the resulting cell pellet was frozen in a −80°C freezer. After weighing the frozen cell pellet, the cells were resuspended in 4 ml Ni2+ buffer [25% (v/v) glycerol, 1 M NaCl, 30 mM Tris, pH 8.0] per gram of harvested cells. The resuspension was sonicated (Heat Systems-Ultrasonics, Inc.) twice with the following settings: 2 × 30 s, output control 5 and 50% duty cycle. Cellular debris was removed by centrifugation at 20 000 g for 30 min at 4°C and the supernatant was filtered through a 0.2-µm pore size Acrodisc 25 mm syringe filter (Pall Life Sciences, Ann Arbor, MI, USA).
Rnt1p was purified from the resulting supernatant with one 1-ml HisTrap HP column (GE Healthcare) on an AKTA fast protein liquid chromatography (FPLC) machine (GE Healthcare). Elution of the protein was performed with an imidazole concentration of 150 mM in Ni2+ buffer and the protein was collected in six 1-ml fractions. Protein purification was confirmed by analyzing an aliquot of each fraction on a SDS–PAGE gel (NuPAGE 4–12% Bis–Tris Gel, Invitrogen) and protein function was confirmed by incubating an aliquot of each fraction with a control Rnt1p substrate and analyzing the resulting cleavage products on an 8% denaturing polyacrylamide gel. Positive fractions were pooled and concentrated to <3-ml volume using a Centricon Centrifugal Filter Device (10 000 MWCO; Millipore) according to the manufacturer's instructions. The concentrated protein was then injected into a Slide-A-Lyzer Dialysis Cassette (10 000 MWCO; Pierce Biotechnology) and buffer-exchanged twice with Rnt1p Storage Buffer [50% (v/v) glycerol, 0.5 M KCl, 30 mM Tris, pH 8.0, 0.1 M DTT, 0.1 M EDTA] at 4°C. The first buffer exchange took place for 4 h and the second buffer exchange occurred overnight. The purified Rnt1p was stored in aliquots at −20°C.
In vitro Rnt1p substrate cleavage assay
Cleavage assays were performed on Rnt1p substrates as previously described (44,45). Briefly, a 10-µl mixture of RNA and Rnt1p were incubated at 30°C for 15 min in Rnt1p reaction buffer [30 mM Tris (pH 7.5), 150 mM KCl, 5 mM spermidine, 20 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA (pH 7.5)]. RNA concentrations were varied from 0.2 to 2.0 µM and the Rnt1p concentration was 2.3 µM. The cleavage reaction products were separated on an 8% denaturing polyacrylamide gel run at 35 W for 30 min. Gels were transferred to filter paper and analyzed for relative substrate and product levels through phosphorimaging analysis on a FX Molecular Imager (Bio-Rad). The levels of cleaved RNA product were determined and fit to a Michaelis-Menten model using Prism 5 (GraphPad), where a relative Vmax was calculated and reported with the standard error determined by the fit of the model.
In vitro Rnt1p substrate mobility shift assay
Mobility shift assays were performed as previously described (44,45). Briefly, a 10-µl mixture of RNA and Rnt1p were incubated on ice for 10 min in Rnt1p binding buffer [20% (v/v) glycerol, 30 mM Tris (pH 7.5), 150 mM KCl, 5 mM spermidine, 0.1 mM DTT, 0.1 mM EDTA (pH 7.5)]. The RNA concentration in all samples was 200 nM and the Rnt1p concentration ranged from 0 to 1.7 µM. The binding reaction products were separated on a 6% native polyacrylamide gel run at 350 V until the samples entered the gel and then at 150 V for 2 h. Gels were transferred to filter paper and analyzed for free RNA and RNA–Rnt1p complex levels through phosphorimaging analysis on a FX Molecular Imager. The fraction of unbound RNA to total RNA was determined and fit to a modified Scatchard model using Prism 5, where a KD value was calculated and reported with the standard error determined by the fit of the model.
RESULTS
Design and selection of an Rnt1p binding library to achieve tunable gene regulatory control
Rnt1p is an RNase III enzyme that cleaves hairpin structures in S. cerevisiae. An Rnt1p substrate can be divided into three critical regions: the IBPB, the BSB and the CEB (36) (Figure 1A). The BSB has a reported structural requirement in which the three nucleotides immediately below the tetraloop must form Watson–Crick base-pairs and the nucleotides in the fourth position must also base-pair, in either a Watson–Crick or wobble conformation, for optimal activity (46). Rnt1p initially binds to the tetraloop and then cleaves the hairpin at two locations within the CEB: between the 14th and 15th nt upstream of the tetraloop and the 16th and 17th nt downstream of the tetraloop. Naturally-occurring Rnt1p hairpins have been identified in numerous non-coding RNAs, where Rnt1p plays a critical role in non-coding RNA processing and editing (47–49) and in transcripts, where Rnt1p was shown to play a role in controlling gene expression (50–52).
Figure 1.
Implementation of Rnt1p hairpins as post-transcriptional genetic control elements and binding library design. (A) Consensus regions of an Rnt1p hairpin. Color scheme is as follows: CEB, red; BSB, blue; initial binding and positioning box (IBPB), green. Black triangles indicate locations of cleavage sites. The clamp region is a synthetic sequence that acts to insulate and maintain the structure of the control element. The regulatory activity of our synthetic hairpins is a function of the modular elements in the CEB and the BSB. (B) Rnt1p hairpins in the 3′-UTR of a gene of interest (goi) reduce protein levels through transcript destabilization by endonucleolytic cleavage. The hairpin contains two tunable modules: the BSB (blue box) and the CEB (red box). Sequence modifications within the BSB and CEB are expected to primarily tune Rnt1p's ability to bind the hairpin (blue dial) and to cleave the hairpin (red dial), respectively. Rnt1p processing of the hairpin results in unstable transcripts and thus reduced protein expression levels. Barrels represent the protein product of the goi; orange oval represents Rnt1p. (C) Sequence and structure of an Rnt1p binding library based on the randomization of 8 nt in the BSB and containing the C13 CEB. (D) Sequences and structures of synthetic CEB sequences (20) used in the screening and characterization of the binding library. ‘B00’ refers to the ‘parent’ BSB used in the cleavage library. (E) Histograms of GFP fluorescence for cell populations containing the Rnt1p constructs based on the ‘parent’ BSB. Numbers above each peak represent the median GFP fluorescence for that sample normalized to a control sample with a construct containing no Rnt1p hairpin.
We previously developed a genetic system in which Rnt1p-mediated cleavage was used to regulate gene expression in yeast through the placement of Rnt1p hairpins in the 3′-UTR of a target transcript (Figure 1B) (20). We developed a set of synthetic Rnt1p hairpins based on sequence modification within the CEB that exhibit a broad range of cleavage rates and thus gene regulatory activities. Previous in vivo work with Rnt1p demonstrated its ability to regulate cellular RNA levels (50,53,54). The CEB library provided the first study linking the modification of a critical region of an Rnt1p substrate to in vivo regulatory activity. Since the BSB and CEB are required elements for Rnt1p binding and cleavage, respectively, we hypothesized that a similar library screening approach could be applied to generate synthetic BSBs exhibiting different Rnt1p binding affinities. While prior in vitro work had established that mutations in the BSB could detrimentally affect in vitro binding and subsequent cleavage (36), no studies have examined a large BSB sequence space for tuning gene regulatory activities in vivo. The synthetic CEB and BSB elements are anticipated to act as modular units such that they could be implemented combinatorially to build synthetic Rnt1p hairpins exhibiting an extended range of gene regulatory activities and more finely-tuned levels of expression.
We designed an Rnt1p binding library based on randomizing the BSB (8 nt) to generate Rnt1p hairpins that exhibit different gene regulatory activities due to altered binding affinity between the hairpin and Rnt1p (Figure 1C). One of the variables in the design of the binding library was the CEB to place within the stem, as we had previously described a set of synthetic CEBs with modified gene regulatory activity (20). As it was unknown how the Rnt1p hairpins would respond to changes in the BSB, we selected a synthetic CEB contained in the hairpin that demonstrated the lowest level of gene expression (C13). All hairpins with active BSBs containing the CEB of C13 were expected to have comparable gene expression levels, improving their probability of being identified in the screen. C13 is also fully base-paired, such that its integration into an Rnt1p substrate stem results in a stable structure that is less susceptible to changes in flanking sequences and has a greater probability of maintaining the desired hairpin structure (Figure 1D). Flow cytometry analysis also indicates that C13 achieves the greatest population separation from a no hairpin control of all synthetic CEBs (Figure 1E). This separation is representative of that expected for the active and inactive binding populations in the binding library, thus, increasing the enrichment of the cell-based sort. We refer to the ‘parent’ BSB that was used in the cleavage library as B00, where synthetic Rnt1p hairpins are identified by their CEB and BSB as Cxx–Bxx (i.e. C13–B00) or Axx–Bxx (i.e. A02–B00).
Synthetic Rnt1p substrates with altered binding affinities were identified through a cell-based fluorescence screen. The designed library (N8) has a diversity of 65 536 different sequences. Due to the rigid structural requirements of the BSB, we predicted that at most 384 sequences (0.586%) would be actively bound and subsequently cleaved by Rnt1p. A manual plate-based screening strategy was previously used to efficiently identify active sequences from the cleavage library. However, due to the loose structural requirements for the CEB, the cleavage library had a substantially higher positive rate than that anticipated for the binding library, such that the plate-based screen was not feasible for the number of colonies that would have to be screened to identify a reasonable diversity of active BSBs. As a higher throughput method we employed FACS to efficiently identify and recover the low percentage of cells in the population with diminished fluorescence.
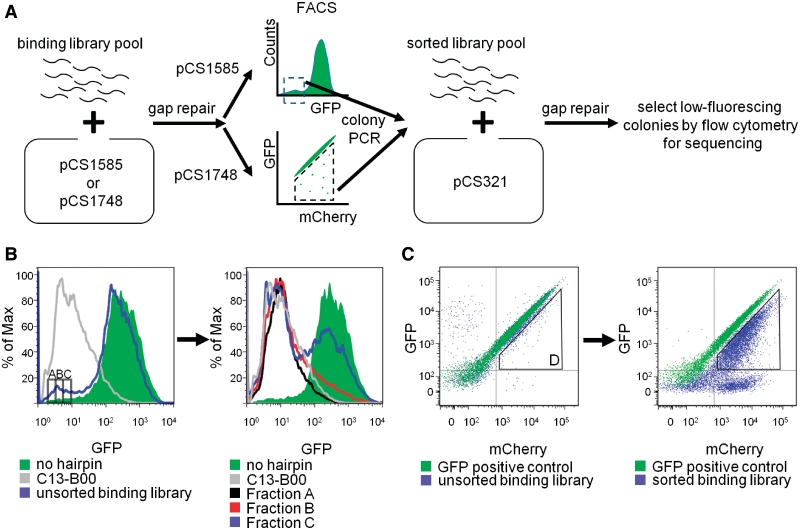
The binding library was transformed into yeast through a library-scale gap-repair strategy and clones exhibiting strong gene regulatory activity were selected through FACS by gating for cells with low fluorescence levels (Figure 2A). We performed two different single FACS screens based on a single-color (pCS1585; yEGFP3) system and a two-color (pCS1748; ymCherry and yEGFP3) system (Liang, J.C. et al., manuscript in preparation). Three fractions (A–C) were collected for the pCS1585 system around the expression level of C13-B00. Following the initial sort, each fraction was regrown and subsequently characterized by flow cytometry (Figure 2B and Supplementary Figure S2). Only fraction A retained a low level of fluorescence, whereas the regrown populations in fractions B and C shifted substantially towards inactive (high fluorescence) levels. The results suggest that sorted fractions B and C contain a large percentage of false positives, or clones harboring plasmids containing inactive Rnt1p hairpins that exhibited low fluorescence levels due to noise in gene expression profiles. Based on the fraction profiles, fraction A was selected for further testing. Most of the false positives due to genetic noise were removed with the two-color pCS1748 system. A gate representing diminished GFP fluorescence was determined based on a GFP positive control lacking an Rnt1p hairpin. All library clones with decreased GFP levels were collected into a single fraction (D). Fraction D was regrown, characterized by flow cytometry and demonstrated to retain low fluorescence levels (Figure 2C and Supplementary Figure S3). Therefore, fraction D was also selected for further characterization.
Figure 2.
In vivo screening of an Rnt1p binding library. (A) A high-throughput, in vivo, fluorescence-based screen for Rnt1p hairpin activity. The library was cloned through gap-repair into yeast in two different plasmid systems. Clones exhibiting low GFP fluorescence were sorted from the population through FACS. A sorted library pool was generated through colony PCR from collected cellular fractions and gap-repaired into the characterization plasmid. Clones that maintained low GFP fluorescence levels were selected for sequencing and further characterization. (B) FACS procedure for the single-color (pCS1585-based) system. Fractions A, B and C were collected based on exhibiting GFP levels similar to the median fluorescence of C13-B00 (left panel). Only fraction A maintained a low level of GFP expression after the fractions were regrown (right panel). (C) FACS procedure for the two-color (pCS1748-based) system. Fraction D was collected based on a gate set to collect all cells exhibiting GFP fluorescence levels below a cells containing a positive GFP control construct lacking an Rnt1p hairpin module (left panel). Fraction D maintained a low level of GFP expression when regrown (right panel).
After the completion of the FACS screens, the sorted constructs were recloned to remove false positives due to mutations in the plasmid or the yeast background that would cause reduced GFP levels independent of Rnt1p activity. We retrieved the selected Rnt1p hairpin sequences from fractions A and D by colony PCR and gap-repaired the recovered hairpin constructs into pCS321. Individual clones were initially characterized for gene regulatory activity by measuring cellular fluorescence through a plate reader assay. Colonies positive for GFP knockdown were sequenced to determine the BSB sequence. In total 16 unique BSB sequences were identified including the ‘parent’ BSB (Supplementary Figure S4 and Supplementary Table S1). The predicted secondary structure of the hairpins was determined by RNAstructure (http://rna.urmc.rochester.edu/RNAstructure.html). The binding library structures deviate from structural requirements that were previously established through in vitro studies (46). All of the BSB structures contain Watson–Crick base pairing in the first 3 nt below the tetraloop; however, in the fourth position certain sequences exhibit mismatching. The results from our cell-based BSB library screen suggest that the in vivo structural requirements for the BSB are not as stringent as previously described.
A synthetic Rnt1p binding library exhibits a range of gene regulatory activities in vivo
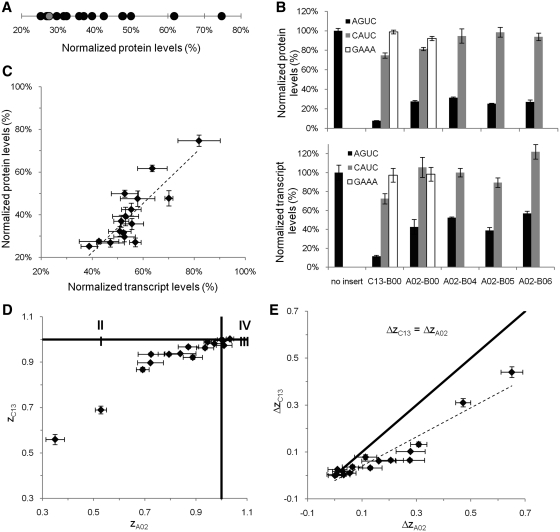
The initial screen for active BSB sequences was performed in the context of a CEB exhibiting high cleavage activity (C13). As a result, most of the recovered binding library hairpins exhibited low gene expression levels, such that differences in activity between the synthetic BSBs were difficult to resolve with the flow cytometry assay at low fluorescence levels (Supplementary Table S1). To gain better resolution on the differences in BSB activity and examine the activity of the synthetic BSBs in the context of a different CEB, we integrated the selected BSB sequences within the context of an Rnt1p hairpin containing a different synthetic CEB (A02) (Figure 1D). The range of regulatory activities spanned by the binding library in the context of the A02 CEB was measured at the protein and transcript levels. Flow cytometry analysis of the synthetic Rnt1p hairpins indicated that the selected set of hairpins spanned an intermediate gene regulatory range—from 25% (A02–B05) to 75% (A02–B01) (Table 1 and Figure 3A). The regulatory activities of the selected hairpins are not evenly distributed across this range, with the majority exhibiting activities in the range of 25–45% relative protein levels. The results suggest that the binding library achieves a smaller regulatory range than that observed with the cleavage library and may be more appropriate for fine-tuning gene expression (20). We built negative controls for several binding library hairpins and for the ‘parent’ hairpins (A02–B00 and C13–B00) by mutating the tetraloop sequence (CAUC or GAAA) to impede Rnt1p activity while maintaining the secondary structure of the hairpins. The negative controls demonstrated that the majority of knockdown observed from each hairpin is due to Rnt1p processing (Figure 3B).
Table 1.
In vivo characterization data for the binding library
| Substrate | Normalized protein levels (%) | Normalized transcript levels (%) |
|---|---|---|
| A02–B00 | 28 ± 1 | 43 ± 8 |
| A02–B00 (CAUC) | 81 ± 2 | 106 ± 11 |
| A02–B01 | 75 ± 3 | 82 ± 8 |
| A02–B02 | 62 ± 2 | 64 ± 6 |
| A02–B03 | 50 ± 2 | 53 ± 5 |
| A02–B04 | 32 ± 1 | 52 ± 1 |
| A02–B05 | 25 ± 0 | 39 ± 3 |
| A02–B06 | 27 ± 2 | 57 ± 2 |
| A02–B07 | 37 ± 3 | 51 ± 3 |
| A02–B08 | 30 ± 2 | 53 ± 4 |
| A02–B09 | 36 ± 3 | 56 ± 5 |
| A02–B10 | 42 ± 3 | 55 ± 4 |
| A02–B11 | 32 ± 2 | 51 ± 4 |
| A02–B12 | 27 ± 2 | 47 ± 5 |
| A02–B13 | 39 ± 4 | 53 ± 5 |
| A02–B14 | 48 ± 4 | 70 ± 2 |
| A02–B15 | 48 ± 4 | 58 ± 7 |
| No insert | 100 ± 3 | 100 ± 8 |
All normalized protein and transcript levels were determined as described in Figure 3A and B, respectively.
Figure 3.
In vivo characterization of the selected Rnt1p binding library and demonstration of the modularity of the BSB sequences. (A) The gene regulatory activities of the binding library spans an intermediate range of protein expression levels. Normalized protein expression levels were determined by measuring the median GFP levels from a cell population containing the appropriate construct through flow cytometry analysis and values are reported relative to that from an identical construct lacking a hairpin module. The ‘parent’ BSB is indicated in gray. (B) The transcript and protein levels associated with several binding library members and their corresponding mutated tetraloop (CAUC or GAAA) controls support that the observed gene regulatory activity is due to Rnt1p processing. Transcript levels were determined by measuring transcript levels of yEGFP3 and a house-keeping gene, ACT1, through qRT–PCR and normalizing the yEGFP3 levels with their corresponding ACT1 levels. Normalized transcript levels for each construct are reported relative to that from an identical construct lacking a hairpin module (‘no insert’). (C) Correlation analysis of protein and transcript levels from the binding library members demonstrates a strong correlation between the two measures of gene regulatory activity. (D) The gene regulatory activity of synthetic BSBs is conserved in the context of different CEB modules. The ratio of the knockdown exhibited from a binding library hairpin to that exhibited from the ‘parent’ (B00 BSB), zC, was determined in the context of two different CEBs and plotted against each other. Regions I and IV represent hairpins whose activities relative to ‘parent’ remain consistent when combined with different CEBs. Regions II and III represent hairpins whose activity varies relative to ‘parent’ when combined with different CEBs. (E) Synthetic BSBs modules generally exhibit higher relative activities in the context of weaker CEB modules. A variable representing the departure from ‘parent’ activity, ΔzC, was calculated in the context of two different CEBs and plotted against each other. The solid line indicates where the values of ΔzC are the same for both hairpins.
The reduced protein expression levels observed from the Rnt1p binding library is expected to be due to a reduction in the steady-state transcript levels due to rapid degradation of the transcript following endonucleolytic cleavage by Rnt1p. We measured relative transcript levels for each Rnt1p hairpin by quantitative real-time PCR (qRT–PCR) (Table 1). A plot of normalized yEGFP3 expression levels versus normalized yEGFP3 transcript levels indicates that there is a strong positive correlation (r = 0.847) between the two measures of activity (Figure 3C). A preservation of rank order was also observed between protein and transcript levels as indicated by the Spearman rank correlation coefficient (ρ = 0.668). Specifically, with decreasing transcript levels a similar decrease in protein levels is generally observed, confirming that the fluorescence observed was due to changes in the steady-state transcript levels. The negative controls based on mutating the tetraloop confirmed that Rnt1p cleavage is the cause of the observed transcript knockdown (Figure 3B).
Synthetic BSBs exhibit modular activity with different CEBs in vivo
We next examined the gene expression data for the Rnt1p hairpins harboring the synthetic BSBs in the context of two CEBs (C13, A02) for trends in regulatory activity across the binding library. We defined a new variable zC as the ratio of the knockdown from a binding library member (Bxx) to that of the ‘parent’ BSB (B00) for a specific CEB (Cxx or Axx):
A z-value greater than unity indicates increased knockdown due to the synthetic BSB, whereas a z-value less than unity indicates decreased knockdown due to the BSB. We calculated z-values for each BSB in the context of the CEBs C13 and A02 (zC13 and zA02, respectively) and plotted the two variables against each other (Figure 3D). For ease in interpretation, we divided the graph into four regions with the point (1,1) at the intersection of the quadrants. Regions I and IV indicate BSBs for which activities relative to parent are conserved between the different CEBs, whereas Regions II and III indicate BSBs that exhibit varying activities in the context of different CEBs. Nearly all BSBs are located in Regions I and IV, with the majority falling in Region I. The data indicate that if a BSB causes increased knockdown in the context of one CEB, it will likely exhibit the same activity in the context of another CEB.
To further examine the gene regulatory activities of the synthetic BSBs, we determined a new variable ΔzC, which is the difference between the zC value of the ‘parent’ (which by definition is 1) and the z-value of the BSB:
We calculated Δz for each BSB in the context of the CEBs C13 and A02 and plotted the variables against each other (Figure 3E). Data points that fall on the ΔzC13 = ΔzA02 line would indicate BSBs that have the same proportional effect on knockdown for both CEBs. The data fall beneath the ΔzC13 = ΔzA02 line in the region where ΔzA02 > ΔzC13, indicating that Rnt1p hairpins with weaker CEBs are affected more by changes in binding affinity through modification to the BSBs. The data exhibit a strong positive correlation (r = 0.946) and can be fit with a trendline by linear regression that passes close to the origin (0,0), suggesting that we see a consistent ratio between ΔzC13 and ΔzA02 values, where this ratio is dependent on the CEBs. The data also exhibit a strong preservation of rank order (ρ = 0.929), demonstrating that the relative activity between BSB modules is maintained regardless of the CEB module present in the hairpin. Taken together, the results show the maintenance of BSB activity in connection with different CEB stems and also that the proportional deviation from the ‘parent’ BSB is determined by the strength of the CEB.
We further examined the ability of the BSB modules to tune activities of more complex RNA control devices based on regulating Rnt1p processing. Previous work demonstrated that incorporation of an RNA sensor component, or aptamer, within the CEB region of an Rnt1p hairpin can result in an Rnt1p switch (38). Specifically, a theophylline aptamer was directly integrated into the CEB region of an Rnt1p hairpin and ligand binding was demonstrated to inhibit Rnt1p cleavage activity, resulting in ligand-responsive control over expression levels. Due to the placement of the aptamer in the CEB, the BSB modules become the only library of parts that can be used to tune the ability of Rnt1p to bind and cleave this sensing-actuation device. We demonstrated the activity of the BSB modules within the different stem/CEB context of this Rnt1p switch. We incorporated two BSB modules (B03, B12) that exhibited different activities into the Rnt1p switch hairpin (Supplementary Figure S5A). The switches incorporating the BSB modules (RS-B03, RS-B12) exhibited altered expression levels relative to parent that correlated with the activities observed from these modules in the context of the A02 stem (Supplementary Figure S5B). These results further support that the synthetic BSBs are effective in tuning expression levels in a variety of Rnt1p hairpin stems and devices.
In vitro characterization demonstrates that Rnt1p binding library members achieve differential activity through alterations in Rnt1p cleavage rates and affinity
We hypothesized that the variation in transcript processing and subsequent protein expression levels exhibited by the binding library is due to variations in binding affinity resulting from alterations in the BSB sequence and/or structure. To examine whether the synthetic Rnt1p binding library members exhibit differences in binding affinity to Rnt1p, we performed in vitro binding assays with purified Rnt1p. Binding reactions were ran with 20 nM of in vitro synthesized radiolabeled RNA encoding an Rnt1p hairpin and varying concentrations of purified Rnt1p. The reactions were ran in the absence of magnesium and other divalent metal ions that are essential for cleavage to allow Rnt1p to bind to the substrates without subsequent cleavage (55). Bound products were separated by non-denaturing polyacrylamide gel electrophoresis and quantified through phosphorimaging analysis (Figure 4A). We analyzed the reaction through a modified Scatchard equation in which the fraction of unbound RNA (R) to total RNA (R0) is plotted against the enzyme (E) concentration. The equation is as follows:
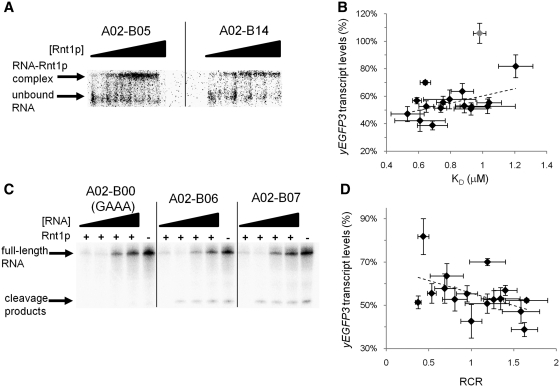
The dissociation constant, KD, for each synthetic Rnt1p hairpin was determined through this analysis method (Table 2). The data indicate that there is a moderate positive correlation (r = 0.486) between KD and in vivo gene regulatory activity (Figure 4B). While we observe several data points demonstrating similar transcript levels for different KD values, hairpins that bind less tightly to Rnt1p (i.e. higher KD) generally tend to have higher transcript levels as anticipated. The binding library has an expanded range of KD values compared to the cleavage library, due to several library members having KD values greater than those previously reported with the cleavage library (20). In vivo, we observe that most binding library members have increased gene expression levels greater than ‘parent’. In vitro, this same phenomenon is experienced as most binding library members have decreased affinity for Rnt1p. Binding library members that have decreased gene expression levels than ‘parent’ in vivo also have KD values comparable or less than ‘parent’ in vitro. However, we also observed that the mutant tetraloop control bound Rnt1p with a similar KD as the library hairpins, although cleavage was not evident (Figure 3B). It has been previously reported that Rnt1p is able to bind its substrates in inactive and active conformations in vitro (45). Therefore, it is plausible that the binding observed in these in vitro assays is due to both types of complexes with Rnt1p. Under this situation, the reported KD may not be solely related to complexes that can be processed. It has also been shown that changes in the BSB affect both binding affinity and hairpin processing by Rnt1p in vitro (36). Therefore, it is important to examine the effects of the binding library on Rnt1p processing rates.
Figure 4.
In vitro characterization of the binding library demonstrates that the observed tuning of gene regulatory activity is achieved through modulation of cleavage rates and binding affinities. (A) Representative mobility shift assays and analyses by non-denaturing polyacrylamide gel electrophoresis of two binding library members: A02–B05 and A02–B14. The top band corresponds to RNA-Rnt1p complexes; the bottom band corresponds to unbound RNA. Rnt1p was added to the following final concentrations in each reaction (left to right; in µM): 0, 0.42, 0.83, 1.25 and 1.66. (B) Correlation analysis of binding affinity (KD) and yEGFP3 transcript levels indicates a moderate positive correlation between binding affinity and gene regulatory activity. The data point for the A02–B00 (GAAA) negative control is indicated in gray. (C) Representative cleavage reaction assays and analyses by denaturing polyacrylamide gel electrophoresis on hairpins A02–B00 (GAAA), A02–B06 and A02–B07. The top band corresponds to unreacted full-length RNA; the bottom band corresponds to the three cleavage products expected from Rnt1p processing. The three cleavage products differ in size by 1 nt and cannot be resolved into individual bands under the conditions used for this assay. RNA was added to the following final concentrations in each reaction (left to right; in µM): 0.2, 0.5, 1.0 and 2.0. Reactions lacking Rnt1p were performed with 0.2 µM of RNA. (D) Correlation analysis of relative cleavage rate (RCR) and yEGFP3 transcript levels demonstrates a moderate positive correlation between cleavage rate and gene regulatory activity.
Table 2.
In vitro characterization data for the binding library
| Substrate | RCR | KD (µM) |
|---|---|---|
| A02–B00 | 1.00 ± 0.12 | 0.61 ± 0.16 |
| A02–B00 (GAAA) | 0* | 0.98 ± 0.04 |
| A02–B01 | 0.44 ± 0.07 | 1.21 ± 0.11 |
| A02–B02 | 0.71 ± 0.20 | 0.87 ± 0.07 |
| A02–B03 | 0.81 ± 0.15 | 1.03 ± 0.17 |
| A02–B04 | 1.65 ± 0.25 | 0.93 ± 0.10 |
| A02–B05 | 1.62 ± 0.15 | 0.69 ± 0.09 |
| A02–B06 | 1.40 ± 0.14 | 0.59 ± 0.03 |
| A02–B07 | 0.37 ± 0.03 | 0.74 ± 0.04 |
| A02–B08 | 1.26 ± 0.12 | 0.65 ± 0.05 |
| A02–B09 | 0.53 ± 0.05 | 0.75 ± 0.04 |
| A02–B10 | 0.95 ± 0.12 | 1.04 ± 0.08 |
| A02–B11 | 1.19 ± 0.26 | 0.93 ± 0.11 |
| A02–B12 | 1.58 ± 0.22 | 0.53 ± 0.10 |
| A02–B13 | 1.34 ± 0.23 | 0.88 ± 0.06 |
| A02–B14 | 1.19 ± 0.21 | 0.64 ± 0.03 |
| A02–B15 | 0.69 ± 0.12 | 0.79 ± 0.16 |
Immeasurable due to lack of product formation is denoted by asterisk.
We analyzed the cleavage reaction between Rnt1p and the binding library through a Michaelis–Menten model, with the substrate (S) being the hairpin transcript, the enzyme (E) being Rnt1p and the product (P) being the cleaved pieces of the transcript. Under these conditions, the following reaction occurs:
The rate of product formation (V) is modeled as:
The maximum rate of product formation (Vmax) is the product of the total enzyme concentration ([E]0) and k2. Alterations in the cleavage efficiency will have an effect on the value of k2 and thus Vmax. We performed in vitro RNA cleavage reactions with a constant concentration of purified Rnt1p against a range of in vitro synthesized radiolabeled Rnt1p hairpins to determine the relative values of k2 for each synthetic Rnt1p hairpin. Reaction products were separated by denaturing polyacrylamide gel electrophoresis and quantified through phosphorimaging analysis (Figure 4C). The resulting data were fit to the Michaelis–Menten model to calculate a relative cleavage rate (RCR), which is directly proportional to Vmax. The RCR value for A02–B00 is set to 1 and the rest of the reported values are normalized to A02–B00.
The RCR values for each synthetic Rnt1p hairpin were determined through this analysis method (Table 2). The results confirm that the mutant tetraloop control is not processed by Rnt1p in vitro (supporting in vivo observations). There is a moderate positive correlation (r = 0.480) between the measured RCR and gene regulatory activity for the synthetic Rnt1p hairpins (Figure 4D). Generally, increases in Rnt1p's ability to cleave a substrate result in greater transcript knockdown. Compared to the cleavage library (20), we observe a smaller range of RCR values with the binding library, due to the observed decreased range in transcript knockdown. In fact, we observe that cleavage library members exhibiting gene regulatory activities within the range exhibited by the binding library members have similar RCR values. However, a correlation analysis between the KD and RCR values indicates that there is no correlation between the binding affinity and relative cleavage rates for the binding library members (data not shown). Changing the BSB does result in changes in binding affinity, but it can also result in variation in the cleavage rate. For any given hairpin, the two properties contribute to the observed transcript levels. In particular, the BSB may affect the hairpin's ability to form active or inactive complexes or the BSB may affect the hairpin's processing rate.
DISCUSSION
We utilized a cell-based library screening approach to develop a set of synthetic BSB sequences to modulate the gene regulatory activity of engineered Rnt1p hairpins. Previous in vitro studies showed that the BSB contains nucleotides critical to Rnt1p binding as mutations in the region resulted in reduced affinity (36). However, this earlier work examined a relatively small sequence space for the BSB region and did not link the in vitro processing activities to in vivo gene regulatory activities. The in vitro studies established a consensus BSB structure, where three Watson–Crick base-pairs were required immediately below the tetraloop followed by another base-pair that could include the wobble guanine–uracil pair. Based on these reported structural requirements, we estimated that a small percentage (~0.6%) of the randomized BSB library would contain hairpins cleavable by Rnt1p. Therefore, a high-throughput FACS-based screen was employed to enrich the library for hairpins resulting in reduced GFP fluorescence. The BSB library was designed in the context of a synthetic CEB from the cleavage library that produced the greatest amount of knockdown (C13) (20). This design biased the library such that any positive hits would exhibit the lowest possible expression levels to enhance the separation, and thus the selection, of the population of cells containing active hairpins versus the larger population containing inactive hairpins (~99.4% of the population). In total, 16 unique BSBs were identified. However, in contrast to previous in vitro work (36), several of the synthetic BSBs did not contain a base-pair in the fourth position from the tetraloop, suggesting that this structural requirement is relaxed in vivo.
The selected BSB sequences were further characterized in the context of a CEB that exhibited weaker gene regulatory activity (A02) (20) to better resolve differences in the BSB activities. The regulatory activities of the BSB sequences are distributed across an intermediate range of 25–75% relative protein levels with the majority exhibiting activities in the range of 25–45%. Interestingly, the binding library exhibited a decreased range of activity relative to the cleavage library (20). The relative effects of each region on Rnt1p-based gene regulatory activity would not have been predicted a priori from prior in vitro characterization studies. Our results suggest that the binding library may be more appropriate for the fine-tuning of gene expression. For example, due to its wide range of gene expression, the cleavage library can be employed initially to first identify regulatory ranges of interest. More focused regulatory activities can then be explored through implementation of the synthetic BSBs with appropriate synthetic CEBs, which will allow a more careful searching of activity space around a selected CEB. As such, the combinatorial application of the synthetic BSB and CEB elements can be used to extend and tune the regulatory range accessible through the engineered Rnt1p hairpins. To demonstrate the ability to combine BSB and CEB sequences, we examined the synthetic BSB sequences in the context of two synthetic CEBs generated in the cleavage library. Our experimental results show that the synthetic CEB and BSB elements act as modular units, that the BSBs maintain their activity under the context of different CEBs, and that the proportional deviation from the ‘parent’ BSB is determined by the strength of the CEB (Figure 3D and E).
In vitro characterization studies determined the relationship between binding affinity, cleavage rate and gene-regulatory activity for the binding library. Changes in the BSB sequence are expected to result in changes in Rnt1p cleavage rate as well as to affect Rnt1p binding (36). We observed a moderate correlation between the binding affinity and transcript levels and between the cleavage rate and transcript levels for the binding library (Figure 4B and D). In contrast, although slight changes in affinity were detected with the cleavage library, these changes were not correlated with gene regulatory activity (20). However, there was a stronger correlation observed between cleavage rate and transcript levels for the cleavage library. The data indicate that nucleotide modifications in the BSB cause changes in affinity and cleavage rate; however, the exact contribution of each of these variables to the observed gene regulatory activity is unclear. In addition, Rnt1p is known to bind in active and inactive complexes where the inactive complex is magnesium-independent (45,56). The inactive complex may be more prevalent in the binding assay due to the absence of magnesium in the reaction buffer, which is also critical to the proper folding of RNA molecules (57). The changes in KD are reflective of how much total RNA is bound regardless of conformation. Thus, it is possible that the changes to the BSB are affecting the partitioning between inactive and active states.
This work extends the regulatory capacity of the first set of post-transcriptional stability control elements in yeast by developing a set of BSB modules that can be integrated with a previously described set of CEB modules to rationally design synthetic Rnt1p substrates. With 16 CEB and 16 BSB modules developed, 256 different Rnt1p hairpins can be generated to achieve finely-tuned levels of gene expression in S. cerevisiae. The ability to tune gene expression levels has been demonstrated to be critical to the optimization of encoded behaviors in complex genetic networks in synthetic biology (58–61) and in metabolic engineering, where in the latter applications such tuning strategies play a key role in minimizing metabolic burden (1–2) and accumulation of toxic intermediates (3,5) and redirecting metabolic flux from native pathways required for cell growth and viability (8). In addition, having well-characterized sets of modules that correspond to different regions of the control element is critical to extending this modular design strategy to more complex sensing-actuation devices (38). For example, the BSB modules become the critical tuning modules in this device architecture, as the sensor component is located within the CEB region and cannot be replaced with the synthetic CEB modules (Supplementary Figure S5). Thus, in combination with other RNA components as well as engineered or native promoter systems, the engineered Rnt1p control modules provide a powerful tool for programming genetic regulatory networks in yeast, and thus advancing the application of this cellular chassis in biomanufacturing and biosynthesis processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation (CAREER award CBET-0917705 to C.D.S.); Alfred P. Sloan Foundation, fellowship (to C.D.S.). Funding for open access charge: National Science Foundation (CBET-0917705).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Liang and A. Chang for assistance with FACS and for providing the pCS1585 and pCS1748 plasmids, K. Hoff for assistance in the expression and purification of Rnt1p and S. Bastian and F. H. Arnold for assistance in sonication and FPLC.
REFERENCES
- 1.Jin YS, Ni H, Laplaza JM, Jeffries TW. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl. Environ. Microbiol. 2003;69:495–503. doi: 10.1128/AEM.69.1.495-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KL, Kim SW, Keasling JD. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab. Eng. 2000;2:328–338. doi: 10.1006/mben.2000.0161. [DOI] [PubMed] [Google Scholar]
- 3.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 4.Zhu MM, Lawman PD, Cameron DC. Improving 1,3-propanediol production from glycerol in a metabolically engineered Escherichia coli by reducing accumulation of sn-glycerol-3-phosphate. Biotechnol. Prog. 2002;18:694–699. doi: 10.1021/bp020281+. [DOI] [PubMed] [Google Scholar]
- 5.Zhu MM, Skraly FA, Cameron DC. Accumulation of methylglyoxal in anaerobically grown Escherichia coli and its detoxification by expression of the Pseudomonas putida glyoxalase I gene. Metab. Eng. 2001;3:218–225. doi: 10.1006/mben.2001.0186. [DOI] [PubMed] [Google Scholar]
- 6.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 8.Paradise EM, Kirby J, Chan R, Keasling JD. Redirection of flux through the FPP branch-point in Saccharomyces cerevisiae by down-regulating squalene synthase. Biotechnol. Bioeng. 2008;100:371–378. doi: 10.1002/bit.21766. [DOI] [PubMed] [Google Scholar]
- 9.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc. Natl Acad. Sci. USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U, Stephanopoulos G. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006;72:5266–5273. doi: 10.1128/AEM.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins KM, Smolke CD. The regulatory roles of the galactose permease and kinase in the induction response of the GAL network in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:13485–13492. doi: 10.1074/jbc.M512317200. [DOI] [PubMed] [Google Scholar]
- 13.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrier TA, Keasling JD. Library of synthetic 5′ secondary structures to manipulate mRNA stability in Escherichia coli. Biotechnol. Prog. 1999;15:58–64. doi: 10.1021/bp9801143. [DOI] [PubMed] [Google Scholar]
- 18.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 19.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 20.Babiskin AH, Smolke CD. A synthetic library of RNA control modules for predictable tuning of gene expression in yeast. Mol. Syst. Biol. 2011;7:471. doi: 10.1038/msb.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 22.Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16:1313–1323. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen HT, Dieterich A, Athenstaedt K, Truong NH, Stahl U, Nevoigt E. Engineering of Saccharomyces cerevisiae for the production of l-glycerol 3-phosphate. Metab. Eng. 2004;6:155–163. doi: 10.1016/j.ymben.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Ostergaard S, Olsson L, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2000;64:34–50. doi: 10.1128/mmbr.64.1.34-50.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 27.Szczebara FM, Chandelier C, Villeret C, Masurel A, Bourot S, Duport C, Blanchard S, Groisillier A, Testet E, Costaglioli P, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- 28.Veen M, Lang C. Production of lipid compounds in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2004;63:635–646. doi: 10.1007/s00253-003-1456-7. [DOI] [PubMed] [Google Scholar]
- 29.Louis M, Becskei A. Binary and graded responses in gene networks. Sci. STKE. 2002;2002:pe33. doi: 10.1126/stke.2002.143.pe33. [DOI] [PubMed] [Google Scholar]
- 30.Lautz T, Stahl U, Lang C. The human c-fos and TNFalpha AU-rich elements show different effects on mRNA abundance and protein expression depending on the reporter in the yeast Pichia pastoris. Yeast. 2010;27:1–9. doi: 10.1002/yea.1726. [DOI] [PubMed] [Google Scholar]
- 31.Vasudevan S, Peltz SW. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell. 2001;7:1191–1200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhou W, Edelman GM, Mauro VP. Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl Acad. Sci. USA. 2001;98:1531–1536. doi: 10.1073/pnas.98.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Edelman GM, Mauro VP. Isolation and identification of short nucleotide sequences that affect translation initiation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2003;100:4457–4462. doi: 10.1073/pnas.0437993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl Acad. Sci. USA. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H, Yang PK, Butcher SE, Kang S, Chanfreau G, Feigon J. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 2001;20:7240–7249. doi: 10.1093/emboj/20.24.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamontagne B, Ghazal G, Lebars I, Yoshizawa S, Fourmy D, Elela SA. Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 2003;327:985–1000. doi: 10.1016/s0022-2836(03)00231-6. [DOI] [PubMed] [Google Scholar]
- 37.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babiskin AH, Smolke CD. Engineering ligand-responsive RNA controllers in yeast through the assembly of RNase III tuning modules. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr090. doi:10.1093/nar/gkr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 40.Gietz R, Woods R. In: Guide to Yeast Genetics and Molecular and Cell Biology, Part B. Guthrie C, Fink G, editors. Vol. 350. San Diego, CA: Academic; 2002. pp. 87–96. [Google Scholar]
- 41.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 42.Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng R, Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1980;77:3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamontagne B, Elela SA. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 2001;342:159–167. doi: 10.1016/s0076-6879(01)42543-2. [DOI] [PubMed] [Google Scholar]
- 45.Lamontagne B, Elela SA. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 2004;279:2231–2241. doi: 10.1074/jbc.M309324200. [DOI] [PubMed] [Google Scholar]
- 46.Lamontagne B, Hannoush RN, Damha MJ, Abou Elela S. Molecular requirements for duplex recognition and cleavage by eukaryotic RNase III: discovery of an RNA-dependent DNA cleavage activity of yeast Rnt1p. J. Mol. Biol. 2004;338:401–418. doi: 10.1016/j.jmb.2004.02.059. [DOI] [PubMed] [Google Scholar]
- 47.Chanfreau G, Elela SA, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elela SA, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 50.Ge D, Lamontagne B, Elela SA. RNase III-mediated silencing of a glucose-dependent repressor in yeast. Curr. Biol. 2005;15:140–145. doi: 10.1016/j.cub.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 52.Lee A, Henras AK, Chanfreau G. Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol. Cell. 2005;19:39–51. doi: 10.1016/j.molcel.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 53.Lamontagne B, Abou Elela S. Short RNA guides cleavage by eukaryotic RNase III. PLoS ONE. 2007;2:e472. doi: 10.1371/journal.pone.0000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larose S, Laterreur N, Ghazal G, Gagnon J, Wellinger RJ, Elela SA. RNase III-dependent regulation of yeast telomerase. J. Biol. Chem. 2007;282:4373–4381. doi: 10.1074/jbc.M607145200. [DOI] [PubMed] [Google Scholar]
- 55.Lamontagne B, Tremblay A, Abou Elela S. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 2000;20:1104–1115. doi: 10.1128/mcb.20.4.1104-1115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavoie M, Abou Elela S. Yeast ribonuclease III uses a network of multiple hydrogen bonds for RNA binding and cleavage. Biochemistry. 2008;47:8514–8526. doi: 10.1021/bi800238u. [DOI] [PubMed] [Google Scholar]
- 57.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 58.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 60.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 61.Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl Acad. Sci. USA. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.