Abstract
Hepatitis B virus (HBV) pregenomic RNA contains a hairpin structure (ϵ) located in the preCore region, essential for viral replication. ϵ stability is enhanced by the presence of preCore variants and ϵ is recognized by the HBV polymerase (Pol). Mutations in the retrotranscriptase domain (YMDD) of Pol are associated with treatment resistance. The aim of this study was to analyze the preCore region and YMDD motif by ultra-deep pyrosequencing (UDPS). To evaluate the UDPS error rate, an internal control sequence was inserted in the amplicon. A newly developed technique enabled simultaneous analysis of the preCore region and Pol in the same viral genome, as well as the conserved sequence of the internal control. Nucleotide errors in HindIII yielded a UDPS error rate <0.05%. UDPS study confirmed the possibility of simultaneous detection of preCore and YMDD mutations, and demonstrated the complexity of the HBV quasispecies and cooperation between viruses. Thermodynamic stability of the ϵ signal was found to be the main constraint for selecting main preCore mutations. Analysis of ϵ-signal variability suggested the essential nature of the ϵ structural motif and that certain nucleotides may be involved in ϵ signal functions.
INTRODUCTION
Hepatitis B virus (HBV) is a double-stranded DNA virus, whose genome (3.2 kb) contains four Open Reading Frames (OPFs) encoding seven proteins: surface antigens (preS1, preS2 and HBsAg), polymerase (Pol), core protein (HBcAg), X protein (HBx) and the soluble ‘e’ antigen (HBeAg). During the viral cycle, HBV-DNA conformation shows different phases: relaxed-circular, partially double-stranded DNA in circulating viruses and covalently closed circular DNA (cccDNA) within the hepatocyte nucleus. cccDNA acts as a mini-chromosome within hepatocytes and transcribes five messenger RNA: preS1, preS2, X, preCore and pregenomic RNA (pgRNA). pgRNA is a greater than full-length HBV genome (3.5 kb) that codes for the Pol and HBcAg proteins. Pol has two catalytic activities, reverse transcriptase (RT) and RNaseH, and contains a spacer sequence and a terminal protein (1).
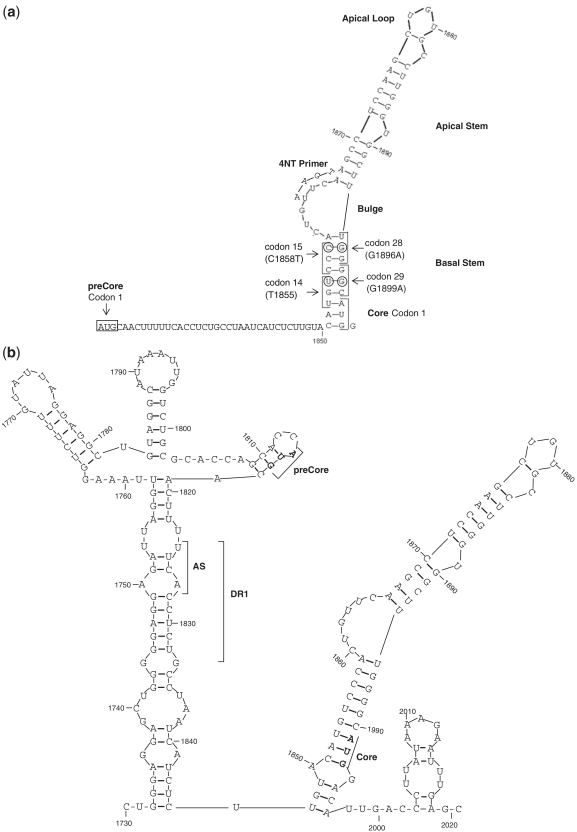
In both the 5′- and 3′-ends of pgRNA, there is a redundant sequence of 60 nt that adopts a stem loop structure (Figure 1a and b) and acts as encapsidation signal (ϵ). The 5′ sequence is crucial for Pol protein recognition and binding to the 5′ ϵ structure (2). Pol triggers pgRNA encapsidation by core proteins, and initiates reverse transcription (3). Pol employs an N terminal tyrosine residue of the terminal protein domain as a primer to generate the first 4 nt (4 nt primer) of the DNA negative strand. This 4 nt primer is covalently attached to Pol and uses the bulge of the ϵ signal as template (Figure 1a) (4). The 4 nt primer and Pol protein complex switches from the 5′- to 3′-end of pgRNA and anneals to a 4-nt acceptor site within the 3′ copy of direct repeat region 1 (Figure 1b) (5).
Figure 1.
(a) Stem-loop structure of the 5′ HBV pgRNA. Main structural motifs (loops and stems), main codons studied (1, 14, 15, 28 and 29 of preCore and 1 of Core genes), and the location of 4-nt primer annealing in the 5′ pgRNA are indicated in the figure. (b) Stem-loop structure of the 3′ HBV pgRNA. The acceptor site (AS), direct repeat region 1 (DR1), and the preCore and Core start codons are indicated in the figure.
pgRNA and preCore mRNA share the same preCore region, but code for different antigens: HBcAg (starting at Position 1901 of the HBV genome) and HBeAg (starting at Position 1814), respectively. Several preCore region mutations abolish HBeAg expression. The most common is G to A at Position 1896 (G1896A), which changes a tryptophan (W) in codon 28 (TG1896G) to an amber stop codon (TA1896G): pcW28stop. This substitution is often associated with the G1899A change in codon 29 (pcG29D) (6). Other minor mutations have been observed in codon 2 (CAA to TAA, Q2stop) and in codon 1 (7). The pcW28stop change is reported to be responsible for 95% of HBeAg-negative patients in our area, mainly in association with viral genotype D (8,9). The G1896A and G1899A mutations have been associated with secondary ϵ structure stability and therefore, with viral replication (8,10–12).
The main mutations conferring resistance to nucleoside analogues, such as lamivudine (LMV), are predominantly detected in the RT catalytic motif, YM204DD, in which methionine (M) is changed to valine (V) or isoleucine (I) (rtM204V and rtM204I) (13,14). Prolonged LMV therapy is associated with a high probability (>70% after 5 years) of resistance to treatment (15). After the first year of therapy, anti-HBe-positive patients have a significantly greater response than HBeAg-positive cases; however, long-term response rates are compromised by the reappearance of HBV DNA in serum (16). Several factors can have an effect on the selection rates of LMV-resistant mutants: ethnic background, viral load, HBV genotype, core promoter variants and preCore variants (17).
Ultra-deep pyrosequencing (UDPS) by GS-FLX platform (Roche) has been used to detect clinically relevant minority drug-resistant variants in treatment-naïve and experienced HIV-1 (18,19) and HBV patients, thereby confirming its ability to identify individual, well-known mutations present in the Pol and HBsAg genes at frequencies of 0.1–1% of the HBV quasispecies (20,21). UDPS by GS-FLX platform is currently the only technology that allows simultaneous quantitative analysis of thousands of clonally amplified sequences of 200–250 nt in length for standard GS-FLX (longer fragments since last year).
The aim of the present study was to analyze by UDPS the HBV preCore region (thermodynamic structural restriction and variability of essential structural motifs of the ϵ signal) and its association with the RT catalytic YMDD motif variants, with analysis of both regions in the same sequence. However, these two regions are separated by a distance of 1 kb in the HBV genome, making it difficult to simultaneously study both regions by classic cloning methodology (22,23). Therefore, our methodological hypothesis was based on executing a PCR that would encompass the preCore and YMDD regions, using primers that include the HindIII site on their 5′-ends. After induction of intramolecular ligation, a circular molecule was produced, which included the YMDD Pol motif and the preCore region flanking a HindIII sequence. The integrity of the restriction site sequence was used as the internal control to verify the fidelity of UDPS analysis.
PATIENTS AND METHODS
Patients
Seven chronically infected HBV patients, all males and with a mean age of 36 years (range 17–50), were included in the study. Patients were selected because they had complete clinical documentation and known viral characteristics by conventional techniques. They had been treated with LMV for a mean of 25 months because of high HBV DNA levels and elevated hepatic biochemical markers. Two patients were HBeAg-positive at baseline and e antigen loss did not occur during antiviral treatment. Genotype changes were not observed during the study period. Baseline biochemical and serological markers, and treatment characteristics are described in Table 1. All patients tested negative for HCV and HIV markers. HBV serology, HBV DNA and LMV-resistant mutations were tested every 3–6 months. The circular construct used to analyze the preCore and YMDD was obtained from four baseline samples (Patients 1–4) and from four samples corresponding to a time of viral breakthrough with mutations in the YMDD motif (Patients 4–7).
Table 1.
Baseline biochemical and serological markers, and treatment characteristics of the seven patients included in the study
| ID | Baseline |
Lamivudine treatment |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HBV DNA (IU/ml) | HBeAg | ALT (IU/ml) | HBV genotype | Type response | Months | HBV DNA (IU/ml) | ALT (IU/ml) | Type mutation | Rescue treatment | |
| 1 | >108 | N | 392 | D | VBK | 48 | 3.6 × 105 | 31 | M204V | LAM+ADV |
| 2 | 6.6 × 106 | N | 167 | A2 | SVR | 24 | <20 | 30 | – | – |
| 3 | 1.2 × 106 | N | 54 | A2 | SVR | 24 | <20 | 33 | – | – |
| 4 | >108 | P | 52 | D | VBK | 12 | 6.2 × 106 | 281 | M204IV | LAM+TDF |
| 5 | 3.3 × 106 | N | 78 | A2 | VBK | 34 | 8.3 × 104 | 401 | M204MIV | LAM+ETV |
| 6 | >108 | P | 214 | A2 | VBK | 24 | 1.3 × 107 | 93 | M204V | LAM+TDF |
| 7 | 2 × 106 | N | 117 | D | VBK | 12 | 2.2 × 104 | 103 | L180LM | LAM+ADV |
SVR, sustained virologic response.
Serology and HBV DNA assays
Serological markers for HBV (HBsAg, HBeAg and anti-HBe), HCV (anti-HCV), hepatitis delta (anti-HDV) and HIV (anti-HIV) were tested by commercial enzyme immunoassays. HBV-DNA was quantified using real-time PCR with a detection limit of 20 IU/ml (COBAS TaqMan HBV V2.0, Roche Diagnostics GmbH, Mannheim, Germany). Main preCore mutations were detected by Sanger sequencing, as previously described (8). HBV genotype and mutations conferring treatment resistance were detected with LiPA (INNO-LiPA, Innogenetics, Ghent, Belgium) (sensitivity 1–5%) and direct sequencing (sensitivity 20%) (24).
Generation of a circular construct containing the YMDD motif and preCore region
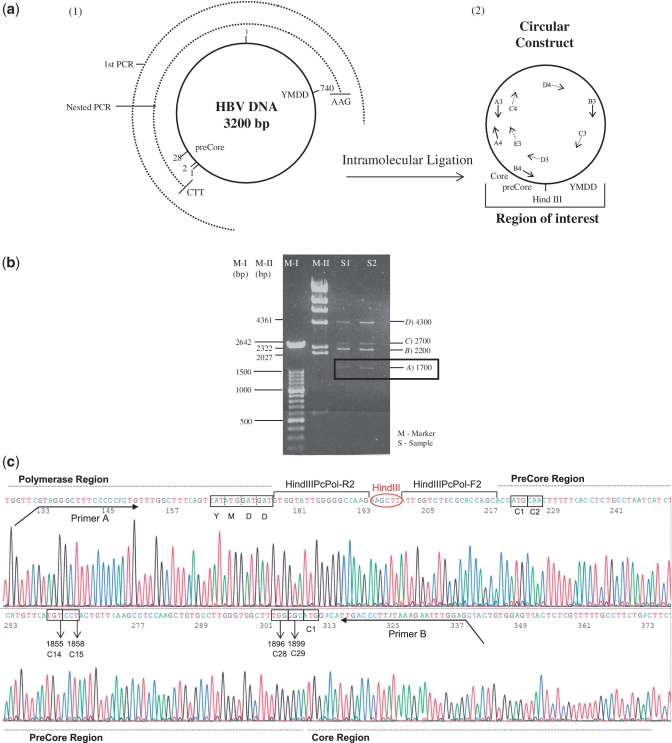
HBV DNA was extracted from 200 μl of serum with the QIAamp DNA MiniKit (QIAGEN, Hilden, Germany). With the aim of analyzing greatly separated regions in the same genome, a 2.7-kb HBV DNA fragment including the YMDD Pol motif and preCore region was PCR amplified (Figure 2a), using the forward primer F1 (Position 1703) and reverse primer R1 (Position 1018) (Table 2). The PCR reaction mix included 200 µM of dNTPs and 400 nM of each primer, and the method was performed as follows: 30 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 3 min, followed by 10 cycles at 94°C for 30 s, 58.6°C for 30 s and 72°C for 3 min and a final hold at 72°C for 7 min. The amplicon obtained was amplified by nested PCR with primers that included the HindIII restriction site sequence (AAGCTT) at the 5′-end (forward HindPcPol-F2, Position 1792, and reverse HindPcPol-R2, Position 764) (Table 2) under the same conditions as the first PCR run, yielding a 2194-kb product in genotype A2 samples and a 2155-kb product in genotype D samples. To minimize amplification errors, Pfu UltraII (Stratagene, Agilent Technologies, La Jolla, USA) polymerase was used in both PCR runs. The nested amplicon was purified through 0.9% agarose gel, digested for 1 h at 37°C with HindIII (NEB, Ipswich, USA), and purified again through 0.9% agarose gel. The next step involved ligation of both HindIII ends of the same fragment (intramolecular ligation) so that the two regions of interest would come close enough in the same molecule to be analyzed by UDPS (Figure 2a). Optimal results were obtained with 0.5 ng/µ1 of the HBV DNA-PCR fragment, and 1 U of T4 DNA ligase (Roche Diagnostics GmbH). The ligation product was loaded onto 0.9% agarose gel; the band pattern obtained is shown in Figure 2b. The band corresponding to the HindIII circular construct (apparent size ~1700 bp) (band a in Figure 2b) was cut from the gel and purified. The identity of the circular constructs was confirmed by sequencing two overlapping PCR amplicons that cover the complete circular construct sequence: a 1.2-kb amplicon obtained with primers A3 and B3 and sequenced with C3, D3 and E3 and a 1.8-kb amplicon obtained with primers A4 and B4 and sequenced with A4, C4, D4 and C3 (primers described in Table 2). The fragment obtained from sequencing the 1.3-kb amplicon, which includes the preCore region, the YMDD motif and the HindIII sequences with C3 is shown in Figure 2c.
Figure 2.
(a) Schematic representation of the intramolecular ligation technique to obtain the preCore and YMDD Pol motif close to each other. (1) The circle in a continuous line represents the complete HBV genome (3200 bp). The semicircles in discontinuous lines represent the first (first PCR, 2700 bp) and the nested (2194 bp for genotype A2 sequences and 2155 bp for genotype D sequences) PCR products. (2) The product obtained after intramolecular ligation and primers used to sequence the circular molecule (Table 2) are indicated in the figure. (b) Agarose gel obtained after intramolecular digestion. Two markers (M-I and M-II) were loaded in the gel; band lengths are shown at the left side of the image. Two samples (S1 and S2) were loaded and four bands were detected; band lengths are indicated at the right side of the image. A band with an apparent size of 1700 bp was the circular molecule, with the preCore and YMDD Pol motif close to each other. Band B was the nested PCR product without ligation, and band D were dimers of nested PCR products without ligation. Band C corresponded to the ligated dimers. B, C and D bands were not used, because the main interest was to analyze regions from the same genome. (c) Sequence of the circular construct including the preCore region, YMDD polymerase motif (in box), and HindIII sequence. PreCore (1, 2, 14, 15, 28 and 29) and Core (1) codons analyzed in the text are also enclosed in boxes.
Table 2.
Sequences of primers used in the study
| Name | Position of the primer and direction in HBV genome (bold characters) | Sequence (5′–3′) |
|---|---|---|
| F1 | 1703, Fw | YATAAGAGGACTCTTGGACT |
| R1 | 1018, Rv | GCAAANCCCAAAAGACCCA |
| HindPcPol-F2 | 1792, Fw | TACAAGCTTATTGGTCTGCGCACCAGC |
| HindPcPol-R2 | 764, Rv | TACAAGCTTCTTGGCCCCCAATACCAC |
| A3 | 2489, Rv | CCCMGTAAARTTYCCCACCTTAT |
| B3 | 245, Fw | GTCTAGACTCGTGGTGGACTTCTCTC |
| C3 | 550, Fw | ATGTWTCCCTCWTGTTGCT |
| D3 | 1864, Fw | TCAAGCCTCCAAGCTGTGCC |
| E3 | 2267, Fw | GGAGTGTGGATTCGCACT |
| A4 | 2270, Fw | GTGTGGATTCGCACTCCTCCAG |
| B4 | 1927, Rv | AAATTCTTTATAAGGGTCAATGT |
| C4 | 2817, Fw | TCACCATATTCTTGGGAACA |
| D4 | 57, Fw | TGCTGGTGGCTCCAGTTC |
| UDPS-Primer A | 703, Fw | GCCTCCCTCGCGCCATCAGGTAGGGCTTTCCCCCACTG |
| UDPS-Primer B | 1931, Rv | GCCTTGCCAGCCCGCTCAGCTCCAAATTCTTTATAWGGGTCRA |
| Fw T1855C | 1834, Fw | CTAATCATCTCTTGTACATGcCCCRCTGTTCAAGCCTCCAAGCTG |
| Rv T1855C | 1879, Rv | CAGCTTGGAGGCTTGAACAGYGGGgCATGTACAAGAGATGATTAG |
| C3-Rv | 569, Rv | AGCAACAWGAGGGAWACAT |
| pTriEx-Fw | – | TCTGCGCTCTGCTGAAGCCAGTTA |
| pTriEx-Rv | – | ACGATCGGAGGACCGAAGGAGCT |
The position and direction of the 5′ primer in the HBV genome (bold characters in the sequence), and the mutagenized position (bold lower cases) are indicated
Simultaneous UDPS analysis of the YMDD motif and preCore region
A 207-bp nucleotide fragment was PCR amplified from the DNA recovered from band a (Figure 2b). All HBV-specific primers included 5′ adaptors as binding sites for the pyrosequencing reaction (left 5′ GCCTCCCTCGCGCCA, right 5′ GCCTTGCCAGCCCGC; both ending with the TCAG barcode). The complete sequence of UDPS-Primer A as forward and UDPS-Primer B as reverse is shown in Table 2. Briefly, PCR was performed using 1× Pfu UltraII polymerase buffer, 10 mM of each dNTP, 20 pmol of forward and reverse primers and 2.5 U of Pfu UltraII DNA polymerase (Stratagene, Agilent Technologies) in a final volume of 100 µl. After a single denaturation step of 2 min at 95°C, samples underwent 30 cycles for 30 s at 95°C, 30 s at 60°C and 2 min at 72°C, and a single final 10-min step at 72°C. Before UDPS, the quality and length of the HBV DNA amplicons was verified with the Agilent 2100 bioanalyzer (Agilent Life Science, Santa Clara, CA, USA). The 207-bp fragment, which included 163 bp of the HBV-DNA sequence, underwent UDPS with the 454 Life Science platform (GS-FLX, Roche Applied Science), according to the manufacturer’s protocol. Base calls and quality scores were determined using the GS-Amplicon Variant Analysis software (included in the GS-FLX platform).
Algorithm for interpreting sequence data
Data accuracy was validated according to previously reported procedures (18,25,26). Briefly, reads with insertion–deletions, reads in which the primer did not match the reference primer by at least 75%, those that did not reach a fixed length and those containing ambiguous calls were discarded. A complementary reverse sequence was obtained from the reverse sequence. The program selected nucleotide sequences that passed the filter, and the sequences were translated to amino acids for the polymerase and preCore regions (both regions are in-frame in the circular construct). Several files were obtained and a matrix of the frequency of changes at each amino acid position in comparison with the master from each patient was used. As was mentioned above, the circular construct used as the target for UDPS analysis was the result of intramolecular ligation of the HindIII 5′- and 3′-ends. Therefore, the integrity of this 6-bp restriction enzyme sequence (AGGCCT) was a requirement for correct ligation, and any nucleotide substitution in this sequence would be a UDPS error. For this reason, instead of using a more complex Poisson filter (19,25,26), which needs UDPS processing of additional clonal sequences (external control), the changes detected in this 6-bp sequence were used to establish the cut-off fidelity of the UDPS process (internal control).
Analysis of RNA folding stability in the HBV pgRNA encapsidation signal
Computer models of RNA folding and structure representations were obtained from the RNA Structure Fold program (version 5.3), developed by Dr David H. Mathews (27,28). All calculations were performed at a temperature of 37°C. In keeping with Dr Mathews’ kindly provided suggestion, a script adaptation of the RNA Structure Fold program (by command line interfaces) was used to calculate the lowest free energy of the ϵ structure. Briefly, each nucleotide sequence was automatically converted to a ‘seq’ file, with the input format required for the program. A ‘bat’ file was created for each ‘seq’ file, enabling execution of the Fold module of the program for each sequence and creating the output ‘ct’ files, which contained the free energy of the structure. The circular construct analyzed included nucleotides from Positions 1814 to 1912, corresponding to the main sequence of the 5′ ϵ signal (8,11,12). The sequences obtained were used to calculate the epsilon energy of the RNA ensemble. Because the epsilon energy data are not normally distributed, between-group comparisons were done using a permutation test based on the standard t score for two groups. The Monte Carlo approach to compute P-values was carried out, with n = 2000 permutated samples. P-values in these comparisons were corrected for multiple testing problems following the false discovery rate procedure. The standard RNA-Structure Fold (5.3) program was used to predict the secondary structures of the 5′ (Figure 1a) and 3′ (Figure 1b) (Position 1730–1930) ϵ signals. Both predicted structures showed a pseudo-triloop at the edge of the apical stem, as has been demonstrated by magnetic resonance (MR) analysis (29).
Phenotyping, mutagenesis and cell culture
A more than full-length HBV (1.1 genome unit) (able to produce functional replicative pgRNA) from a patient chronically infected with HBV genotype A was cloned in pTriEx-mod vector (kindly provided by Dr F. Zoulim), as described by Durantel et al. (30). This construct was used to simulate HBV variants by site-directed mutagenesis. In the present study, a thymine to cytosine mutation at Position 1855 (T1855C) was generated, using the Multi-Site-Directed Mutagenesis Kit (Agilent Technologies), following the manufacturer’s instructions. This is a silent change that does not induce an amino acid substitution (both code for a cysteine). The forward primer was Fw-T1855C, and the reverse primer Rv-T1855C (Table 2). Because of the 60-nt redundancy in HBV pgRNA, Position 1855 is repeated at the 5′- and 3′-ends. Thus, the mutagenesis process yielded three different constructs that contained the T1855C change at the 5′-end, the 3′-end or simultaneously at both ends. Generation of these mutations was confirmed by direct sequencing of the clones with primers annealing pTriEx-mod-1.1 HBV just before the HBV ligation (promoter-Fw for the 5′-end, and C1D1-Rv for the 3′-end, shown in Table 2).
The three pTriEx-mod-1.1 HBV constructs were transfected in Huh7 human hepatoma cells cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% calf serum. Plasmid transfections were performed using Fugene-HD (Roche Diagnostics GmbH). Cells were seeded in 12-well plates at a density of 106 cells/ml and were transfected at 80–90% confluence, following the manufacturer’s instructions. The supernatant was used to quantify production of HBsAg (Architecht, Abbot Diagnostics, Sigo, Ireland), HBeAg (Vitros, Ortho-Clinical Diagnostics, High Wycombe, UK) and HBV DNA (COBAS TaqMan HBV V2.0, Roche Diagnostics GmbH). HBV DNA was extracted from the supernatant (QiagenAMP DNA Mini Kit, Qiagen, Hilden, Germany) according to the manufacturer’s instructions and used to evaluate HBV DNA production. The HBV viral particle origin of HBV-DNA detected in culture supernatants was confirmed by differential PCR. Briefly, consecutive dilutions (1/1, 1/10, 1/102, 1/103 and 1/104) of extracted DNA were performed, and specific regions of HBV and pTriEx-mod were amplified, using the paired primers C4 and C3-Rv for HBV, and pTriEx-Fw and pTriEx-Rv for a 592-bp region of the plasmid (Table 2). Two negative controls were performed in the study, one that contained only Fugene, which enabled evaluation of Huh7 replication in the presence of the transfection reagent, and another that contained only HBV DNA without reagent. HBsAg and HBeAg tested negative, but quantification of HBV DNA was positive (2.15 × 102 UI/ml). Differential PCR analysis, using primers for both HBV and pTriEx amplification, were positive only at a dilution of 1/10. HBV PCR of wild-type and mutant clones remained positive up to 1/103 to 1/104 dilutions, indicating production of HBV DNA resulting from HBV transfection of Huh7 cells. Amplification of the pTriEx region in HBV transfection reactions was only detectable at a dilution up to 1/10; in further dilutions, PCR was negative.
RESULTS
HindIII restriction sequence as internal control of UDPS
In the 200 817 (Table 3) sequences analyzed, only 189 nt changes were observed at the HindIII restriction enzyme site. Sixteen HindIII mutated sequences were found at a frequency ranging from 1 to 24 (average, 22) and all carried only one amino acid change. The single nucleotide error rate was 1.6 × 10−4 and the codon error rate was 4.7 × 10−4. These values were considered the UDPS error rates, and the cut-off for the sequence analysis was set at 0.03% (twice the nucleotide error rate).
Table 3.
Percentage of changes in the polymerase, precore and core positions of each sample processed
| Sample | Pt ID | HBeAg | HBV Genotype | Total sequences | Amino acid at codon 204 of polymerase (YMDD motif) |
preCore Region |
Core Region |
Free energy (kcal/mol) in consensus sequence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Codon 1 |
Codon 2 |
Codon 28 |
Codon 1 |
||||||||||||||||
| M | Va | Ia | Other | WT (M) | Otherb | WT (Q) | Stopb | Otherb | WT (W) | Stopb | Otherb | WT (M) | Other | ||||||
| Baseline | 1 | N | D | 19 421 | 99.70 | 0.11 | 0.09 | 0.09 | 99.73 | 0.27 | 99.80 | 0.03 | 0.17 | 0.14 | 99.80 | 0.06 | 99.26 | 0.74 | −26.1 |
| Baseline | 2 | N | A2 | 29 024 | 99.63 | 0.08 | 0.20 | 0.10 | 0.13 | 99.87 | 99.82 | 0.02 | 0.16 | 99.64 | 0.25 | 0.11 | 99.53 | 0.47 | −27.6 |
| Baseline | 3 | N | A2 | 15 585 | 99.89 | 0.02 | 0.05 | 0.04 | 99.88 | 0.13 | 99.94 | 0.02 | 0.04 | 0.02 | 99.96 | 0.02 | 99.89 | 0.11 | −25.9 |
| Baseline | 4 | P | D | 34 182 | 99.33 | 0.08 | 0.47 | 0.12 | 99.48 | 0.52 | 99.71 | 0.06 | 0.23 | 91.59 | 8.31 | 0.10 | 99.49 | 0.51 | −25.2 |
| VBK | 4 | P | D | 25 175 | 0.25 | 93.90 | 5.63 | 0.23 | 99.51 | 0.49 | 99.73 | 0.11 | 0.16 | 98.56 | 0.21 | 0.23 | 99.59 | 0.41 | −25.2 |
| VBK | 5c | N | A2 | 20 683 | 0.08 | 0.20 | 99.46 | 0.27 | 99.54 | 0.46 | 99.65 | 0.14 | 0.21 | 0.10 | 99.85 | 0.05 | 99.09 | 0.91 | −25.5 |
| VBK | 6 | P | A2 | 25 175 | 1.06 | 98.69 | 0.01 | 0.24 | 90.19 | 9.81 | 99.75 | 0.07 | 0.18 | 97.61 | 1.31 | 1.08 | 99.21 | 0.79 | −27.1 |
| VBK | 7c | N | D | 31 572 | 0.07 | 99.69 | 0.00 | 0.24 | 99.49 | 0.51 | 99.79 | 0.09 | 0.12 | 0.14 | 99.70 | 0.16 | 99.30 | 0.70 | −26.1 |
Bold values indicate percentages higher than 1%.
aLMV-resistant variants. Mutations other than V and I were grouped as ‘Others’.
bHBeAg-abolishing variants.
cSamples in which polymerase and precore-mutated variants are predominant.
Analysis of the main preCore codons
PreCore codons 1, 2 and 28 were analyzed; changes in these codons are the main cause of HBeAg non-expression. The percentage of changes in each sample processed is shown in Table 3. In four of the five HBeAg-negative patients (Patients 1, 3, 5 and 7), HBeAg-negative biochemical status was due to a stop mutation in codon 28 (W28stop, main preCore mutation). In Patient 2, HBeAg-negative status was due to mutations in codon 1 of the preCore region (99.87%). This mutation was also detected at a lower frequency in Patient 6 (9.81%). Patients 2 and 6 were both infected with genotype A2.
The circular construct included the ϵ structure sequence of the 5′ pgRNA end. The base pairing rate between Position 1896 in codon 15 and Position 1858 in codon 28 was analyzed. Among a total of 90 386 sequences with TA1896G in codon 28, 89 837 (99.39%) showed CCT1858 in codon 15 and among all sequences with CCT1858 in codon 15, 99.76% showed TA1896G. These results indicate that correct base pairing between Positions 1896 and 1858 is an extremely important constraint in the G1896A mutation. The second most frequent nucleotide substitution, G1899A (28.35%), was paired with T1855 in 99.3% of cases, and was observed in the presence of G1896A in 99.6% of cases.
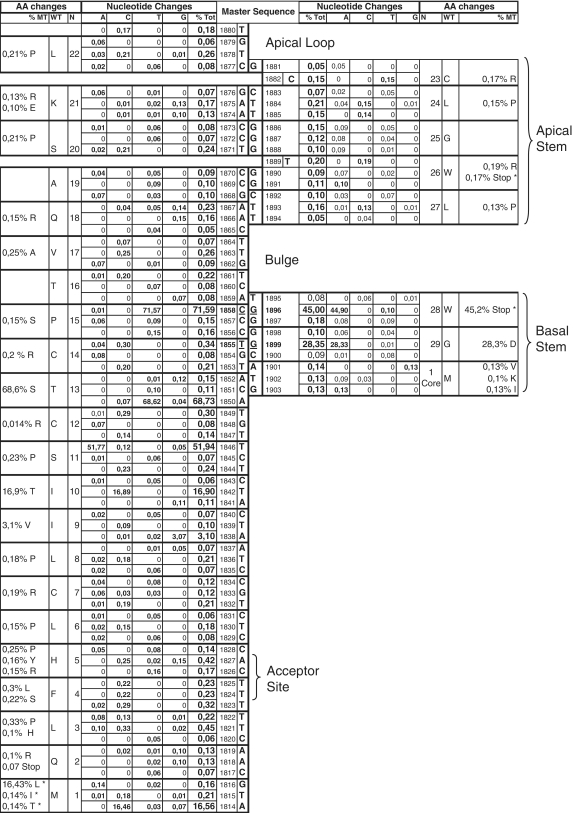
Conserved positions in the preCore region and maintenance of base pairing in the basal and apical stems
A schematic representation of base-pairing between the preCore positions analyzed, corresponding to ϵ, is shown in Figure 3, which includes the percentage of nucleotide and amino acid changes for all sequences. Although a high level of conservation was observed, there was a low percentage of variability (≥0.05%, close to the UDPS error rate). Interestingly, the base-paired positions in the basal (1851–1859 and 1895–1903) and apical (1866–1877 and 1881–1894) stems showed similar percentages of changes to maintain base-pairing.
Figure 3.
Schematic representation of base pairing between the preCore positions analyzed, which correspond to the stem–loop structure. The columns in the center indicate the consensus sequence obtained from the laboratory’s entire collection of genotypes A sequences (bold characters). The ‘Nucleotide changes’ column indicates the percentage of each nucleotide change (%A, %C, %T and %C) and the total changes (%Tot) for all positions, attending to the master sequence. The ‘Amino acid (AA) changes’ column indicates the codon representing the position (N), the amino acid coded from the master sequence (WT), and the percentage of sequences that code for a different amino acid (%MT).
Attending to the upper stem positions, a T to C change in 1884, 1885 and 1893 was observed in >0.1% of sequences (0.15, 0.14 and 0.13%, respectively), with a complementary change of A to G in their base-paired Positions (1875, 1874 and 1867) in very similar percentages (0.13, 0.1 and 0.13%, respectively). Additionally, the low percentage (0.1%) of G to A changes in Position 1891, which represent a new stop codon in the preCore sequence (W26stop), were also compensated by a complementary C to T change in a similar percentage (0.09%) in the base-paired C1869T position. In all the apical stem positions, base-pairing of the structure was maintained.
The hexanucleotide edge of the upper stem (Positions 1877–1882) was also analyzed, with emphasis on the TGT nucleotides (1878–1880), which form a pseudo-triloop structure essential for pgRNA packaging (31). The pseudo-triloop was found to be highly (but not completely) conserved (99.5% of cases). Four variants were detected at a frequency of >50 relative to the master: 403 CGT, 330 TGC, 118 TAT and 66 AGT. Among the three nucleotides, the most highly conserved was G1879 (0.06% variability), and this was one of the most highly conserved nucleotides in the complete ϵ signal. Outside the pseudo-triloop, C1882 showed 0.15% variability, and the single nucleotide bulge T1889, located in the middle part of the apical stem, had 0.2% variability (Figure 3).
Interestingly, the most frequent changes in the ϵ structure were located in the basal stem (G1896A and G1899A), where the rule of base-pairing maintenance was also observed (see ‘Analysis of main preCore codons’). The low percentage of T to C changes (0.3%) in Position 1855 was surprising considering that this change did not induce an amino acid substitution. Its complementary base, Position 1899, showed a higher rate of G to A mutations (28.3%), despite that fact that a significant amino acid change (G29D) was produced. Study of Position T1855 to C by mutagenesis is described in the next section.
Stability of the pgRNA 5′ ϵ signal: free energy of the stem loop structure
The free energy of the thermodynamic ensemble of the stem loop structure of the pgRNA 5′ ϵ signal was calculated for all sequences obtained by UDPS analysis (energy values for master sequences shown in Table 3). The distribution obtained, attending to HBV genotype and the presence (mutated) or absence (wild-type) of mutations in preCore codons 1 (pc1) and 28 (pc28) is shown in Table 4. An analysis was performed between values from the same genotype group; however, comparisons including pcC1 were not taken into account because the first preCore codon (pc1) is not present in the pgRNA 5′-end (it starts at Position 1818). The presence of a mutation in pc28 destabilized the ϵ structure in genotype A2 sequences (higher energy in codon 28 mutations), whereas in genotype D, the opposite was observed, indicating that in this case, pc28 mutations stabilize the ϵ structure (lower energy in pc28 mutations).
Table 4.
Epsilon energy distribution attending to HBV genotype and the presence or absence of mutations in the preCore (codons 1 and 28)
| Genotype | 28 | 1 |
|
|---|---|---|---|
| WT | MT | ||
| A2 | WT | −25.44a | −25.98c |
| MT | −24.30b | −23.79d | |
| D | WT | −23.76e | −24.81g |
| MT | −24.39f | −25.48h | |
a versus b: P = 0.001; a versus c: P = 0.006; a versus d: P = 0.018; b versus c: P = 0.001; b versus d: P = NS; c versus d: P = 0.001;
e versus f: P = 0.0015; e versus g: P = 0.0210; e versus h: P = 0.0015; f versus g: P = NS; f versus h: P = 0.0210; g versus h: P = NS
a versus e: P = 0.002; b versus f: P = NS; c versus g: P = 0.007; d versus h: P = 0.013.
The low percentage of T to C changes in Position 1855 mentioned above was even more surprising when stabilization energy was calculated. The rare T1855C (0.30%) variant showed the lowest energy value (−30.1 kcal/mol in genotype A2) and therefore, the highest theoretical thermodynamic stability. This observation, in addition to the finding that T1855C did not induce an amino acid change, prompted us to mimic the T1855C mutation by site-directed mutagenesis. Two clones that are found in natural HBV replication were used to transfect Huh7 cells: the WT clone and another with the 1855 C mutation in both the 3′- and 5′-ends. Two more clones with the mutation in only one of the two pgRNA ends were selected to analyze possible differences in the nucleotide depending on its location in pgRNA. Each transfection was performed at least six times and detailed results of the average expression of HBsAg, HBeAg and HBV DNA are shown in Table 5. In the presence of the double 5′ plus 3′ mutation, HBsAg, HBeAg and HBV-DNA had systematically lower values than T1855 WT, indicating a possible essential nature of the nucleotide. Surprisingly, quantification of HBsAg, HBeAg and HBV DNA yielded higher levels in clones carrying only one mutation in either the 5′- or 3′-end.
Table 5.
Average expression of HBsAg, HBeAg, and HBV DNA in the different transfection experiments
| Experiment | HBsAg (IU/ml) | HBeAg (arbitrary units) | HBV DNA (log IU/ml) |
|---|---|---|---|
| Wild-type (T1855) | 65.89 | 5.52 | 6.5 |
| 1855C mutation in 5′ and 3′ | 55.47 | 3.58 | 6.1 |
| 1855C mutation in 5′ | 115.90 | 6.96 | 6.3 |
| 1855C mutation in 3′ | 175.37 | 11.70 | 6.8 |
The 4-nt bulge and the acceptor site
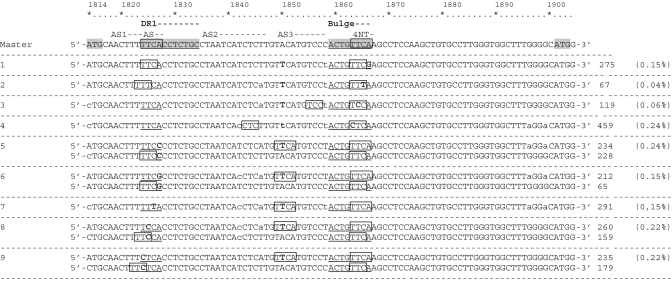
The ϵ structure bulge contains 4 nt (TTCA) that act as the template for synthesizing the 4 nt primer to start minus DNA strand synthesis. This small sequence shifts from the 5′ bulge to an acceptor site (AS) located in the 3′ direct repeat region 1 (DR1) (Figure 1b). Conservation of the 4-nt bulge and AS in DR1 seemed to be essential for HBV replication; hence, both regions were analyzed. Variants occurring at a frequency of >50 (2-fold the most prevalent variants in the HindIII control sequence: >0.03%) were considered significant. A total of 189 737 sequences were included, and the vast majority (98.53%) showed the same sequence in the bulge and DR1 regions. The remaining sequences (N = 2783, 1.47%) contained changes that prevented complete annealing of 4 nt primer in the AS DR1 site, due to a single nucleotide substitution. Less than a third of mutated sequences (N = 920, 0.48%) contained the change only in the bulge sequence, while in the other cases (N = 1863, 0.98%), the change was only present in the AS of DR1.
The 2783 sequences without correct homology between the 4 nt primer (TTCA) and the AS DR1 site were classified into nine different variants by homology of the bulge sequence (Figure 4): variants 1–4 were defined by mutations within the bulge (Positions 1863–1866) and variants 5–9 by mutations within the DR1 AS (Positions 1824–1827). Sequences from variants 5–9, which represented 1232 cases (44 0.65% of total sequences), showed complete 4-nt homology with a sequence located in Positions 1849–1852 (TTCA corresponding to HBV genotype D), which was considered a putative acceptor site (AS3). Variant 3, with 119 sequences (4, 0.06% of the total) showed 3-nt homology with an upstream AS3 sequence. The sequence groups of variants 2, 4, 8 and 9 (N = 864 cases; 31, 0.46% of the total) showed 3-nt homology with alternatives to the AS sequences located at Positions 1818–1823 [‘ω’ or AS2 in Positions 1835–1846 (27)]. Lastly, 568 sequences (20.5, 0.28% of the total) had an AS canonical region. In summary, when sequences of the 4 nt bulge primer and its AS DR1 canonical site were not completely complementary, 44% of cases showed 4-nt homology with the putative AS3 acceptor site (HBV genotype A2 characteristic sequence) while the remaining 56% showed only 3-nt homology, mainly located in previously described alternative acceptor sites (32).
Figure 4.
Sequences without correct homology between the 4 nt primer in the bulge (1863–1866) and acceptor site (AS) in direct repeat region 1 (DR1) (1824–1827). The genotype A2 consensus sequence from our laboratory collection used as the pattern is presented in the first line, and variants are added below. The number of sequences of each variant and the percentage of the total represented is indicated at the end of each line. Bold characters indicate mutations from the master, and lower cases indicate a mixture with the nucletotide of the consensus sequence. Boxes indicate homologous sequences and suggest alternative ASs.
YMDD analysis
The YMDD motif was analyzed in four baseline samples and in four samples containing LMV-resistant mutations, detected by InnoLipa. Percentages of the sequences obtained with different nucleotides are shown in Table 3; the columns indicate the amino acid detected. The wild-type form was represented as methionine (M), and mutants conferring resistance were valine (V) and isoleucine (I). Amino acids other than M, V or I, detected in a lower percentage, were grouped as ‘Others’. As was seen in the baseline samples, the wild-type form of Position 204 was highly predominant (>99%), but in the four samples, a low percentage of the main rtM204V/I mutant forms (0.07–0.2%) and a significant percentage of other variants (0.09–0.12%) were detected. In the four samples corresponding to a time of viral breakthrough (VBK) analyzed, the LMV-resistant variants rtM204V/I were the main populations (98.7–99.7%). However, low percentages of wild-type sequences were also detected (0.07–1.06%). In three patients with VBK (cases 4, 6 and 7) the rtM204V mutation was predominant, but Patient 4 had the predominant mutation in combination with a considerable percentage of rtM204I (5.6%). The rtM204I mutation alone predominated in Patient 5, but a small percentage (0.2%) of rtM204V was also detected. These results agree with those obtained by Lipa (Table 1). Interestingly, the percentage of ‘Other’ variants in Position 204 (0.23–0.27%) was higher in VBK samples than in baseline samples, suggesting that these variants might also be involved in LMV resistance.
Simultaneous detection of preCore and polymerase gene mutations in the same HBV genome
The possibility of simultaneous mutations in the YM204DD Pol motif (Position 204) and preCore (codon 1, 2 and 28) genes was analyzed in the eight circular constructs processed. Sequences were grouped according to the presence or absence of mutations in the four regions (preCore codons 1 and 2 were analyzed together), and the percentages found are shown in Table 6. In Patients 5 and 7, who were both HBeAg-negative and had mutations against LMV, the sequences showing simultaneous mutations in the preCore and Pol genes comprised the major populations. Interestingly, sequences carrying mutations in the preCore and Pol genes were detected in significant percentages in all cases, particularly Patient 6, in whom 9.9% of sequences showed simultaneously occurring LMV mutations and preCore codon 1 mutations.
Table 6.
Percentage of sequences, depending on the presence or absence of mutations in YMDD and preCore positions
| YMDD | preCore |
Baseline Samples |
VBK samples |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pt 1 | Pt 2 | Pt 3 | Pt 4 | Pt 4 | Pt 5 | Pt 6 | Pt 7 | |||
| HBe N | HBe N | HBe N | HBe P | HBe P | HBe N | HBe P | HBe N | |||
| 204 | Codon 1-2a | Codon 28 | Gen D | Gen A2 | Gen A2 | Gen D | Gen D | Gen A2 | Gen A2 | Gen D |
| Wt | Wt | Wt | 0.20 | 0.05 | 0.04 | 90.93 | 0.25 | 0.00 | 0.05 | 0.00 |
| Wt | Mt | Wt | 0.00 | 99.41 | 0.00 | 0.55 | 0.00 | 0.03 | 0.00 | 0.00 |
| Wt | Wt | Mt | 99.21 | 0.01 | 99.71 | 7.82 | 0.00 | 0.04 | 1.01 | 0.07 |
| Wt | Mt | Mt | 0.30 | 0.16 | 0.14 | 0.03 | 0.00 | 0.01 | 0.00 | 0.00 |
| Mt | Wt | Wt | 0.01 | 0.00 | 0.00 | 0.21 | 97.94 | 0.13 | 88.78 | 0.29 |
| Mt | Mt | Wt | 0.00 | 0.29 | 0.00 | 0.00 | 0.59 | 0.00 | 9.86 | 0.01 |
| Mt | Wt | Mt | 0.29 | 0.08 | 0.11 | 0.46 | 1.20 | 99.24 | 0.28 | 99.03 |
| Mt | Mt | Mt | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.56 | 0.02 | 0.60 |
The negative (N) or positive (P) status of HBe antigen expression and HBV genotype (Gen A2 or D) are indicated in each patient column.
Bold values indicate percentages higher than 1%.
aWT codon 1 and 2 are not mutated. MT codon 1 and/or codon 2 are mutated. However, mutations are mainly found in codon 1.
Sequential study of the preCore region and YMDD motif
In Patient 4, the circular construct was obtained from samples taken at baseline and at VBK. Mutations conferring LMV resistance were detected in the baseline sample (0.55%), and were much more prevalent at VBK (99.54%) (Table 3). Although the rt204I mutation was detected in a higher percentage than rt204V at baseline (0.47% versus 0.08%), rt204V was more frequent (93.9%) than rt204I (5.63%) at VBK. As to preCore mutations, the percentage of stop codon 28 significantly decreased at VBK (0.12%) relative to the baseline sample (8.31%) (Table 3).
Start codon of the core gene
The circular construct included the start codon of the Core gene, and significant percentages (0.11–0.91%) of mutations in this codon were detected in all samples processed (Table 3). It must be kept in mind that these mutations abolished Core protein production; hence HBV genomes carrying these changes would be defective for viral capsid production.
preCore fragment overlapping the HBx gene
The HBx gene spans Position 1374–1838 of the HBV genome, overlapping the preCore gene in the last eight codons (Positions 1814–1838). For this reason, mutations in the HBx stop codon (Positions 1836–1838) and its implications on the preCore ORF were analyzed. Although most sequences (194 423; 96.82%) had the expected TAA stop codon, 5837 sequences (2.9%) carried the TAG amber stop codon, which represents an isoleucine to valine amino acid substitution in the overlapping preCore region (ATC to GTC). Of note, a significant percentage of sequences (324, 0.16%) showed a T1836C substitution, which changes the ochre stop codon TAA to CAA (Gln). After this substitution, the HBx protein would be translated to the next in-frame stop codon TAG at Position 1992, inducing expression of 51 additional amino acids at the COOH-terminal region. In the preCore ORF, the T1836C mutation also induces a change of leucine to proline. The remaining 233 sequences showed other amino acid substitutions, which were not analyzed because they occurred at a frequency below our established cut-off.
DISCUSSION
The circular construct developed in the present study provided information about the HBV RT YMDD Pol and preCore regions, thus enabling analysis of simultaneous mutations in both regions of the same genome. The obligate presence of the 6-bp HindIII sequence, which linked the two regions, was used as an internal control of fidelity of the UDPS process. The calculated nucleotide error rate (1.6 × 10−4), with more transitions than transversions (60% versus 40%), was similar to reported error rate values obtained with clonal DNA as the external control (18–20,26,25), thus validating the use of HindIII as the exclusive error rate control. Nonetheless, to be more precise and concise, only variants in a percentage higher than twice this error rate (>0.03%) were taken into consideration.
Analysis of the YMDD motif of the Pol gene confirmed the presence of low but significant percentages (0.07–0.55%) of LMV resistance mutations in baseline samples from all patients studied, thus illustrating the complexity of the HBV quasispecies. Interestingly, significant populations of other YMDD variants (0.09–0.12%) were also detected at baseline. At VBK, the rtM204V/I variants were the main populations detected (99.7–93.9%), but wild-type sequences were also found in small percentages (0.07–1.06%). Once again, the presence of YMDD variants was associated with LMV resistance (14). Variant rtM204V was predominant in three of four cases of VBK, whereas rtM204I was predominant in only one. In the sequentially studied case, rt204I was detected in a higher percentage than rt204V at baseline (0.47% versus 0.08%), but after VBK, rt204V was more prevalent (93.9%) than rt204I (5.63%). This suggests that the type and relative frequency of mutations at baseline does not predict the mutations selected during treatment (33).
The high percentages of viral populations with preCore and LMV-resistant mutations observed in LMV-resistant anti-HBe cases seem to indicate that mutations in these regions simultaneously occur in the same genome. In the limited available studies directly investigating this simultaneous presence (22,23), there were no correlations of LMV-resistant mutations with HBV genotype or the presence (or not) of the preCore stop codon. Furthermore, in vitro study has shown that progeny DNA levels were restored in constructs with preCore and LMV mutations (34,35). Only a small number of clones were analyzed in the previous studies (22,23) because the preCore and YMDD regions are more than 1 kb apart, a fact that makes their study difficult. When the present research was performed, clonal analysis by UDPS was limited to a 250-bp sequence length (~400 bp in its current version). Nonetheless, the methodology reported here enabled ultra-deep clonal study of both types of variants in the same genome. The results obtained with this novel PCR-based technique support the idea that simultaneous mutations in the preCore region and YMDD Pol motif are common in the HBV quasispecies in CHB patients, regardless of the viral genotype (A2 or D) or HBeAg status.
The presence of genomes without the Core antigen start codon suggests the existence of minor viral populations that are defective for viral capsid production, another indication of the complexity of HBV viral quasispecies. The presence of these genomes in HBV infection can be explained by possible collaboration between viruses from the same infection by a trans-complementation mechanism: if wild-type and Core-defective genomes coinfect the same liver cells, Core proteins produced in excess by the wild-type viral genome might be used to encapsidate Core-defective HBV genomes. Possible reduction of HBV core proteins in the cytoplasm pool due to the presence of Core-defective genomes could decrease their expression in the cell membrane, thus modulating host immune pressure and maintaining the infection. Therefore, the presence of defective genomes might establish cooperation with wild-type viruses, thus favoring viral fitness. This mechanism of trans-complementation has been described in in vitro studies analyzing defective HBV Core clones (36). Further mutagenic studies are needed to evaluate the possible decrease in host immune response in this situation. Research evaluating possible roles of the minor (0.16%) mutated sequences detected (additional amino acid in the HBx COOH-terminal peptide due to a mutation in the HBx stop codon) would also help to clarify the role of the HBx gene in HCC pathogenesis.
In agreement with previous studies (37), the two HBeAg-positive cases had significantly higher percentages of preCore stop mutations in codon 28 (8.31 and 1.31%), suggesting easy selection of preCore mutations when the host immune response increases during viral infection. Likewise, in HBeAg-negative patients, wild-type codon 28 preCore genomes were detected in lower percentages (0.02–0.14%), explaining cases of natural or LMV-related seroreversion (38,39). An interesting observation in the longitudinally studied patient was the significant decrease in the main preCore mutation at VBK, which coincides with reports suggesting that HBV strains carrying preCore stop codon 28 and no Pol resistant variants are more sensitive to LMV treatment than preCore wild-type sequences (35,38,39). However, additional longitudinal UDPS analyses, as reported in the present study, must be performed to further substantiate this possibility.
Epsilon elements are present twice on pgRNA, one located near the 5′ terminus and the other at the 3′ terminus. Correct base-pairing between Position 1896 (codon 28) and 1858 (codon 15) from the lower stem of the encapsidation signal is one of the main constraints for preCore variability and ϵ thermodynamic stability, as has been reported previously (8) and was highly reinforced by our results. Regarding HBV genotypes, the main preCore mutation (A1896G variant) was clearly predominant in genotype D, while in genotype A2, significant percentages of preCore mutation were located at codon 1. This thermodynamic restriction between HBV genotypes corroborates results from previous studies using clonal or direct sequencing techniques (8,11).
After initial ϵ-Pol binding, the HBV apical stem loop of ϵ has to open up to form a primer synthesis-competent complex, but the highly stable HBV upper stem is a barrier. The free energy to overcome this barrier may come from interaction with RT and by capping the TGT pseudo-triloop with capsid proteins (31). Highly conserved positions and maintenance of correct base pairing in the upper stem were observed in our results. A change of T to C in Positions 1884, 1885 and 1893 was found in 0.15, 0.14 and 0.13%, respectively, but interestingly, the complementary change of A to G in their base-paired Positions (1875, 1874 and 1867) showed very similar percentages (0.13, 0.1 and 0.14%, respectively). Additionally, the low percentage (0.1%) of G to A changes detected in Position 1891, which in fact represent a new stop codon in the preCore sequence, was also compensated with complementary C to T changes in a similar percentage (0.09%) in the 1869 base-paired position. Observation of correct base pairing of the apical and upper stem of ϵ might reinforce the idea that structural rather than sequence restrictions are involved in conservation of the ϵ encapsidation signal.
The hexanucleotide sequence located at the top of the apical stem (from 1877 to 1882) was found to be highly conserved among 1200 HBV strains (40). MR analysis (41) has shown that the 1878TGT1880 sequence folds in a pseudo-triloop (Figure 1), a structure correctly predicted by the RNA folding program used in the present study (27,28). The pseudo-triloop is not needed for RT binding, but is required for pgRNA encapsidation, suggesting its interaction with capsid proteins (29,42). High conservation of T1878, G1879 and T1880 nucleotides was observed in our study, particularly G1879, which suggests that they have a predominant role in the ϵ-capsid interaction (43). Structural and functional experiments must be performed to analyze the effect of these changes on the HBV replication process.
The mutation combination of G1899A plus G1896A was frequently detected in our study. Guarnieri et al. (36) reported that G1899A does not significantly increase in vitro viral replication, despite significant theoretical stabilization of the ϵ structure. G1899 is found in the Kozac sequence and precedes the start codon of HBcAg. The reason why G1899A is a highly prevalent change, in contrast to its paired stem position, the conserved T1855C, remains unknown. According to our results, the 1855 T:G1899 to 1855 T:A1899 change was 100 times more frequent than the 1855 T:G1899 to 1855 C:G1899 change. Furthermore, in contrast to previously reported results (36), the site-directed mutagenesis and the transfection studies showed that presence of the T1855C mutation in both the 5′- and 3′-ends led to a slight, but systematic, reduction in HBsAg, HBeAg and HBV-DNA levels. If T1855 were to have an essential role, selection of the C substitution would be limited and explain the low prevalence of this change. The most striking findings of our study were that chimerical (but not natural) constructs with only one 1855 C mutation at the 5′- or 3′-end were associated with higher HBsAg, HBeAg and HBV-DNA levels than wild-type clones. Although the 5′ and 3′ structures have differing functions in the singular HBV replication cycle (4,5), enhancement of viral replication activity in chimerical constructs with a single mutation remains unexplained. Specific experiments using primary hepatocyte cells, such as macaque hepatocytes or other in vitro cell systems are needed to explore the possible involvement of T1855.
Analysis of nucleotide variability in the 4-nt bulge primer (4 nt primer) used as a template for synthesis of the minus DNA strand and its corresponding AS in the DR1 region confirmed the high conservation of both regions. It has been speculated that the 4 nt primer is more essential than DR1 AS. The existence of alternative acceptor sites (32) is suggested, as well as the idea that 3-nt sequences would be completely functional for minus strand DNA synthesis (36). Our results support these concepts because the 4 nt bulge primer was slightly more conserved than AS DR1 (99.5% versus 98.9%). As was expected, the ϵ structural motifs were strongly conserved in our study; very few cases (1.47%) had different sequences in the 4 nt primer and its canonical DR1 AS. However, 44% showed complete 4-nt homology with the putative acceptor site, AS3 (Positions 1849–1852), whereas the remaining 56% showed only 3-nt homology with canonical DR1 AS or with sequences located in previously reported alternative acceptor sites (32). Therefore, our results seem to agree with the existence of an alternative to the DR1 acceptor sites for the 4 nt primer (32). Analysis of the theoretical secondary structure of the 3′-end ϵ signal (Figure 1b) showed that the AS3 sequence has tighter Watson–Crick pairing than canonical DR1 AS. This fact could represent an energy barrier for 4 nt annealing, and explain the low prevalence of mutated bulge variants to enable correct annealing of 4 nt with AS3.
The high cost of the pyrosequencing method used in this research is a limit to the number of patients that can be studied, but the large number of sequences obtained for each patient provides deeper information on the HBV viral quasispecies than has been previously reported. The construct was designed to evaluate specific positions in the preCore and polymerase regions, but not the entire gene. Thus, our conclusions are restricted to the YMDD and preCore regions. Because of the same cost concerns, only LMV-treated patients were selected; other RT positions associated with LMV and other antiviral treatments were not included. The circular construct designed for this study was useful for analyzing two widely separated regions. This method can be applied to other genes if a fragment with the two regions of interest at the ends can be amplified by PCR.
In conclusion, UDPS study confirmed the feasibility of detecting simultaneously occurring mutations in the preCore region and YMDD polymerase motif. The presence of LMV-resistant variants in baseline naïve samples, detection of preCore mutations in HBeAg-positive patients and the presence of WT preCore variants in HBeAg-negative cases were also demonstrated. Low percentages of defective Core genomes and mutated HBx strains were found, illustrating the complexity of the HBV quasispecies. Viral strains present in small percentages can act as reservoirs and be selected in response to external changes (e.g. administration or elimination of drug pressure, or an increased host immune response). Genomes with mutations in the start codon of the Core gene might result in defective particles, whose presence might be allowed due to cooperation between viruses. The thermodynamic stability of the ϵ signal was confirmed to be the principal restriction for selection of the main preCore mutation, which is responsible for abolishment of HBeAg expression. Furthermore, analysis of ϵ signal variability revealed the essential role of structural ϵ motifs and possible involvement of some nucleotides in ϵ signal function. Lastly, we found that correct annealing between the 4 nt primer of the bulge and the AS does not seem to be an absolute condition for the 4 nt primer switch needed for minus DNA strand synthesis, suggesting that alternative cis-acting sequences, such AS3 defined in the present report, might contribute to define the normal acceptor site in the 3′ of pgRNA.
FUNDING
This study was funded by a grant from the Spanish Ministry of Health and Consumer Affairs (FIS PS09/00899). CIBERehd is funded by Instituto Carlos III, Ministry of Health and Consumer Affairs. Funding for open access charge: Spanish Ministry of Health and Consumer Affairs (FIS PS09/00899).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr F. Zoulim et al. from INSERM for kindly providing the Huh7 cells used in this study and detailed information on HBV phenotyping, and Celine Cavallo for English language support and helpful suggestions.
REFERENCES
- 1.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Lee J, Ryu WS. Four conserved cysteine residues of the hepatitis B virus polymerase are critical for RNA pregenome encapsidation. J. Virol. 2009;83:8032–8040. doi: 10.1128/JVI.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang GH, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck J, Nassal M. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 1998;18:6265–6272. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rieger A, Nassal M. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5 ϵ encapsidation signal and the 3′-proximal direct repeat DR1. J. Virol. 1996;70:585–589. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris ATH. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;9:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J. Virol. 1990;64:1298–1303. doi: 10.1128/jvi.64.3.1298-1303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Frias F, Buti M, Jardi R, Cotrina M, Viladomiu L, Esteban R, Guardia J. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology. 1995;22:1641–1647. doi: 10.1002/hep.1840220605. [DOI] [PubMed] [Google Scholar]
- 9.Jardi R, Rodriguez F, Buti M, Costa X, Valdes A, Allende H, Schaper M, Galimany R, Esteban R, Guardia J. Mutations in the basic core promoter region of hepatitis B virus. Relationship with precore variants and HBV genotypes in a Spanish population of HBV carriers. J. Hepatol. 2004;40:507–514. doi: 10.1016/j.jhep.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Pollack JR, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lok AS, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl Acad. Sci. USA. 1994;91:4077–4081. doi: 10.1073/pnas.91.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd AH, Kidd-Ljunggren K. A revised secondary structure model for the 3ϵ-end of hepatitis B virus pregenomic RNA. Nucleic Acids Res. 1996;24:3295–3301. doi: 10.1093/nar/24.17.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dienstag JL, Perrillo RP, Schiff ER, Bartholomew M, Vicary C, Rubin M. A preliminary trial of lamivudine for chronic hepatitis B infection. N. Engl. J. Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 14.Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL, Brown N, Condreay LD. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 15.Lok ASF, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA, et al. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714–1722. doi: 10.1053/j.gastro.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Chien RN, Yeh CT, Tsai SL, Chu CM, Liaw YF. Determinants for sustained HBeAg response to lamivudine therapy. Hepatology. 2003;38:1267–1273. doi: 10.1053/jhep.2003.50458. [DOI] [PubMed] [Google Scholar]
- 17.Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- 18.Zagordi O, Klein R, Däumer M, Beerenwinkel N. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 2010;38:7400–7409. doi: 10.1093/nar/gkq655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solmone M, Vincenti D, Prosperi MCF, Bruselles A, Ippolito G, Capobianchi MR. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J. Virol. 2009;83:1718–1726. doi: 10.1128/JVI.02011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margeridon-Thermet S, Shulman NS, Ahmed A, Shahriar R, Liu T, Wang C, Holmes SP, Babrzadeh F, Gharizadeh B, Hanczaruk B, et al. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J. Infect. Dis. 2009;199:1275–1285. doi: 10.1086/597808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lok AS, Hussain M, Cursano C, Margotti M, Gramenzi A, Grazi GL, Jovine E, Benardi M, Andreone P. Evolution of hepatitis B virus polymerase gene mutations in hepatitis B e antigen-negative patients receiving lamivudine therapy. Hepatology. 2000;32:1145–1153. doi: 10.1053/jhep.2000.19622. [DOI] [PubMed] [Google Scholar]
- 23.Cho SW, Hahm KB, Kim JH. Reversion from precore/core promoter mutants to wild-type hepatitis B virus during the course of lamivudine therapy. Hepatology. 2000;32:1163–1169. doi: 10.1053/jhep.2000.19618. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Frias F, Jardi R, Schaper M, Gimferrer M, Elefsiniotis I, Tabernero D, Esteban R, Buti M. Redetection of HBV lamivudine-resistant mutations in a patient under entecavir therapy, who had been treated sequentially with nucleos(t)ide analogues. J. Med. Virol. 2007;79:1671–1673. doi: 10.1002/jmv.20982. [DOI] [PubMed] [Google Scholar]
- 25.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl Acad. Sci. USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter JS, Mathews DH. RNA structure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flodell S, Petersen M, Girard F, Zdunek J, Kidd-Ljunggren K, Schleucher J, Wijmenga S. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 2006;34:4449–4457. doi: 10.1093/nar/gkl582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durantel D, Carrouée-Durantel S, Werle-Lapostolle B, Brunelle MN, Pichoud C, Trépo C, Zoulim F. A new strategy for studying in vitro the drug susceptibility of clinical isolates of human hepatitis B virus. Hepatology. 2004;40:855–864. doi: 10.1002/hep.20388. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Boyer M. Hepatitis B virus reverse transcriptase and ϵ RNA sequences required for specific interaction in vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraham TM, Loeb DD. The topology of hepatitis B virus pregenomic RNA promotes its replication. J. Virol. 2007;81:11577–11584. doi: 10.1128/JVI.01414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuen MF, Tanaka Y, Fong DYT, Fung J, Wong DK-H, Yuen JCH, But DYK, Chan AOO, Wong BCY, Mizokami M, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J. Hepatol. 2009;50:80–88. doi: 10.1016/j.jhep.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Chen RYM, Edwards R, Shaw T, Colledge D, Delaney WE, Isom H, Bowden S, Desmond P, Locarnini SA. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology. 2003;37:27–35. doi: 10.1053/jhep.2003.50012. [DOI] [PubMed] [Google Scholar]
- 35.Tacke F, Gehrke C, Luedde T, Heim A, Manns MP, Trautwein C. Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of lamivudine-resistant mutants. J. Virol. 2004;78:8524–8535. doi: 10.1128/JVI.78.16.8524-8535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guarnieri M, Kim K-hwan, Bang G, Li J, Zhou Y, Tang X, Wands J, Tong S. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J. Virol. 2006;80:587–595. doi: 10.1128/JVI.80.2.587-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volz T, Lutgehetmann M, Wachtler P, Jacob A, Quaas A, Murray JM, Dandri M, Petersen J. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology. 2007;133:843–852. doi: 10.1053/j.gastro.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 38.Chen CH, Lee CM, Lu SN, Changchien C, Wang JC, Wang JH, Hung CH, Hu TH. Comparison of sequence changes of precore and core promoter regions in HBeAg-positive chronic hepatitis B patients with and without HBeAg clearance in lamivudine therapy. J. Hepatol. 2006;44:76–82. doi: 10.1016/j.jhep.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Kuwahara R, Kumashiro R, Murashima S, Ogata K, Tanaka K, Hisamochi A, Hino T, Ide T, Tanaka E, Koga Y, et al. Genetic heterogeneity of the precore and the core promoter region of genotype C hepatitis B virus during lamivudine therapy. J. Med. Virol. 2004;72:26–34. doi: 10.1002/jmv.10558. [DOI] [PubMed] [Google Scholar]
- 40.Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 2002;30:4803–4811. doi: 10.1093/nar/gkf603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girard FC, Ottink OM, Ampt KAM, Tessari M, Wijmenga SS. Thermodynamics and NMR studies on duck, heron and human HBV encapsidation signals. Nucleic Acids Res. 2007;35:2800–2811. doi: 10.1093/nar/gkm131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ampt KAM, van der Werf RM, Nelissen FHT, Tessari M, Wijmenga SS. The unstable part of the apical stem of duck hepatitis B virus epsilon shows enhanced base pair opening but not pico- to nanosecond dynamics and is essential for reverse transcriptase binding. Biochemistry. 2009;48:10499–10508. doi: 10.1021/bi9011385. [DOI] [PubMed] [Google Scholar]
- 43.Petzold K, Duchardt E, Flodell S, Larsson G, Kidd-Ljunggren K, Wijmenga S, Schleucher J. Conserved nucleotides in an RNA essential for hepatitis B virus replication show distinct mobility patterns. Nucleic Acids Res. 2007;35:6854–6861. doi: 10.1093/nar/gkm774. [DOI] [PMC free article] [PubMed] [Google Scholar]