Abstract
Alternative pre-mRNA splicing allows dramatic expansion of the eukaryotic proteome and facilitates cellular response to changes in environmental conditions. The Saccharomyces cerevisiae gene SUS1, which encodes a protein involved in mRNA export and histone H2B deubiquitination, contains two introns; non-canonical sequences in the first intron contribute to its retention, a common form of alternative splicing in plants and fungi. Here we show that the pattern of SUS1 splicing changes in response to environmental change such as temperature elevation, and the retained intron product is subject to nonsense-mediated decay. The activities of different splicing factors determine the pattern of SUS1 splicing, including intron retention and exon skipping. Unexpectedly, removal of the 3′ intron is affected by splicing of the upstream intron, suggesting that cross-exon interactions influence intron removal. Production of different SUS1 isoforms is important for cellular function, as we find that the temperature sensitivity and histone H2B deubiquitination defects observed in sus1Δ cells are only partially suppressed by SUS1 cDNA, but SUS1 that is able to undergo splicing complements these phenotypes. These data illustrate a role for S. cerevisiae alternative splicing in histone modification and cellular function and reveal important mechanisms for splicing of yeast genes containing multiple introns.
INTRODUCTION
Pre-messenger RNA splicing involves the removal of non-coding sequences (introns) and ligation of the remaining exon sequences to produce a mature mRNA. Through the process of alternative splicing, numerous protein isoforms can be produced from a single gene depending upon the cell’s need to respond to particular environmental conditions. In higher eukaryotes, whose genes are often interrupted by multiple introns, alternative splicing occurs through alternative splice site (SS) usage such that a vast array of alternative isoforms are produced through skipping or retention of exons. For example, over 90% of mammalian genes contain multiple introns, and ~80% of the mammalian alternative splicing events appear to encode alternative open reading frames (ORFs) and eventually encode proteins of different functions (1).
A much smaller proportion of genes in the yeast Saccharomyces cerevisiae contain introns. However, it appears that in yeast, as in other eukaryotes, changes in splicing patterns occur in response to changes in the cell’s environment or changes in cellular needs. In fact, genome-wide analysis of pre-mRNA splicing in yeast shows large-scale changes in pre-mRNA splicing patterns in response to environmental stress (2). There are also a few specific examples of regulated splicing. The gene encoding a meiotic regulator MER2 is transcribed in mitosis as well as meiosis but is only efficiently spliced during meiosis. MER2 contains a non-canonical 5′ splice site (5′ SS) that contributes to its regulation (3). Another example is the regulated splicing of RPL30, whose gene encodes an essential ribosomal protein. When the Rpl30 protein is in excess, it binds to the RPL30 transcript and inhibits the transcript’s splicing (4). This unspliced transcript subsequently makes it to the cytoplasm and is targeted for nonsense-mediated decay (5). A third transcript, YRA1, encodes an mRNA export factor. The intron in YRA1 contains a non-canonical branchpoint (BP) sequence that contributes to intron retention and degradation of this transcript to maintain the appropriate levels of Yra1 protein (6–8). Intriguingly, Yra1 protein, like Rpl30, regulates its own transcript; Yra1 protein appears to inhibit YRA1 splicing and facilitate export of the unspliced pre-mRNA (7). Finally, a recent example of alternative splicing in yeast illustrates that intron-containing RNAs can be translated to affect cellular function. The transcript PTC7 contains an intron that can be either removed or retained (9). Both transcripts can be exported and translated, and the proteins generated from the two isoforms localize to different cellular compartments and mediate different cellular responses (9).
We previously demonstrated that histone H2B ubiquitination was increased in cells harboring a deletion of known splicing factors (10). A closer mechanistic analysis revealed that H2B deubiquitination was dependent upon the activity of Sus1, a component of the SAGA complex, and the gene encoding Sus1 contained two introns. Removal of both introns was required to generate a mature SUS1 mRNA encoding the Sus1 protein responsible for deubiquitination of histone H2B. Intriguingly, SUS1, like YRA1, RPL30 and MER2, contains non-canonical sequences in the first intron, and even a wild-type strain shows some accumulation of a partially-spliced pre-mRNA with this intron retained. The presence of these multiple isoforms suggested to us that SUS1 might undergo ‘alternative splicing’ in the form of intron retention. Furthermore, since SUS1 contains multiple introns, a variety of isoforms can be generated from the SUS1 pre-mRNA, offering a unique opportunity to understand how removal of multiple introns is achieved in this model eukaryote.
Metazoan genes often contain numerous long introns, sometimes on the order of several thousand nucleotides, and relatively short exon sequences (11). Hence, splicing and alternative splicing mechanisms have evolved to remove introns in this particular gene landscape. For example, accurate recognition of SSs involves an ‘exon definition’ model in which SS selection is mediated through interactions between splicing factors across exons (12,13). Furthermore, splicing and alternative splicing involve a host of proteins that interact with core components of the spliceosome, particularly the SR and hnRNP family of proteins, to recognize regulatory sequences throughout the intron-containing genes (14). Intron–exon architecture (nucleosome occupancy) (15) and even intron secondary structure (16–18) can also influence splicing outcomes. With recent indications that most splicing occurs co-transcriptionally (19), these mechanisms must be considered within this co-transcriptional context.
In order to understand how expression of the two-intron gene SUS1 is regulated, we undertook a detailed analysis of SUS1 splicing. These studies reveal that SUS1 is subject to both intron inclusion and exon skipping, that these different splicing events are influenced by suboptimal SSs in the first intron, and that core components of the spliceosome play distinct roles in these mechanisms. The two SUS1 introns do not appear to be removed independently as removal of the 5′ intron influences removal of the downstream 3′ intron, consistent with a role for cross-exon ‘communication’ in the splicing of SUS1. Importantly, SUS1 splicing appears to be regulated, as the SUS1 splicing pattern can be influenced by environmental stress conditions. Regulation of SUS1 splicing is required for complete Sus1 function, as the expression of a SUS1 WT construct that can undergo splicing (and intron retention) fully complements sus1Δ temperature sensitivity and the histone H2B deubiquitination defect, although the SUS1 cDNA does not. This is in agreement with data from the accompanying article by Cuenca-Bono et al. (20) which shows that SUS1 is alternatively spliced under different conditions to yield functionally important protein products. Together these studies illustrate important mechanisms underlying alternative splicing in S. cerevisiae. Furthermore, these findings demonstrate that post-transcriptional regulation of this specific gene product plays a critical role in histone modification.
MATERIALS AND METHODS
Yeast strains and growth analysis
Yeast strains were generated by standard yeast genetics and molecular biology techniques. The yeast strains used in this study are indicated in Supplementary Table S1. Plasmids were transformed into the yeast strains using standard LiAc transformation method, and transformants were selected on selective plates. The cells were grown in selective media to maintain the plasmid. For dilution series experiments, cells were grown in SC-leu media to OD600 0.4–0.5 at 30°C and then spotted on SC-leu plates at 10-fold serial dilution. The plates were kept at the temperatures indicated to observe the growth.
Growth conditions for temperature shift experiments
Wild-type (WT) cells were grown in 150 ml of YPD media (containing 2% w/v glucose) to OD600 0.4 at 30°C. Then the culture was divided into three equal (50 ml) aliquots. One aliquot was returned to 30°C to continue growth. Cells from the second aliquot were collected by centrifugation, suspended in an equal volume of pre-warmed 37°C media, and grown at 37°C. Cells from the third aliquot were also collected, suspended in an equal volume of pre-warmed 42°C media and grown at 42°C. 10 ml of cells from each growth condition were collected at 20 min. intervals to isolate RNA and analyze SUS1 splicing.
Generation of SUS1 constructs
The SUS1 gene was PCR amplified from yeast (Saccharomyces cerevisiae) genomic DNA along with the 5′-UTR (340 bp) and 3′-UTR (325 bp) using the primers SUS1Up-F and SUS1Dw-R (Supplementary Table S2). Restriction enzyme cleavage sites for SalI and BamHI were introduced into the SUS1Up-F and SUS1Dw-R primers, respectively. PCR products were then digested with restriction enzymes BamHI and SalI and cloned into pRS315. Selected clones were verified by sequencing. The resulting construct SUS1-WT (Figure 4A) was used as a template to generate other SUS1 constructs (Figure 4A). The second intron was removed from pRS315-SUS1 WT using the primers I2Δ-F and I2Δ-R. The upstream part of the second intron was PCR amplified using the primers SUS1Up-F and I2Δ-R. The downstream part of the second intron was PCR amplified using I2Δ-F and SUS1Dw-R. The resulting PCR products were purified, mixed at a 1:2 ratio and PCR amplified using the primers SUS1Up-F and SUS1Dw-R. PCR products were then digested with BamHI and SalI and cloned into pRS315. The first intron was removed similarly using the primers I1Δ-F and I1Δ-R (see Supplementary Table S2 for primer sequences). The non-canonical 5′ SS and BP sequences of the first intron were made canonical using pRS315-SUS1 WT as a template for ‘quick change mutagenesis’ (Stratagene). The canonical 5′ SS and BP of the second intron were made non-canonical following the same method. Primers used for mutagenesis are shown in the Supplementary Table S2. All of the constructs were verified by sequencing. The ‘swapped intron’ construct was kindly provided by Bernardo Cuenca-Bono and Susana Rodriguez-Navarro. The 5′-UTR and 3′-UTR sequences were inserted into this construct by recursive PCR (21). This product was then amplified using the end primers SUS1Up-F and SUS1Dw-R, and cloned into pRS315.
Figure 4.
Non-canonical sequences in the first intron affect its removal, and splicing of the first intron affects removal of the second. (A) Schematic representation of the SUS1 constructs used in this study. The asterisks indicate the non-canonical SSs in the SUS1 intron. The SS sequence of the each intron is indicated in each schematic diagram, and the locations of primer pairs used for PCR are shown with arrows. The name given to each construct is indicated to the left of the schematic. Intron deletion is indicated by a Δ. The nc or c indicates that the wild-type SUS1 intron sequences have been mutated to the non-canonical (for intron 2) or canonical (for intron 1) sequences. When no specific mutation of the intron is indicated, the intron is present in its WT form. (e.g. I1c indicates that intron 1 is mutated to contain canonical SSs, and the second intron retains its WT sequence). (B) Splicing of the first intron is affected by its non-canonical sequences and stimulates removal of the second intron in WT cells. The upper panel shows the results of 8% polyacrylamide gel electrophoresis separation of the different RNAs. The stick diagram indicated above each lane represents the construct used for the splicing assay. Asterisks represent non-canonical SS sequences. The identity of the RT–PCR products is indicated schematically next to the gels. SCR1 is the loading control. The second set of lanes 5–7 are a darker exposure of the panel to their left. The bar graph indicates the relative percentage of each of the SUS1 isoforms. Note: The identities of the single-intron products were confirmed by sequencing. Nonetheless, the second intron product consistently shows a higher than expected mobility. (C) Intron 1 affects splicing of intron 2 when it is in the 5′ position. Schematic diagram of the SUS1 gene. For clarification, the first intron (1I) is indicated by a bold line and the second intron (2I) is indicated by a dotted line. The asterisks indicate the non-canonical SSs. The gel picture shows the RT-PCR results depicting the efficiency of SUS1 splicing when the introns are swapped.
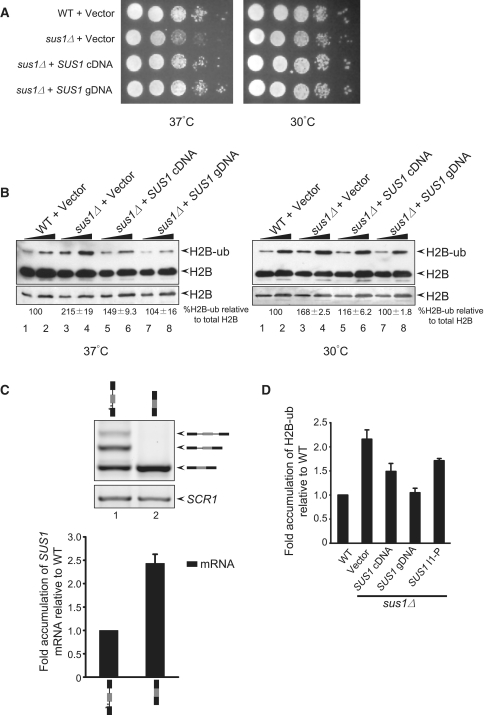
The construct expressing the first exon and part of the first intron (Figure 6) was generated by performing recursive PCR using the primers SUS1-I1P-F and SUS1-I1 P-R. The product was cloned into pRS315 and verified by sequencing.
Figure 6.
SUS1 cDNA does not fully complement sus1Δ growth and H2B deubiquitination defects. (A) The expression of the SUS1 genomic DNA construct (with both introns) complements the growth defects of sus1Δ better than SUS1-cDNA expression. 10-fold serial dilutions of each strain were plated on SC-Leu plates and grown at 37°C and 30°C for 3 days. (B) Expression of the SUS1-gDNA construct complements the overubiquitination phenotype of sus1Δ cells better than expression of the SUS1-cDNA construct. The left panel shows the western blot analysis of histone H2B when cells are grown at 37°C. H2B and ubiquitinated H2B (H2B-ub) are indicated by the arrows to the right of the blot. The lower panel shows a lower exposure of the H2B portion of the upper blot. Two serial dilutions of the whole cell extract are shown. The panel to the right shows the western blot when cells are grown at 30°C. The average percentage of H2B-ub was measured compared to the total H2B (H2B + H2B-ub) and fold accumulation of ubiquitinated H2B was calculated by setting the WT value to 100. The values (%H2B-ub± SD, where n = 3–4) are shown below each gel. Note, no reproducible change in total H2B has been observed under the conditions tested here. (C) SUS1 cDNA expresses high levels of the SUS1 message. RNA was extracted from the sus1Δ cells containing SUS1 gDNA (lane 1) or SUS1 cDNA (lane 2) and analyzed by RT–PCR. The stick diagrams above each lane show the constructs. SCR1 serves as a loading control, and the bar diagram (lower panel) shows quantitation of SUS1 mRNA. The signals were normalized to SCR1, and fold accumulation of mRNA was measured compared to the WT. (D) Expression of the first intron-containing construct up to the PTC partially suppresses the overubiquitination phenotype of sus1Δ. The bar diagram represents the fold accumulation of ubiquitinated H2B compared to the WT.
cDNA synthesis for splicing analysis
Cells were grown in SC-leu media between OD600 0.6 and 0.8. Total RNA was isolated from 10 ml of cells using a hot-phenol extraction method and dissolved in 100 µl of diethylpyrocarbonate (DEPC)-treated water. Total RNA was quantitated by spectrophotometer, and its quality was checked by agarose gel analysis. A total of 4 µg of RNA was treated with DNaseI (Promega) according to the manufacturer’s instructions. cDNA synthesis was performed using the MaximaTM First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer’s instructions and using the gene specific SUS1-2 primer as previously described (10).
Quantitative radioactive PCR
The quantitative radioactive PCR was performed as described in (10). Briefly, SUS1-Forward primer was end labeled with gamma-32P-ATP. The PCR was performed with labeled SUS1-Forward and SUS1-2 primer for 25 cycles using 1 µl of cDNA (diluted1:10). The loading control SCR1 was amplified using the primers SCR1-F and SCR1-R from 2.5 µl of cDNA (diluted 1:50). For high resolution analysis of the splicing products (Figure 4), the radioactive PCR products from the different strains were run on an 8% polyacrylamide 1XTBE gel. The gel was exposed to the phosphorimager screen and visualized by scanning using a Typhoon Phosphorimager (GE Healthcare). Signals were analyzed and quantitated using Image Quant 5.2 software. To visualize the skipped product for the different strains, the lower portion of the same gel was exposed longer and then scanned using a Typhoon Phosphorimager.
SUS1 RNA (non-radioactive PCR) was analyzed by performing PCR for 25 cycles using the primers SUS1-Forward and SUS1-2 and 1 µl undiluted cDNA. For SCR1, PCR was performed using primers SCR1-F and SCR1-R taking 2.5 µl of cDNA (diluted 1:50). PCR products were then analyzed on a 2.5% agarose gel and stained with ethidium bromide (EtBr). The gel image was captured and scanned using a Typhoon Phosphorimager (GE Healthcare) and quantitated using Image Quant 5.2 software.
Characterization of the SUS1 Exon 2 skipped RNA
PCR was performed using the primers SUS1-F and SUS1-R and 1 µl of undiluted cDNA in a 25 µl reaction volume for 25 cycles. 60 µl of PCR products were pooled and purified using a PCR purification column (Qiagen). PCR products were then run on an 8% polyacrylamide gel and stained with EtBr. The gel image was then captured by phosphorimager (shown in Supplementary Figure S1). The gel was visualized by UV, and the band containing the exon 2 skipped product was excised, eluted and characterized by sequencing using primers SUS1-Forward and SUS1-2. For SYBR green staining, 40 µl of PCR products was concentrated and run on an 8% polyacrylamide gel. PCR for the SCR1 loading control was carried out similarly. After staining (SYBR® Green I, Lonza), the gel was scanned using a Typhoon phosphorimager to capture the image. Signals were quantitated using Image Quant 5.2 software.
Western blot analysis to detect the ubiquitination status of histone H2B
Western blotting was performed as described previously (10). The WT and sus1Δ cells were transformed with the pZS145 (22) expressing FLAG-tagged histone H2B. WT cells containing pZS145 vector were transformed with pRS315 and transformants were selected on SC-his-leu plates. The sus1Δ cells containing pZS145 were transformed with either pRS315, pRS315-SUS1WT, pRS315-SUS1cDNA, or pRS315-SUS1-I1P. Cells were grown at 30°C overnight to saturation in SC-his-leu media, to maintain the plasmids, then cultures were diluted to OD600 0.1 with fresh media. The diluted cultures were then equally divided into two tubes and grown at either 30 or 37°C up to OD600 between 0.4 and 0.6. Equal numbers of cells were pelleted from each strain and lysed in 1× loading dye (50 mM Tris–HCl, pH 6.8, 100 mM BME, 2% sodium dodecyl sulfate (SDS), 0.1% bromophenol blue, 10% glycerol) at 95°C with 0.5 mm glass beads (BioSpec Products, Inc.). Following centrifugation at 13 000 rpm for 10 min, equal amounts of lysate from each strain were loaded onto a 12% SDS polyacrylamide gel. The gel was then transferred onto a polyvinylidene fluoride (PVDF) membrane and probed with anti-FLAG (Sigma) antibody at 1:2000 dilution in 5% milk. The blot was then processed and developed using the ECL plus kit (Amersham) as per manufacturer instructions. The signals were detected using a Typhoon Phosphorimager and quantitated using Image Quant 5.2 software.
RESULTS
The SUS1 gene has an unusual two-intron structure
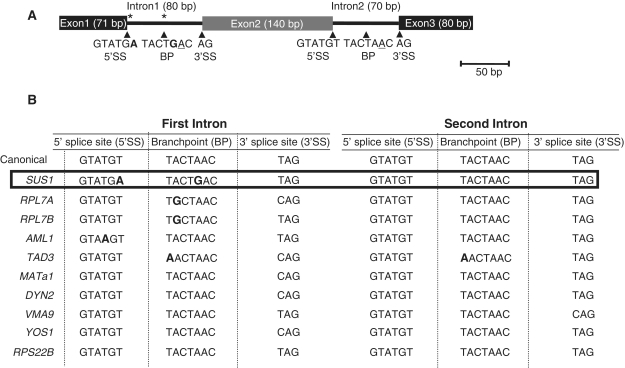
SUS1 has a number of features that make it unique. Not only is it only one of 10 genes in S. cerevisiae with multiple introns, but the first intron contains a non-canonical 5′ SS and BP sequence (Figure 1A) (10,23). The non-canonical BP sequence (TACTGAC) is unique among all the known intron-containing genes in S. cerevisiae, and the combination of a non-canonical 5′ SS and BP sequence (GTATGA+TACTGAC) is unique to SUS1. Relative to metazoans, there is generally little divergence from canonical recognition sequences in S. cerevisiae. These observations led us to explore whether SUS1 may reveal underappreciated splicing mechanisms in yeast consistent with alternative splicing and whether, based upon sequence comparison, other yeast genes containing multiple introns might be subject to similar splicing control.
Figure 1.
SUS1 and other two-intron genes in S. cerevisiae contain unusual sequence features. (A) Schematic diagram of SUS1 gene structure. The asterisks indicate the non-canonical SSs sequences. The non-canonical bases within the first intron are indicated in bold. The conserved adenosine at the BP is underlined. (B) Table illustrating the 10 two-intron genes in S. cerevisiae. The non-canonical nucleotides around the 5′ SS and BP are indicated in bold.
Although in general yeast introns show little divergence from the canonical recognition sequences, when we examined the prevalence of non-canonical sequences in the first intron of other two-intron genes, we find that half of them share this unusual property with SUS1. While SUS1 is the most striking example, with non-canonical sequences at both the 5′ SS and the BP (Figure 1A), RPL7A, RPL7B and TAD3 all have non-canonical sequences in the BP region, while AML1 has a non-canonical A (instead of T) at the +4 position of the 5′ SS. In fact, when the two-intron genes are compared to single intron-containing genes, the tendency toward non-canonical sequences, particularly at the BP, is enriched in two-intron genes. About 50% of two-intron genes contain non-canonical branch region sequences, compared to 7.5% of the single intron genes (24). In this way, first introns of two-intron yeast genes are more similar to metazoan genes (in their lack of adherence to strict intron sequences) than to other yeast genes with only one intron. This feature appears to be specific to first introns of two-intron genes, as only one of these genes, TAD3, has a non-canonical nucleotide within its second intron—an A at the first position of the BP sequence (Figure 1B). Even though two-intron genes in general tend to have non-canonical sequences in the first intron, SUS1 is striking in that it has both a non-canonical 5′ SS and BP sequence, which would be predicted to significantly decrease the efficiency with which this intron is removed.
While the presence of non-canonical BP sequences is itself rare—a total of 22 non-canonical BPs among all the available S. cerevisiae intron sequences, the majority of the non-canonical BP sequences vary only at the first position (16/22: CACTAAC for 6, GACTAAC for 5, AACTAAC for 4) (24,25, Hossain and Johnson, unpublished). SUS1 is the only gene with a BP sequence TACTGAC, where the non-canonical nucleotide is next to the adenosine that takes part in the nucleophilic reaction with the 5′ SS. An analysis of the SUS1 intron by Cuenca-Bono et al. (see accompanying paper) illustrates that the unusual SUS1 intron structure is evolutionarily conserved with its homologs from different species, suggesting a conserved biological role for a non-canonical first intron in SUS1.
The pattern of SUS1 splicing is altered upon heat stress
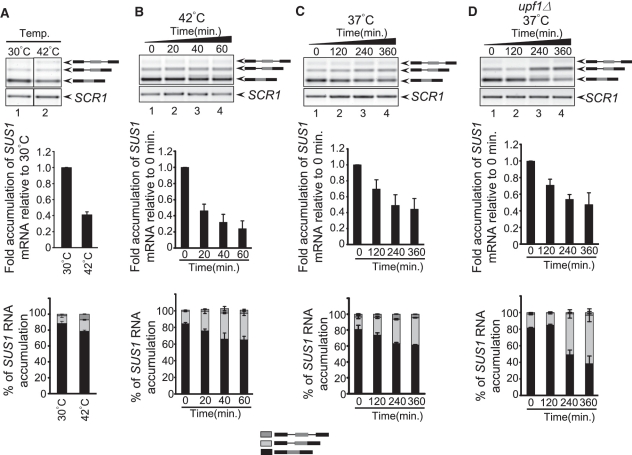
We previously reported that in wild-type cells, mature SUS1, which encodes a component of the histone H2B deubiquitination machinery, is the predominant isoform produced. However, we consistently observe that approximately 10% of SUS1 is partially spliced, with intron 1 retained. In light of the unusual and conserved intron features of SUS1, we predicted that there might be conditions under which retention of one or both of the SUS1 introns would be favored. So we analyzed whether the relative levels of the different SUS1 isoforms changed when cells were exposed to short 42°C heat shock. Cells were grown at 30°C to an OD600 of 0.4, then spun down and resuspended in pre-warmed media (42°C) and incubated at 42°C for 20 min. When RNA was analyzed from these samples, we consistently observe approximately 50% decrease in mRNA accumulation compared to growth at 30°C and an increase in unspliced SUS1 pre-mRNA. Not only do we observe a change in the absolute amounts of the isoforms, but we also note that the ratios of mRNA: partially spliced: unspliced RNA change, such that there is a greater proportion of the SUS1 unspliced and partially-spliced RNA relative to total RNA (Figure 2A). When cells are exposed to high temperature for longer time periods, the levels of SUS1 mRNA continue to decrease to 25% of the levels observed at T0. Additionally, the proportion of SUS1 RNA that is in the unspliced/partially-spliced forms is ~30% of total SUS1 RNA, compared to ~15% for cells grown at T0 (Figure 2B). Since long exposure to 42°C compromises cell viability, we next determined if cells grown at 37°C for several hours also led to changes in splicing, and we observe that mature mRNA levels decrease by >50% after 4 h of growth at 37°C. Furthermore, the partially spliced message increases, and the ratio of this product relative to the other SUS1 isoforms increases >2-fold (Figure 2C). Cells grown at 30°C do not show changes in SUS1 splicing over these time periods (data not shown). These studies demonstrate that under temperature stress conditions, cells show a rapid change in SUS1 isoforms. This is consistent with studies by Cuenca-Bono et al. (20) in the accompanying article showing that prolonged exposure to elevated temperature causes decreased production of the Sus1 protein. These results suggest that SUS1 splicing is regulated to cause changes in the SUS1 RNA isoforms and Sus1 protein levels.
Figure 2.
SUS1 pre-mRNA splicing generates different splicing patterns when cells are grown at elevated temperature. (A) Analysis of SUS1 RNA by RT–PCR when WT cells are shifted from 30°C to 42°C for 20 min. The stick diagrams on the right show the different isoforms of SUS1 RNA. RT–PCR analysis of SCR1 serves as a loading control. The bar graph (middle panel) represents fold accumulation of SUS1 mRNA relative to WT cells at 30°C. The amount of mRNA was first normalized to SCR1 and fold accumulation was measured relative to 30°C. The lower panel shows the percentage of each SUS1 isoform relative to total SUS1 RNA. The bars indicate the average of two to three independent experiments with standard deviation.(B) SUS1 splicing when WT cells are shifted from 30°C to 42°C. The stick diagrams on the right show the different isoforms of SUS1 RNA. SCR1 serves as a loading control. The middle panel represents the fold accumulation of SUS1 mRNA relative to 0 min, and the lower panel shows the relative amount of each SUS1 isoform. Fold accumulation of mRNA was calculated relative to 0 min following normalization to SCR1.(C) SUS1 splicing pattern when cells are shifted to 37°C for prolonged period of time. Bar diagrams are described in (B). (D) The retained intron products that accumulate at elevated temperature are targeted by the NMD degradation pathway. The gel image shows the SUS1 RNA in upf1Δ cells when shifted from 30°C to 37°C, as described in (C). Bar diagrams are described in (B).
The product that accumulates at elevated temperature is degraded by the nonsense-mediated decay pathway
It has been demonstrated that in yeast, changes in splicing that cause intron retention can target these messages for decay by the nonsense-mediated decay (NMD) pathway (26), and deletion of UPF1, a key component of the NMD pathway, can reveal products of splicing that are normally degraded (27). The SUS1 isoform containing the retained first intron encodes a premature termination codon within the first 16 nucleotides of the first intron, leading us to consider the possibility that the full extent of intron 1 retention at elevated temperatures was being masked by degradation of this product in the NMD pathway. To test this possibility, we performed the same experiment as is shown in B, except that the cells that were shifted to 37°C were deleted of UPF1. In upf1Δ cells, there is a striking accumulation of the product containing the first intron at elevated temperature over time (Figure 2D), indicating that the actual level of intron retention under these conditions is even higher when degradation of this product is prevented. These data reinforce that there are dramatic changes in SUS1 splicing in response to environmental conditions and suggest that the levels of SUS1 mRNA are down-regulated by a combination of alternative splicing and NMD. Since NMD in yeast is a cytoplasmic process, these results also indicate that at least some of these partially spliced messages are able to move into the cytoplasm. Interestingly, work described in the accompanying manuscript by Cuenca-Bono et al. supports the idea that partially-spliced SUS1 moves into the cytoplasm, since a truncated peptide is generated from the partially spliced SUS1 construct. These studies led us to examine more closely the mechanisms by which the splicing machinery contributes to splicing outcomes.
Components of the spliceosome differentially affect SUS1 intron accumulation and exon skipping
Alternative splicing in mammals is largely determined by the activities of accessory protein factors that bind to exonic and intronic silencing or enhancing sequences (E/ISE and E/ISS sequences) (28). These protein families, the SR family of proteins and hnRNPs, have well characterized motifs that allow them to bind to pre-mRNAs and alter the use of specific splicing signals, particularly weak SSs (29,30). Despite the fact that these proteins do not exist in yeast, the presence of genes with multiple introns and the tendency of these genes to have non-canonical splicing signals suggest that, just as metazoan introns are differentially sensitive to accessory factors (leading to alternative splicing), yeast introns within a single pre-mRNA may be differentially sensitive to the activities of core splicing factors.
To test this, we analyzed splicing of the SUS1 transcript in cells harboring deletions of various components of the splicing machinery and compared the intron accumulation to that in WT cells. Consistently, we observe that WT cells accumulate some pre-mRNA containing the first intron, a result that we reported previously and attributed to the non-canonical sequences at the 5′ SS and BP of the first intron (10). When different factors involved in pre-mRNA splicing are deleted, we observe differential effects on intron accumulation (Figure 3A). Deletion of either subunit of the CBC (cbp20Δ shown here) primarily affects removal of the first intron, leading to accumulation of a product in which the first intron is retained [Figure 3A, lane 2 and (10)], whereas, deletion of components of the U2 snRNP, Msl1 or Lea1, or the commitment complex protein Mud2 affects both the first and the second intron, such that a completely unspliced pre-mRNA accumulates (Figure 3A, lanes 4−6). Deletion of NAM8, a component of the U1 snRNP, leads to a phenotype that is intermediate between WT and cbp20Δ cells. For each of these RNAs, the RNA samples were subjected to polyacrylamide gel electrophoresis, and the bands were extracted and sequenced to confirm the identities of the products (Supplementary Figure S1).
Figure 3.
The pattern of SUS1 splicing is dependent upon the activity of core components of the spliceosome. (A) The pattern of SUS1 splicing is altered in splicing factor mutants. RNA was isolated from the indicated strains and reverse transcribed to make cDNA, followed by PCR with [32P]-labeled primers and separated by polyacrylamide gel electrophoresis. The top panel shows the unspliced transcript, partially-spliced pre-mRNA and mature mRNA, while the panel below shows a longer exposure of the bottom of the same gel in order to highlight the product of exon 2 skipping. The percentage of the exon 2-skipped product is indicated for each strain below each lane. The schematic diagram of each product is shown on the right. The panel with SCR1 depicts the loading control. (B) NPL3 deletion does not affect SUS1 splicing. RNA from WT and npl3Δ cells were analyzed to see the SUS1 splicing as described in A. (C) The retained intron product is subject to Upf1-dependent RNA decay. The upper panel shows the splicing pattern of SUS1 pre-mRNA in WT, upf1Δ, cbp20Δ and cbp20Δ upf1Δ cells. The schematic diagram of each product is shown on the right. RT–PCR analysis of SCR1 from the RNA sample is shown as a loading control. The bar graph indicates the relative proportion of each of the SUS1 isoforms.
When the gel used to analyze SUS1 splicing was overexposed, we consistently observe a small amount of spliced product in which the middle exon is skipped, which we confirm by extracting the band from the gel and sequencing it (Figure 3A, bottom panel and Supplementary Figure S1). This product is found in low abundance—approximately 5% of the total; it remains low and does not appear to change in response to temperature changes (data not shown). While it is not yet clear if there are biological consequences to SUS1 exon skipping, it is interesting to note that the degree of exon skipping is significantly affected by components of the splicing machinery. The U2 snRNP components and Mud2 suppress exon skipping, as deletion of any of these factors leads to an increase in exon skipping relative to WT. This may not be so surprising since these factors are probably required for optimal recognition of the non-canonical BP sequence, and without them the downstream BP is used. Interestingly, deletion of components of the CBC have the opposite effect. The CBC contributes to exon skipping such that its deletion causes a decrease in the skipped product relative to WT. This indicates that the CBC normally reinforces the use of the cap proximal splicing signals, which is consistent with other studies showing that the CBC is particularly important for recognition of suboptimal SSs (31). Without the CBC, the non-canonical sequences in the first intron are not efficiently recognized, leading to inefficient splicing. Meanwhile, the downstream splicing event is not so severely affected, resulting in accumulation of the partially spliced RNA and low levels of the mature SUS1 mRNA. It should be noted that previous studies from both mammals and yeast show that changes in transcription elongation can lead to a decrease in exon skipping since slowed pol II elongation allows more time for joining of the two exons flanking the first transcribed intron (32,33). Since the CBC is recruited to genes co-transcriptionally and most splicing occurs co-transcriptionally (19), we cannot rule out that deletion of the CBC may affect transcription in a manner that decreases elongation and contributes to decreased exon-skipping, a hypothesis that we are currently exploring.
Exon skipping is a well-documented alternative splicing (AS) event across eukaryotes; intron retention on the other hand is thought to be a rare AS event in vertebrates and invertebrates (34), while it is the most prevalent type of AS in fungi, protozoa and plants (1). Since patterns of exon and intron inclusion in metazoans and plants are often associated with the activities of SR proteins, we determined whether the one known SR-like protein in S. cerevisiae, Npl3, could also affect the splicing pattern of SUS1. Unlike the core components of the splicing machinery or the CBC, NPL3 deletion has little, if any, effect on SUS1 splicing (Figure 3B). Together these data demonstrate that the capacity for alternative splicing is intrinsic to the basal, ‘core’ spliceosome
When UPF1 was deleted from WT cells we observed an accumulation of the intron 1 containing pre-mRNA and, to a lesser extent, the fully unspliced pre-mRNA even at 30°C. This stabilization enhances the splicing effect observed by deletion of the CBC. We do not observe a change in the skipped product (Supplementary Figure S1D) when UPF1 is deleted from WT cells. From these results, we can make two conclusions. First, as we observed at 37°C, retention of the first intron preferentially targets these messages for NMD; and secondly, retention of the first intron provides a mechanism whereby the cell can down-regulate levels of SUS1 mRNA.
The non-canonical 5′ SS and BP sequences decrease the efficiency of first intron removal
In order to assess whether the presence of non-canonical sequences in the first intron affects its removal, SUS1 mutant constructs were generated (Figure 4A), and their splicing efficiencies were analyzed. The non-canonical 5′ SS and BP in the first intron (WT) were mutated to consensus (I1c) and the splicing efficiencies of these constructs were compared. Consensus sequences in the first intron lead to significantly more spliced product compared to WT SUS1 (Figure 4B, compare lanes 1 and 5, Supplementary Figure S2, lanes 1 and 4). To determine the relative contribution of each, the 5′ SS and BP were mutated separately, and we find that each contributes equally to first intron retention in WT cells (Supplementary Figure S2, lanes 2 and 3). To determine if non-canonical sequences in the second intron would have a similarly deleterious effect on removal of this intron, the intron 2 5′ SS and BP sequences were made non-canonical (I1c-I2nc). This construct accumulates significantly more partially-spliced SUS1 (with intron 2 retained) compared to the construct in which both introns are consensus (I1c) (Figure 4B, lanes 3 and 5). These data confirm our prediction that the 5′ SS and BP contribute to retention of the SUS1 intron.
Efficient splicing of the first intron influences splicing of the second intron
In metazoan cells where alternative splicing is a common feature of regulation of genes containing multiple introns, interactions that occur across the exon can influence efficiency of intron removal—an ‘exon definition’ model. In this model, binding of U1 snRNP to the 5′ SS promotes recognition of the upstream 3′ SS across the exon (12). Furthermore, for many metazoan genes there are multiple regions of alternative splicing within the same gene, and several reports indicate that there can be tight coordination between AS events at two regions within the same gene (35,36). Hence, there are indications in higher eukaryotes that interactions across exons can affect removal of each intron. We were interested to know whether removal of each of the SUS1 introns was independent or whether the two splicing events, intron 1 (I1) removal and intron 2 (I2) removal, could influence one another.
To test this, we generated a series of SUS1 constructs in which either the first or the second intron was removed (ΔI1 or ΔI2, respectively), and to determine how the non-canonical sequences in SUS1 may contribute to any effects that we observed, the SSs in intron 1 were changed to canonical sequences (I1c-ΔI2) and the 5′ SS and BP in intron 2 were replaced with the non-canonical intron 1 sequences (ΔI1-I2nc) (Figure 4A). First, we measured intron retention in cells harboring the ΔI2 construct, and we find that deletion of the second intron still leads to accumulation of intron 1 containing pre-mRNA. We observe little, if any, increase in the levels of mature SUS1 mRNA compared to WT, even when the precursor contains only the first intron (Figure 4B, lanes 1 and 2). However, ΔI1 leads to significantly more mature SUS1 (Figure 4B, lane 7, see quantitation below). These findings indicate that intron 1, with its non-canonical SSs, limits splicing. The more surprising result came when ΔI1 was compared to a SUS1 construct in which the non-canonical SS sequences in I1 were mutated such that intron 1 contained the canonical 5′ SS and BP sequences. When intron 1 is canonical, it is removed efficiently such that the only partially spliced product that we observe contains the second intron (Figure 4B, lane 5). However, when intron 1 is artificially removed from this pre-mRNA, we observe an increase in the partially-spliced, intron 2-containing product relative to I1c, suggesting that the presence of intron 1 facilitates intron 2 removal (Figure 4B, compare lanes 5 and 7 and quantitation). Interestingly, we do not find strong evidence that deleting the second intron has a significant effect on removal of the first intron. Intron 1 appears to be spliced as efficiently from I1c-ΔI2, in which the second intron is deleted, as it is from I1c (compare lanes 5 and 6, see quantitation).
In order to address the apparent effect of intron 1 on intron 2 splicing in a different way, we analyzed constructs in which the first intron was mutated to the consensus sequence and the second intron was mutated to include the non-canonical 5′SS and BP sequences (I1c-I2nc). Splicing of this construct was compared to splicing of ΔI1-I2nc, containing the same non-canonical I2 but with the first intron deleted (Figure 4B, lanes 3 and 4). We predicted that if intron 1 facilitated splicing of intron 2 (as we observe with the ‘all canonical’ construct), artificial removal of I1 would lead to an increase in partially-spliced SUS1 and less mature SUS1 mRNA compared to the construct in which the first intron was present. Again, this is what we observe (Figure 4B, lanes 3 and 4, see quantitation in lower panel); in the absence of the first intron, the second intron is less efficiently spliced. No such effect on mRNA levels is observed when the second intron is artificially removed (Figure 4B, lanes 1 and 2). Taken together, these data indicate that the presence of the first intron stimulates removal of the second and that there are splicing effects that are exerted across exon 2 so that removal of one intron influences removal of the other. These observations suggest that this cross-exon ‘communication’ is an important feature of SUS1 intron removal.
Interestingly, the cross-exon effect in SUS1 shows some directionality. When the second intron contains non-canonical sequence, effectively making it more difficult to remove, the removal of the first intron is unaffected as no fully unspliced pre-mRNA is detected (Figure 4B, lane 3). On the other hand, when non-canonical sequences are in the first intron, making I1 more difficult to remove, we consistently observe some fully unspliced pre-mRNA, suggesting that I1 with non-canonical sequence influences the second intron’s removal (Figure 4B, lane 1). To test this further, the two introns were swapped such that I1 was in the second position and vice versa (Figure 4C). If the cross-exon effects were independent of intron position, we expected that both SUS1 configurations would lead to equal amounts of unspliced SUS1 pre-mRNA. However, we found the non-canonical intron only influences splicing of the other when it is in the upstream position, as almost no unspliced pre-mRNA is observed in the ‘swapped’ construct (Figure 4C). Had the cross-exon effects been independent of position, we might have expected the two constructs to have similar levels of unspliced pre-mRNA. Taken together, the data shown in Figure 4 suggest that assembly of the spliceosome at sequences in the 5′ intron enforce splicing of the second intron. Previous studies of splicing in mammalian cells showed tight coordination between two alternatively spliced regions within the same gene. In this case splicing of the 5′ most intron influenced removal of the 3′ intron, similar to our observations (35). Intriguingly, this ‘polar’ effect was promoter specific and was affected by transcriptional elongation rates (changes in transcription decreased the polarity of the coordinated removal of the introns), indicating that assembly of the spliceosome at the sequences that were the first to become co-transcriptionally available affected downstream events (35). We are currently exploring the possibility that a similar mechanism underlies SUS1 splicing and the splicing of other yeast genes containing multiple introns.
Intrinsic features of the second intron affect the efficiency of its removal
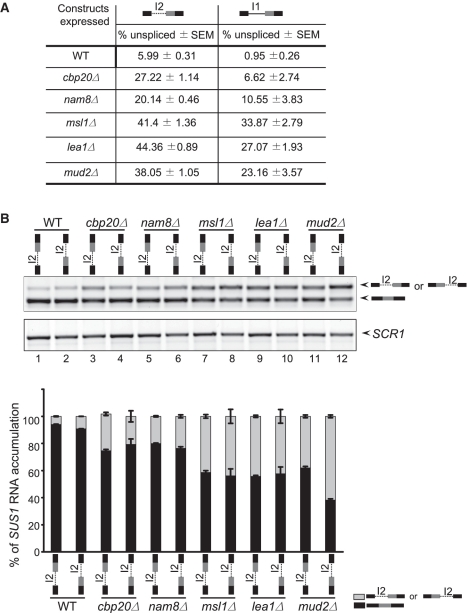
The analyses of SUS1 mutants confirm that the non-canonical SSs influence splicing efficiency. Interestingly, even when both introns are canonical, we consistently observe some partially spliced message containing the second intron (Figure 4B, lane 5). Furthermore, for the two-intron construct in which only the first intron is non-canonical (WT), we always observe some accumulation of fully unspliced RNA, indicating that some of the second intron remains unspliced (Figure 4B, lane 1) and suggesting that some intrinsic feature of the second intron other than SS signals may mitigate its removal.
Since the presence of two introns could complicate our analysis of the features of the second intron that affect its removal, we assessed splicing of either of the single intron constructs with canonical sequences (I1c-ΔI2 or ΔI1, Lanes 6 and 7) and consistently observe that the second intron is retained more than the first. To rule out the possibility that the difference in the efficiency of intron 2 removal was due to the difference in its location (e.g. I1 closer to the cap than I2), single intron constructs were generated in which there was no 3′ intron and either of the introns, I1 (with canonical SSs) or I2, were placed in the same 5′ intron position (Figure 5A). As we observed in Figure 4B, the single intron construct in which the only intron present is the second always accumulates some unspliced RNA (between 5% and 7% unspliced). This is in contrast with the mutated intron that normally sits in the first position, in which the unspliced pre-mRNA is nearly undetectable (Figure 5A). This was unexpected, since both introns contain canonical sequences. When splicing was analyzed in splicing factor mutants, we also observe a stronger splicing defect with the construct containing I2 alone. Interestingly, efficient removal of this intron is particularly sensitive to the activity of the U2 snRNP proteins and the commitment complex protein Mud2, all of which help to recognize sequences around the BP before the first catalytic step of splicing; when these factors are deleted, there is a significant increase in unspliced RNA (~40% unspliced, Figure 5A and B). In the absence of Mud2, splicing of this intron becomes dependent on its proximity to the 5′-end of the RNA (Figure 5B). Splicing of this intron is also sensitive to the presence of CBC and Nam8, but to a lesser extent: ~27 and 20% unspliced, respectively (Figure 5A). All of these results indicate that some intrinsic feature of this intron limits its splicing. It is notable that when the two introns are present in the WT context, we do not detect a partially spliced product containing I2, despite these indications that some intrinsic feature of the intron abrogates its splicing (10) (sequencing results from Supplementary Figure S1). This observation, along with the data presented in Figure 4, supports the model that the presence of the 5′ intron I1 stimulates removal of intron 2.
Figure 5.
Intron 2 (I2) is inefficiently removed from a single intron context. (A) The efficiency of splicing of the second intron is lower than removal of the first intron when both contain consensus sequences and are in the same sequence context. Table shows the accumulation of the SUS1 pre-mRNA when I1c (bold line) is in the first position (second column) or I2 (dotted line) is in first position (first column). (B) The distance from the 5′ cap affects the efficiency with which the second intron is removed, particularly in the absence of Mud2. The constructs used for the splicing assay are indicated above each lane. The lower panel shows SCR1 as a loading control. The bar diagram shows the accumulation of SUS1 RNA isoforms for the different mutants. The data presents the average of the three different experiments with standard error mean (SEM).
While we do not know exactly what feature of the second intron affects its splicing, we have noted that SUS1 from four closely-related yeast species share a number of conserved features, particularly in the second intron. The four species we have examined Saccharomyces paradoxus, S. mikatae, S. pastorianus and S. kudriavzevii all contain two introns; they all have a first intron with non-canonical sequences (Cuenca-Bono et al.), and the second intron of each of the SUS1 genes is of a similar size (~70 nt). These ~70 nt sequences share small regions of identity (Supplementary Figure S3A), and when these sequences are subjected to folding prediction, each is predicted to have two stable stem loop structures in which the 5′ SS is predicted to lie partially in the double-stranded region, and the BP sequence is predicted to lie in a loop of the second stem-loop structure, similar to S. cerevisiae (Supplementary Figure S3B). Notably, there are precedents in the literature that show that in S. cerevisiae, RNA secondary structure can influence intron removal (18,37). However, further experiments are necessary to confirm these putative structures and determine if they or the conserved sequences in SUS1 play any role in the efficiency of second intron removal. Overall, the evolutionary conservation of the two-intron nature of SUS1, raises the intriguing possibility that splicing efficiency for each of the introns may play an important role in SUS1 regulation.
The ability to produce a partially spliced SUS1 RNA is required for optimal cellular Sus1 function
To understand the role of SUS1 splicing in its in vivo function, we undertook a viability analysis in which either the SUS1 cDNA or the genomic sequence was expressed in sus1Δ cells. The cells were grown to a comparable OD and plated in a series of 10-fold dilutions. Cell viability was monitored at 30°C and 37°C. We consistently observe that sus1Δ cells show a temperature-sensitive growth defect on plates, as previously reported (38), and our own results suggest that SUS1 splicing is regulated in response to increased temperature. So, SUS1 was expressed under the control of its own promoter with 340 bp of 5′-UTR and with the 325 bp 3′-UTR, either containing both introns (SUS1 gDNA) or with both introns removed (SUS1 cDNA). We find that expression of the SUS1 cDNA does not complement the sus1Δ growth phenotype to the same extent as the genomic DNA construct (Figure 6A), a result also reported by Cuenca-Bono et al.
We previously showed that mis-splicing of SUS1 and the concomitant decrease in SUS1 mRNA causes a defect in histone H2B ubiquitination, consistent with its function as part of the H2B deubiquitination module of SAGA. Suppression of the overubiquitination defect was observed when the SUS1 cDNA was expressed (10). To determine whether the suppression observed with the SUS1 cDNA was equivalent to suppression conferred by the genomic DNA whose RNA products can undergo splicing, we analyzed the ability of each construct to restore proper H2B-ub levels in a sus1Δ strain. In sus1Δ cells, H2B ‘overubiquitination’ is apparent at both 30°C and 37°C but was more striking at 37°C (Figure 6B, lanes 1–4, left and right panels). The SUS1 gDNA restores H2B ubiquitination to WT levels (lanes 7 and 8), while the SUS1 cDNA shows slightly lower levels of suppression of the sus1Δ H2B overubiquitination defect (lanes 5 and 6, see quantitation below), consistent with the slightly weaker suppression of the 37°C growth defect by SUS1 cDNA. As described above, the partially spliced SUS1 is subject to NMD to control SUS1 mRNA and protein levels. However, this level of control is not possible when the cDNA encoding the fully-spliced message is expressed. Notably, the level of mRNA from the cDNA construct is 2.5-fold higher than that from the gDNA construct (Figure 6C). It is likely that the different RNA levels could contribute to differences in the growth suppression and H2B overubiquitination suppression conferred by the two constructs, i.e. there is an optimal range of SUS1 required for proper cellular function, such that too little (empty vector) or too much (cDNA) is deleterious to Sus1-dependent functions.
This does not rule out the possibility that the partially spliced SUS1 RNA is exported and translated to produce a peptide that is itself functional. Since retention of the first intron produces a premature termination codon (PTC), we analyzed whether expression of a DNA expressing the truncated SUS1 RNA might affect H2B ubiquitination. As shown in Figure 6B, gDNA restores H2B-ub to near WT levels, while the cDNA is unable to fully suppress the H2B overubiquitination phenotype. Interestingly, SUS1 I1-P containing the first intron up to the PTC suppresses H2B overubiquitination better than the empty vector, but not as well as SUS1 cDNA (Figure 6D). To summarize, suppression of H2B overubiquitination in sus1Δ follows the trend: vector<SUS1-I1-P<cDNA<gDNA. These data are consistent with reports from Cuenca-Bono et al. showing that a SUS1 mutant (SL-9) that produces a truncated protein can partially suppress the growth defect of sus1Δ cells. While we do not know precisely how the truncated Sus1 product or the tight control of SUS1 mRNA levels affect H2B ubiquitination, it is clear from these studies that formation of alternative isoforms is important for Sus1 function and that this alternative isoform production is regulated at multiple levels.
DISCUSSION
While it has long been appreciated that alternative splicing plays a central role in regulating eukaryotic gene expression, the relatively small number of intron-containing genes and even smaller number of genes with multiple introns in S. cerevisiae has called into question the importance of alternative splicing in this organism. Here we show that the two-intron gene SUS1 undergoes changes in its splicing in response to a change in environmental conditions (e.g. prolonged temperature elevation) (Figure 2). Intriguingly, cells in which Sus1 is completely absent (sus1Δ) show a growth defect at elevated temperatures and although the SUS1 cDNA can partially complement the defect, cells that are able to undergo splicing of SUS1 (gDNA) can fully complement sus1Δ growth and H2B ubiquitination, suggesting that splicing provides a mechanism for generating an isoform (or isoforms) that allows optimal function of the cell (Figure 6). We propose two non-mutually exclusive possibilities: that formation of a partially spliced product that can be targeted to the NMD pathway (Figures 2 and 3) allows an important mechanism by which to modulate Sus1 protein levels and/or that the partially spliced pre-mRNA can be translated into a functional polypeptide. Consistent with this latter possibility, data from the accompanying Cuenca-Bono et al. paper provides evidence of production of a short polypeptide from the transcript in which intron 1 of SUS1 is retained. Furthermore, our data demonstrate that expression of a construct that includes sequence up to the predicted PTC can partially suppress the sus1Δ H2B deubiquitination defect. Some combination of these mechanisms is likely to be important to regulate Sus1 activity in the cell, and work that is currently under way is aimed at distinguishing between the relative contributions of these mechanisms for optimum cellular function under a variety of stress conditions. Nonetheless, our observations that half of all multi-intron yeast genes have sequence features that would be predicted to differentially affect intron retention (Figure 1) and all of these introns, if retained, would be predicted to encode premature termination codons, suggest that alternative splicing may be a more important form of regulation in S. cerevisiae than previously appreciated. Hence, an understanding of the different mechanisms by which alternative SUS1 isoform production contributes to regulated gene expression is likely to inform our understanding of the regulation of these other genes.
These studies with SUS1 also reveal a number of unexpected features of the yeast splicing reaction. Our observation that specific introns within a single gene are differentially sensitive to the activities of specific splicing factors (Figures 3 and 5) suggests that the core machinery in yeast, and likely other eukaryotic organisms, could provide a mechanism for alternative splicing. Indeed, Pleiss et al. (39) utilized splicing-sensitive microarrays to show that deletion of different core components of the spliceosome showed distinct profiles of genome-wide splicing, illustrating that splicing can be regulated by the activity of the core machinery. One implication of these results is that components of the splicing machinery may themselves be regulated in response to stress to influence splicing of particular introns. Indeed, analyses of gene expression changes in response to stress illustrate that the splicing proteins that we have analyzed here all change in their expression levels under a variety of stress conditions (40). Work that is currently under way will elucidate whether these changes in environmental conditions and splicing factor levels lead to concomitant changes in SUS1 splicing patterns. Although we have not found a role for the other SUS1 isoform described here, in which the middle exon is skipped (Figure 3), the formation of this product is also sensitive to the activities of core components of the spliceosome and the cap binding complex. It is possible that formation of this isoform may be regulated and that its regulation may be important under some conditions—a possibility that we are also exploring.
Recently, bioinformatic analyses estimate that 40% of human genes contain multiple regions of alternative splicing, and analysis of EST sequences suggest that for many of these genes, these alternative splicing events are not independent but are, in fact, coordinated, such that removal of one intron affects removal of another more distal intron (35). When a specific transcript was analyzed, it was found that each of the individual alternative splicing events was mediated by exon definition, suggesting that interactions across several exons were important for splicing. Interestingly, splicing of the 5′ intron specifically influenced removal of the 3′ intron. The results of these studies support a model in which a splicing event ‘primes’ spliceosome assembly to occur at a distal sequence as soon as the appropriate signals are transcribed and recognized. The 5′→3′ nature of transcription makes this effect polar, and consistent with this, changes in transcription relax the polarity of the coordinated intron removal (35). Our studies of SUS1 splicing show that removal of one intron can affect removal of the other (Figure 4), and these data also suggest that the 5′ intron has a stronger effect on removal of the 3′ intron than the other way around. We propose a model in which interactions that occur across the middle exon influence removal of the two introns (Figure 7). Furthermore, the co-transcriptional nature of pre-mRNA splicing (19) would explain why the effects that we observe also appear to be polar. While we do not know the exact nature of these cross-exon interactions, our data suggest that components of the upstream spliceosome facilitate assembly of the downstream spliceosome.
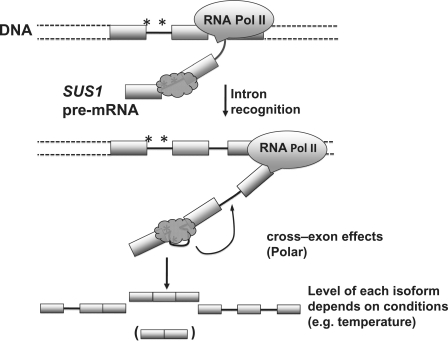
Figure 7.
The proposed model of SUS1 splicing. Under WT conditions, the 5′ intron is recognized by the splicing machinery (dark gray shape). The efficiency of removal of this intron is limited by the non-canonical sequences found at the 5′ SS and BP (Indicated by asterisks). When the signals in the 3′ intron are synthesized, splicing events at the 5′ intron stimulate spliceosome assembly and/or splicing catalysis for removal of the 3′ intron. The primary outcome of these ‘cross-exon’ effects is the removal of both introns to generate the mature SUS1 mRNA. Some product in which the first intron is not efficiently removed and some completely unspliced SUS1 transcripts are also observed. The relative amount of each of these products is dependent upon environmental conditions such as temperature. A small amount of the skipped product is also observed and shown in parenthesis, although its levels do not appear to be affected by the environmental conditions tested. Boxes represent exons; lines between boxes represent introns. The DNA and SUS1 pre-mRNA are labeled.
Previous studies with the intron-containing YRA1 transcript showed that retention of the YRA1 intron provided a mechanism for regulating Yra1 protein levels. In this case, Yra1 protein facilitated the export of the unspliced YRA1 message such that it could be targeted to the Xrn1 (but not NMD)-dependent decay pathway (7), thus forming a tight regulatory loop. Since Sus1 is also a component of the RNA export machinery (as part of the TREX-2 complex) and, like Yra1, it contains sequences that would favor intron retention, it is possible that SUS1 splicing regulation also provides a mechanism for control of the RNA export machinery.
One of the features of Sus1 that distinguishes it from Yra1 is that it has a separate role in histone modification (histone H2B deubiquitination) as a component of the SAGA complex. Our previous studies demonstrated that histone modification via another component of the SAGA complex, Gcn5, can have striking effects on co-transcriptional spliceosome assembly (41,42). It is appealing to speculate that Sus1-mediated H2B deubiquitination could contribute to regulation of SUS1 splicing, adding an additional layer of autoregulation. Moreover, the observation that a polypeptide can be generated from the partially-spliced product (Cuenca-Bono et al.) raises the possibility that this product may play a role in some aspect of the SUS1 regulation and putative autoregulation.
Since the Sus1 protein is part of two large multi-subunit complexes that are critical for proper gene expression, SAGA and the TREX-2 export complex, it is not surprising that there are multiple mechanisms involved in tight regulation of Sus1 levels and functions. The studies described here demonstrate that regulation of RNA splicing is critical for SUS1 regulation. Furthermore, splicing regulation is likely to be a more common feature of S. cerevisiae gene regulation than previously appreciated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation awards (NSF CAREER MCB-0448010 and MCB-1051921 to T.L.J.); National Institutes of Health (GM085474 to T.L.J.). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs Susana Rodriguez-Navarro and Josep Vilardell and colleagues for sharing unpublished results and for useful comments and suggestions. We would also like to thank Erik Soule for technical assistance and Julia Claggett for careful reading of the manuscript.
REFERENCES
- 1.Kim E, Goren A, Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- 2.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 4.Vilardell J, Warner JR. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 5.Vilardell J, Chartrand P, Singer RH, Warner JR. The odyssey of a regulated transcript. RNA. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preker PJ, Kim KS, Guthrie C. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA. 2002;8:969–980. doi: 10.1017/s1355838202020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juneau K, Nislow C, Davis RW. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics. 2009;183:185–194. doi: 10.1534/genetics.109.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hossain MA, Claggett JM, Nguyen T, Johnson TL. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15:1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 13.Schellenberg MJ, Ritchie DB, MacMillan AM. Pre-mRNA splicing: a complex picture in higher definition. Trends Biochem. Sci. 2008;33:243–246. doi: 10.1016/j.tibs.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 15.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2011;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Cuenca-Bono B, García-Molinero V, Pascual-García P, Dopazo H, Llopis A, Vilardell J, Rodríguez-Navarro S. SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr496. doi:10.10.93/nar/gkr496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prodromou C, Pearl LH. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 22.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 23.Pascual-Garcia P, Rodriguez-Navarro S. A tale of coupling, Sus1 function in transcription and mRNA export. RNA Biol. 2009;6:141–144. doi: 10.4161/rna.6.2.7793. [DOI] [PubMed] [Google Scholar]
- 24.Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis CA, Grate L, Spingola M, Ares M., Jr Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawashima T, Pellegrini M, Chanfreau GF. Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA. 2009;15:2236–2247. doi: 10.1261/rna.1736809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 30.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 31.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fededa JP, Petrillo E, Gelfand MS, Neverov AD, Kadener S, Nogues G, Pelisch F, Baralle FE, Muro AF, Kornblihtt AR. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol. Cell. 2005;19:393–404. doi: 10.1016/j.molcel.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, Shendure J, Mitra RD, Church GM. Single molecule profiling of alternative pre-mRNA splicing. Science. 2003;301:836–838. doi: 10.1126/science.1085792. [DOI] [PubMed] [Google Scholar]
- 37.Rogic S, Montpetit B, Hoos HH, Mackworth AK, Ouellette BF, Hieter P. Correlation between the secondary structure of pre-mRNA introns and the efficiency of splicing in Saccharomyces cerevisiae. BMC Genomics. 2008;9:355. doi: 10.1186/1471-2164-9-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc. Natl Acad. Sci. USA. 2011;108:2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.