Abstract
CUGBP, Elav-like family member 1 (CELF1) is an RNA binding protein with important roles in the regulation of splicing, mRNA decay and translation. CELF1 contains three RNA recognition motifs (RRMs). We used gel retardation, gel filtration, isothermal titration calorimetry and NMR titration studies to investigate the recognition of RNA by the first two RRMs of CELF1. NMR shows that RRM1 is promiscuous in binding to both UGU and CUG repeat sequences with comparable chemical shift perturbations. In contrast, RRM2 shows greater selectivity for UGUU rather than CUG motifs. A construct (T187) containing both binding domains (RRM1 and RRM2) was systematically studied for interaction with tandem UGU RNA binding sites with different length linker sequences UGU(U)xUGU where x = 1–7. A single U spacer results in interactions only with RRM1, demonstrating both steric constraints in accommodating both RRMs simultaneously at adjacent sites, and also subtle differences in binding affinities between RRMs. However, high affinity co-operative binding (Kd ~ 0.4 µM) is evident for RNA sequences with x = 2–4, but longer spacers (x ≥ 5) lead to a 10-fold reduction in affinity. Our analysis rationalizes the high affinity interaction of T187 with the 11mer GRE consensus regulatory sequence UGUUUGUUUGU and has significant consequences for the prediction of CELF1 binding sites.
INTRODUCTION
CELF1, also known as CUG-BP1 or EDEN-BP, is a member of the CELF/Bruno-like protein family, a group of RNA binding proteins that is widely conserved in the animal kingdom, both at the functional and the sequence level (1–5). CELF1 and its relatives can regulate mRNA function at the level of alternative splicing, translational repression, translational activation, deadenylation and destabilization (6–13). Up-regulation of CELF1 has been linked to muscular dystrophy and ageing in humans, while the CELF1 knockout mice suffer from reduced viability, growth retardation and infertility (14,15). CELF1 is ubiquitously expressed and has been shown to regulate the expression of proteins involved in somitic segmentation, muscle differentiation, cell proliferation and the production of inflammatory cytokines, and is therefore likely to be involved in a variety of important physiological and pathological cellular processes (2,13,16–23).
Unmodified CELF1 has a low affinity for CUG repeats and binding studies indicate that its preferred substrates are GU rich sequences, both in mammalian species and for Xenopus (24–26). Recently, these GU rich elements were identified as abundant targets of the CELF proteins that function both in alternative splicing and in mRNA destabilization (2,12,27,28). By bioinformatic analysis, the sequence UGUUUGUUUGU was derived as the consensus binding element (13,29). However, many well characterized natural binding sequences consist of more dispersed UGU repeats with apparent dissociation constants of 3–100 nM (7,13,18,24,29,30).
There is a different class of natural CELF1 RNA substrates that are characterized by GC rich sequences, similar to the CUG repeat. CELF1 regulates the translation of these mRNAs by binding to their 5′-UTRs. Binding to substrates is enhanced by phosphorylation, which may be why these binding sites have so far not been identified in systematic screens of CELF1 substrates (19,21,31,32).
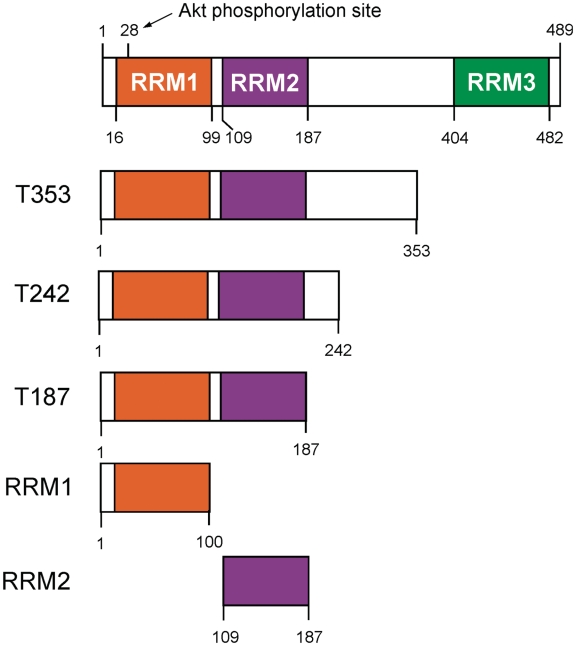
The CELF proteins all have three RNA binding motifs (RRMs), two at the N terminus and one at the C terminus (Figure 1). The domains required for RNA binding of CELF1 have been studied on multiple substrates (25,31,33,34). Using a yeast three-hybrid approach, human CELF1 was found to bind GU rich elements both by the third RRM alone and by a construct from which the third RRM was deleted. Smaller constructs containing RRM1 and RRM2 alone were not capable of binding to the RNA (25). For Xenopus CELF1 [also called EDEN-BP based upon the recognition of the Embryonic Deadenylation ElemeNt (EDEN)], binding to a GU rich substrate in vitro was found to depend solely on the first two RRMs and a portion of the linker region. This linker region has also been implicated in the dimerization or oligomerization of the protein (33,35). CUG binding of CELF1 is also thought to be dependent on the highly conserved first two RRMs indicating that these domains may convey dual specificity (34).
Figure 1.
Schematic representation of the protein constructs used in this study and an indication of domain boundaries. The position of the RRMs are shown shaded. Residue Ser28 represents the proposed Akt phosporylation site in RRM1.
Recently structural studies were published for the third RRM of human CELF1 and Drosophila Bruno (36,37). These indicate that this RRM has an N-terminal extended region and specifically binds UGU, but not CUG, tri-nucleotides. We report here that the first two RRMs also specifically bind UGU tri-nucleotides independently of the linker region between them. For both RRMs to bind in tandem to an RNA molecule, the spacing (x) between the tri-nucleotides in the UGUNxUGU recognition motif has to be ≥1 nt, with x = 2–5 demonstrating effective binding to RRM1 and RRM2. From gel filtration and Nuclear Magnetic Resonance (NMR) titration studies, we are able to show that RRM1 and RRM2 have different affinities and specificities for UGU and CUG/UGC motifs. These data lead to a new consensus binding sequence for CELF1.
MATERIALS AND METHODS
Cloning and expression
Full length coding sequences of Xenopus CELF1 were obtained by RT–PCR on oocyte RNA, using primers on the start and stop codons. The primers added Xho1 and Nhe1 sites to allow cloning in pET28a. Sequencing revealed that the isoform A was identical to the coding region in NM_001090727.1, isoform B to NM_001086501.1 and isoform C had the same insertion as EST DC055829.1, but was otherwise identical to isoform A. The DNA of the T187 construct was cloned into the pET-28 a expression vector (Novagen), in fusion with an N-terminal His-tag and thrombin cleavage site. The RRM1 construct was produced by point mutation of Asn102 to a stop codon. The RRM2 DNA sequence was cloned into the NheI/HindIII site of pET28a. All proteins were overexpressed in Escherichia coli BL21 (DE3) cells grown in Luria Broth in the case of unlabelled material, and M9 minimal media for the production of 15N- and 13C-labelled material. All media was treated with 30 µg/ml kanamycin. Protein expression was induced for 16 h at 30°C. Harvested cells were lysed by sonication in 25 mM potassium phosphate, 50 mM NaCl, pH 7.0 buffer. A protease inhibitor cocktail (Roche) was added immediately before sonication.
After centrifugation to remove cell debris, the lysate was applied to a His-tag binding cobalt column (TALON) and allowed to bind for 2 h. After binding the beads were washed with 25 ml of high salt buffer (2 M NaCl, 25 mM potassium phosphate, pH 7.0), followed by 25 ml of 10 mM imidazole. The protein was eluted with 500 mM imidazole, 25 mM potassium phosphate, 50 mM NaCl). The His-tag was removed by an overnight incubation with thrombin at room temperature. T187, RRM1 and RRM2 were all purified by gel filtration (Superdex 75), followed by desalting into water on a HiTrap desalting column (Amersham Biosciences). Protein was lyophilized immediately after desalting.
UV crosslinking and gel retardation
The maskin 3′-UTR (Msk) and the xxsB1 plasmids, RNA probe synthesis from them and UV crosslinking with RNase digestion have been described previously (38). For gel retardation, the RNAs oligonucleotides synthesized by Dharmacon were end-labelled with [γ-32P]-ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (NEB) in 10 μl for 30 min at 37°C. After denaturation at 90°C for 3 min, 1 μl of RNA was added to 9 μl of protein solution at increasing T187 concentrations in buffer A (12.5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.9, 5 mM MgCl2, 15% glycerol, 1 mM EDTA, 5 µg/ml BSA, 0.025% NP40, 80 mM NaCl, 0.2 mg/ml heparin, 100 ng/µl poly I/C) or B (20 mM HEPES, pH 7.0, 2 mM MgCl2, 5 mM DTT, 100 mM KCl, 0.2 mg/ml heparin, 1 µg/ml tRNA) to achieve ratios of RNA: protein from 1:0 to 1:100. Binding reactions (10 μl) were incubated for 10 min, and native gel electrophoresis was used to separate bound and free RNA species as described previously (39).
Gel filtration
The RNA oligonucleotides (Dharmacon) were dissolved in RNase free H2O, to make a 7-mM solution. The RNAs were denatured for 3 min at 90°C then placed on ice for another 3 min to remove any secondary structure. RNAs were incubated with the protein (1:1 ratio) in buffer for 10 min at room temperature. A 250 μL volume of protein solution, RNA solution or protein–RNA complex were loaded on a Gel filtration Superdex 75 10/300 column (Amersham Biosciences) at 4°C. The final protein concentration was 50 µM for all experiments and the final RNA concentration 50 µM for a 1:1 ratio and was altered to achieve the different ratios in each case. The column was pre-equilibrated and run in 25 mM HEPES pH 7.0, 50 mM NaCl at 0.5 ml/ml.
NMR spectroscopy
NMR samples were prepared by dissolving lyophilized protein in 600 µl of 25 mM potassium phosphate, 50 mM NaCl, pH 7.0, 10% D2O (v/v). All experiments were carried out at 25°C on a 600 MHz Bruker Avance spectrometer. A protein concentration of 1 mM was used in the collection of 3D data. An amount of 400–500 µM protein samples were used in the RNA titrations, with RNA added from a 7 mM concentrated stock over the course of the titration. The RNA was supplied by Dharmacon. 1H/15N-TROSY spectra were collected at each titration point, with the titration being terminated when no further variation was seen. Backbone assignments of the T187, RRM1 and RRM2 constructs were obtained using 1H/15N-HSQCs, and triple resonance experiments (NHCO, NHCACO, CBCANH, CBCACONH). Data was processed using Topspin 2.1 and analysed in CCPNMR Analysis. Heteronuclear 1H/15N Nuclear Overhauser Effect (NOE) data were collected on an 800 MHz spectrometer with ultrashield magnet and cryoprobe facility.
Isothermal titration calorimetry
ITC measurements were collected using a Microcal VP-ITC calorimeter. All experiments were performed at 25°C. Samples were dissolved in degassed RNase free water. An amount of 25 µM RNA solutions were prepared in the cell, and a 250 µM solution of the protein injected from the syringe. The titration consisted of 30 injections of 10 µl each at 5-min intervals. Data was analysed using the Microcal ORIGIN software.
Mass spectrometry
All ESI mass spectra were collected on a Waters SYNAPT instrument with a quadruple time-of-flight mass analyser calibrated using horse heart myoglobin. The MasslynxTM (Waters) software was used to acquire and analyse the data. Samples were prepared by dissolving lyophilized protein in water and then desalting into 50 mM ammonium acetate through a HiTrap desalting column (Amersham Biosciences). RNA was added by injection of small amounts from a stock solution of 7 mM RNA in RNase-free water. Samples were injected at 5 µl/min. Data for some RNA–protein complexes required collection in negative ion mode.
RESULTS
CELF1 constructs and UV cross-linking
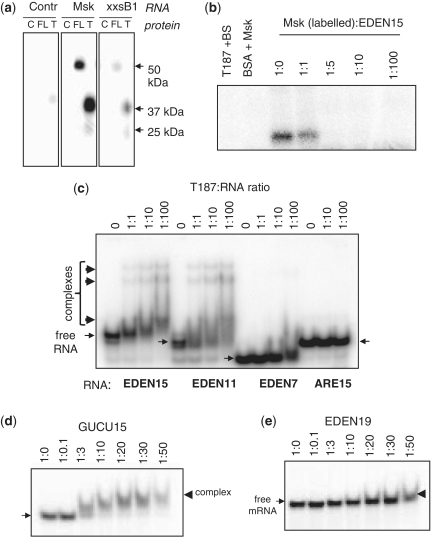
In order to study the structure and RNA binding properties of CELF1, we amplified the Xenopus CELF1 coding region from oocyte cDNA and cloned it into a bacterial expression vector. To define the minimum requirement for specific RNA binding in the N terminal RRMs of CELF1 we prepared His-tagged bacterial expression constructs to produce individual and multiple domains of the original Xenopus CELF1, as indicated schematically in Figure 1 and as SDS–PAGE in Supplementary Figure S1. Earlier reports indicated that the first 353 amino acids were required for binding (termed T353), so we initiated our studies with the T353 construct and the full length expression clone (35). As shown in Figure 2a, the purified proteins showed efficient UV crosslinking to a high affinity natural substrate, the maskin 3′-UTR, as well as reduced efficiency to a low affinity substrate, a short section of the cyclin B1 3′-UTR (40). Under the conditions used, little cross-linking was observed with a control RNA. Surprisingly, a minor degradation product of the T353 protein of ~25 kDa also cross-linked to both the high and the low affinity substrates (data not shown). This demonstrates that the linker region is not an absolute requirement for specific binding and it encouraged us to try some of the shorter constructs. In Figure 2b UV crosslinking experiments show the maskin 3′-UTR interaction with a construct spanning residues 1–187 (T187) was completely competed out using synthetic EDEN15 RNA at ratios of 1:5, while no crosslinking was detected with a control RNA (Bluescript). These data demonstrate that the T187 region containing the first two RRMs binds RNA with a high specificity.
Figure 2.
The first 187 amino acids of CELF1 are sufficient for specific binding to natural and synthetic binding sites. (a) UV-crosslinking of BSA control (C), full length CELF1 (FL), T353 (T) with radioactive maskin 3′-UTR (Msk), a truncated cyclinB1 3′-UTR (xxsB1) and a Control RNA (Bluescript polylinker transcript). Sizes of crosslinked bands are indicated. (b) UV-Crosslinking competition assay. The maskin 3′-UTR was competed with synthetic EDEN15. Radioactive control RNA (Bluescript: BS) or maskin 3′-UTR with EDEN15 in the indicated molar ratios were incubated with T187 and covalently crosslinked using by UV light. (c) Gel retardation assay with T187 EDEN15, EDEN11 and EDEN7, as well as with the AU rich element (ARE15) incubated in buffer A. An amount of 50 nM of RNA end labelled with γ-P32-ATP was incubated with increasing concentration of t187 protein, ratio RNA: protein from 1:0 to 1:100. Slender arrows indicate free RNA, thick arrows complexes induced by the protein. (d) Gel retardation of 45 nM GUCU15 with increasing concentrations of T187 in buffer B. (e) Gel retardation of 45 nM EDEN19 with increasing concentrations of T187 in buffer B.
CELF1 RNA complex formation measured using gel shift and gel filtration
We designed a series of synthetic RNAs (Table 1) based on the reported binding sequences of CELF1 and tested binding using gel shift, gel filtration and mass spectrometry. In a gel shift assay, the T187 protein bound well to the consensus EDEN sequence (EDEN15, also known as the GRE or the UGUU repeat), forming three different complexes (Figure 2c). EDEN11, which lacks one repeat, formed similar complexes at similar protein concentrations. In contrast, EDEN7 (UGUUUGU) only formed small amounts of complex at much higher concentrations of protein indicating that EDEN11 contains the minimal optimal binding site. No complex formation was observed with an AU rich element (Figure 2c). The GUCU repeat, which is present in the Maskin 3′-UTR, also gave efficient gel shifts, while the EDEN19 sequence from the c-Mos mRNA bound only weakly, if at all (Figure 2d and e). These data indicate that T187, as has been reported for the full length protein, binds to UGU rich sequences, but that the context is not optimal in EDEN19 and that EDEN7 does not contain a full binding sequence.
Table 1.
RNA sequences
| RNA name | Sequence |
|---|---|
| EDEN19 | UGUAUGUGUUGUUUUAUGU |
| EDEN15 | UGUUUGUUUGUUUGU |
| EDEN11 | UGUUUGUUUGU |
| EDEN9 | UUGUUUGUU |
| EDEN7 | UGUUUGU |
| EDEN7m | UGUUUAU |
| EDEN6 | UGUUUG |
| GUCU15 | UGUCUGUCUGUCUGU |
| CUG15 | CUGCUGCUGCUGCUG |
| CUG2 | CUGCUG |
| ARE15 | UAUUUAUUUAUUUAU |
| ARE7 | UAUUUAU |
| UG7 | UGUGUGU |
| UG15 | UGUGUGUGUGUGUGU |
| EDEN7 sequences (UGU(U)xUGU) | |
| EDEN7 (x = 1) | UGUUUGU |
| EDEN2U (x = 2) | UGUUUUGU |
| EDEN3U (x = 3) | UGUUUUUGU |
| EDEN4U (x = 4) | UGUUUUUUGU |
| EDEN5U (x = 5) | UGUUUUUUUGU |
| EDEN6U (x = 6) | UGUUUUUUUUGU |
| EDEN7U (x = 7) | UGUUUUUUUUUGU |
Potential RRM binding sites underlined.
We next performed gel filtration experiments using an analytic Superdex 75 column to detect CELF1 RNA complex formation and characterize stoichiometry of binding. The column was calibrated using known molecular weight standards. Table 2 shows the results for the T187 construct and isolated RRM1 and RRM2 domains showing complexes formed with a variety of RNAs which were also analysed in isolation. T187, RRM1 and RRM2 elute at slightly higher values (28.2, 18.5, 13.3 kDa) than their calculated molecular weights (21.7, 12.1, 9.6 kDa respectively) probably reflecting the elongated shape. Consistent with gel shift experiments no complex formation was observed for these constructs when mixed at a 1:1 molar ratio with the ARE7 RNA. The results for T187 and EDEN15 also confirmed the observations from the gel shift assays and two different complexes of weights 42.5 and 59.2 kDa are clearly separated from a 1:1 molar mixture. With an excess of protein (1:0.25 ratio) the larger complex predominates indicating there are likely two binding sites for T187 on EDEN15. An excess of RNA results in the smaller complex in isolation representing a 1:1 complex. Speculative schematic models representing how the RRMs might engage the UGUs on EDEN15 are illustrated together with the gel filtration profile in Figure 3a. We also observed complex formation between T187 and RNAs GU15, CUG15.
Table 2.
Summary of gel filtration experiments with synthetic RNA and recombinant CELF1 constructs
| Protein/RNA sample | Elution volume (ml) | MW (kDa) | Complex | Stoichio- metry |
|---|---|---|---|---|
| T187 (21.7 kDa) | 12.5 | 28.2 | – | – |
| T187/EDEN15 | 11.2,10.3 | 42.5,59.2 | Y | 1:1, 2:1 |
| T187/EDEN11 | 11.4 | 40.3 | Y | 1:1 |
| T187/EDEN9 | 11.5 | 39.0 | Y | 1:1 |
| T187/EDEN7 | 11.7 | 36.4 | Y | 1:1 |
| T187/EDEN7m | 12.0 | 33.4 | Y | 1:1 |
| T187/CUG15 | 11.5 | 38.9 | Y | 1:1 |
| T187/GU15 | 11.2,10.4 | 43.1,56.2 | Y | 1:1, 2:1 |
| T187/ARE7 | 12.5 | 28.2 | N | – |
| RRM1 (12.1 kDa) | 13.7 | 18.5 | – | – |
| RRM1/EDEN7 | 12.3 | 29.5 | Y | 2:1 |
| RRM1/(UG)3 | 13.0 | 23.3 | Y | 1:1 |
| RRM1/EDEN7m | 13.0 | 23.7 | Y | 1:1 |
| RRM1/ARE7 | 13.6 | 19.0 | N | – |
| RRM2 (9.6 kDa) | 14.7 | 13.3 | – | – |
| RRM2/EDEN7 | 13.4 | 20.7 | Y | 1:1 |
| RRM2/ARE7 | 14.6 | 14.0 | N | – |
| EDEN15 | 13.3 | 21.5 | ||
| EDEN11 | 13.9 | 17.4 | ||
| EDEN7 | 14.9 | 12.4 | ||
| EDEN7m | 14.9 | 12.4 | ||
| ARE7 | 14.8 | 12.7 | ||
| (UG)3 | 15.8 | 9.3 |
Figure 3.
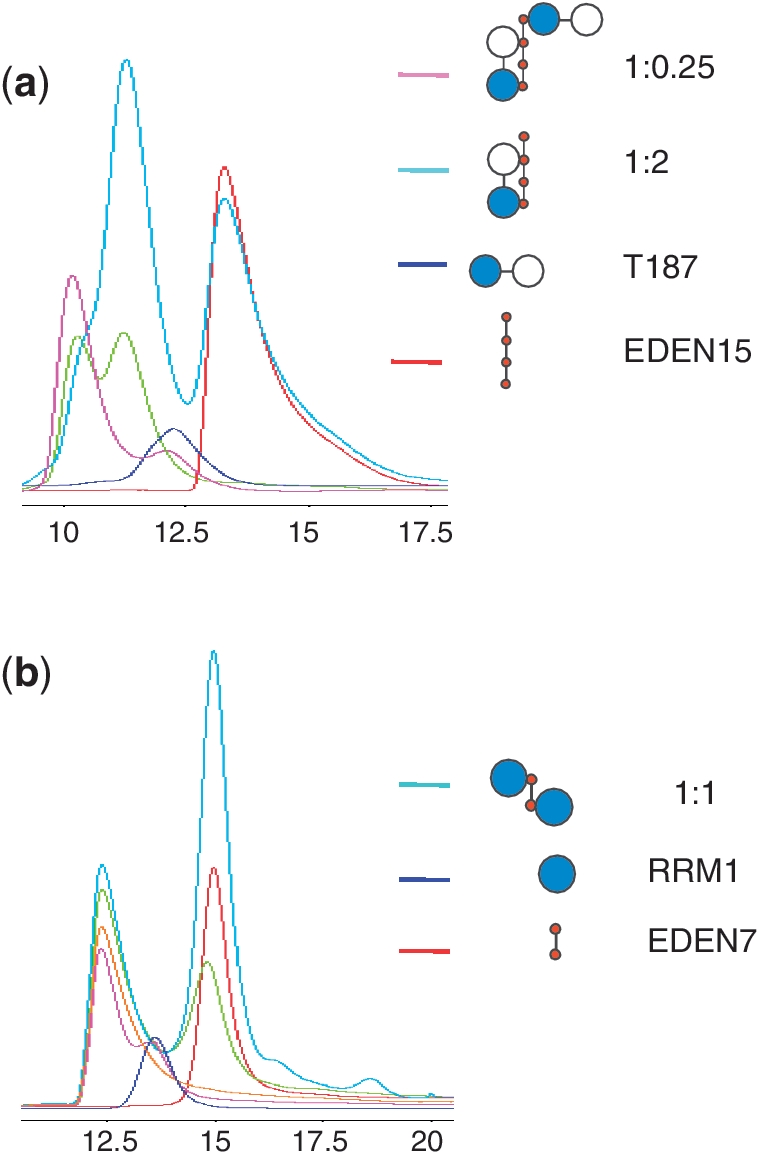
CELF1 RNA complex gel filtration profiles. (a) Plot of UV absorbance versus elution volume for T187 protein (blue) with RNA EDEN15 (red) mixed at two different molar ratios, 1:0.25 (pink); 1:1 (green), 1:2 (turquoise). (b) CELF1 RRM1 RNA complex gel filtration profiles. In blue RRM1 and in red EDEN7 alone. Different ratios RRM1:EDEN7: 1:0.25 (pink); 1:0.5 (orange); 1:1 (green) and 1:2 (turquoise).
Gel filtration also successfully identified complexes for RRM1 and RRM2 with the EDEN7 RNA. In the case of RRM2 the calculated weight of 20.7 kDa is consistent with a 1:1 complex. Unexpectedly, identical experiments with RRM1 resulted in a larger weight of 29.5 kDa suggestive of a 2:1 complex. This is perhaps not so surprising as EDEN7 contains two UGU motifs. To test this we designed RNA with sequence UGUUUAU termed EDEN7m intended to disrupt the second UGU binding site. The size of the RRM1:EDEN7m complex was then significantly reduced to 23.7 kDa consistent with a 1:1 complex. A titration of different ratios of RRM1 and EDEN7 is illustrated in Figure 3b but unlike similar experiments with T187 and EDEN15 no 1:1 complex is detected. Similarly, mass spectrometry of an RRM1/EDEN9 mixture showed two species of mass 12 174 and 27 118. The RRM1 construct has a theoretical mass of 12 182.8, and EDEN9 plus two molecules of RRM1 has a calculated mass of 27 135.6, consistent with the observed species. No peak representing the 1:1 complex was observed (Supplementary Data).
NMR analysis of protein-RNA interactions for the isolated RRM1 and RRM2
We performed 2D and 3D multi-dimensional NMR spectroscopy on both singly (15N) and doubly labelled (13C/15N) samples of recombinant RRM1, RRM2 and T187 all of which were soluble in the 0.2–1.0 mM concentration range and gave concentration-independent NMR spectra. The initial assignment of the two individual RRMs greatly facilitated the analysis of the longer T187 construct which contains 12 proline residues. Ninety-three percent of the non-proline backbone NH resonances (1H and 15N) of T187 could be assigned, with the unassigned residues located largely within the disordered N-terminus of RRM1. We further examined protein dynamics by measuring 1H/15N NOEs (Supplementary Data). Within the folded RRMs of T187 the NOEs are typically ≥0.7, consistent with compact globular structures. However, residues within the loop sequence between strands β2 and β3 of RRM1, and those within the linker sequence between domains (98–105), show a significant reduction in heteronuclear NOE (<0.6), with residues 100–102 in particular showing evidence for considerable flexibility (NOE < 0.3).
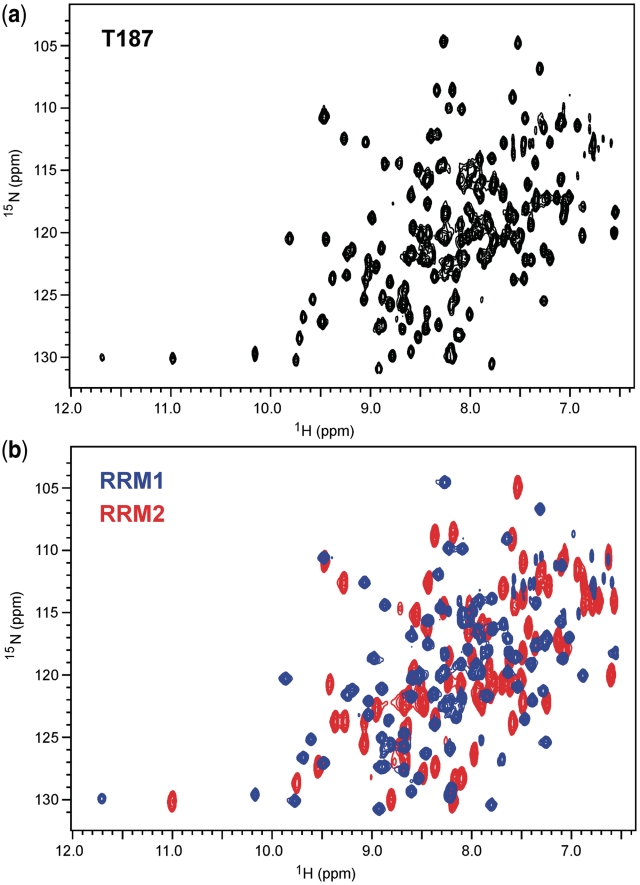
Superposition of the 1H/15N 2D TROSY spectrum of T187 with first RRM1 and then RRM2 shows the spectrum of T187 to be well approximated as the sum of those of RRM1 and RRM2 (Figure 4), with differences localized to residues 100 and 101 in RRM1, 108 and 109 in RRM2 which lie at the domain boundaries. The data are consistent with a structural representation of T187 as two independent folded domains connected via a flexible linker. We also examined a longer construct T242 which contains a portion of the C-terminal linker region between RRMs 2. The TROSY spectrum for T242 is very similar to that of T187, but reveals that the additional ~50 cross peaks have narrow line widths and generally poor chemical shift dispersion, indicative of a random coil, flexible C-terminal tail.
Figure 4.
2D 1H/15N-TROSY NMR spectra of T187 (a) and overlayed spectra of RRM1 (red) and RRM2 (blue) in (b) showing that the longer construct is well represented by the sum of the spectra of the individual domains, except for a few perturbations at the domain boundaries. Spectra were collected at 298 K in 25 mM phosphate buffer, 50 mM NaCl, 10% D2O (v/v), pH 7.0 with protein sample concentrations in the range 400–500 µM.
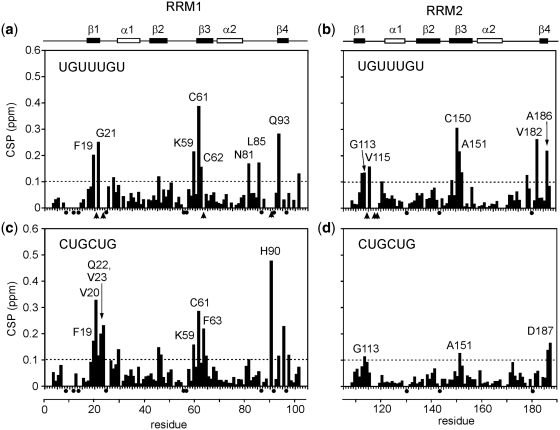
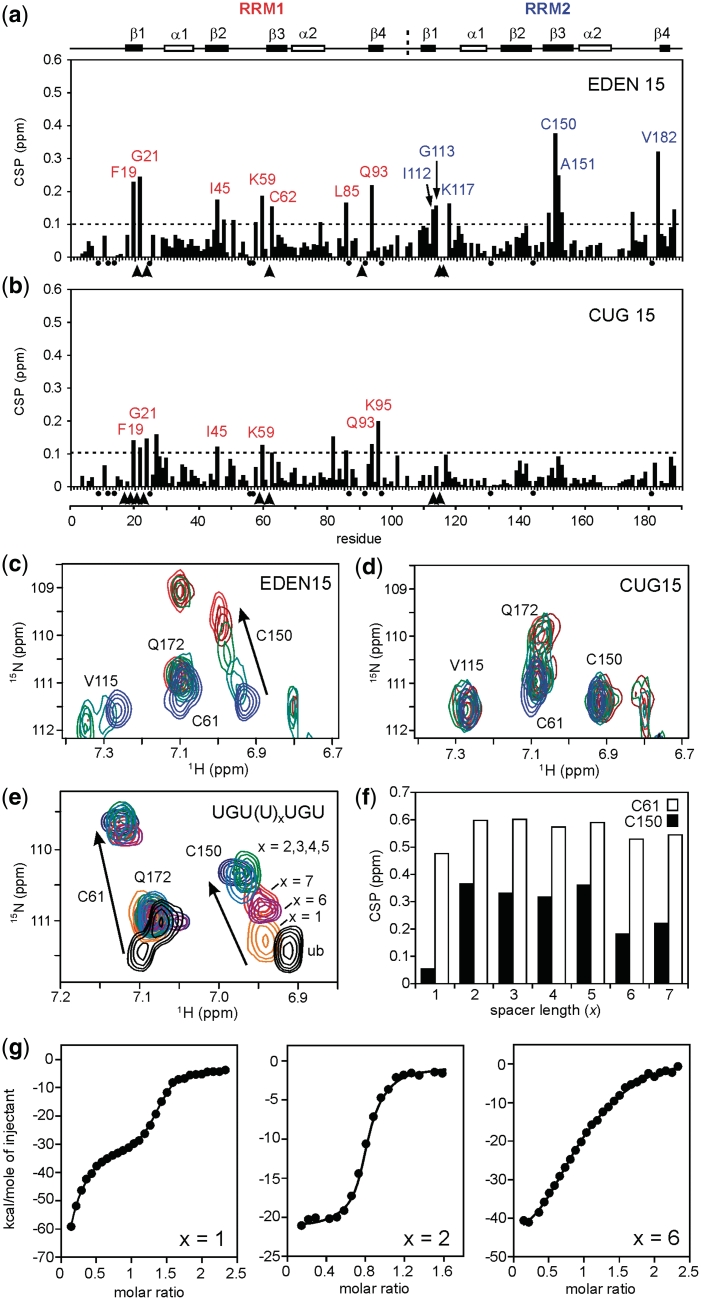
Sequence alignment with other RRMs reveals that RRM1 and RRM2 of CELF1 have some unusual features. Both RRMs have many of the well-conserved aromatic residues on strands 1 and 3 found in other RRMs (F19, F63 and F66 in RRM1 and F110, F152 and F155 in RRM2). However, a unique feature is a Cys substitution in place of a Phe for both RRM1 and RRM2 at residues C61 and C150 respectively. To elucidate the RRM binding specificity, we studied the 1H/15N chemical shift perturbations (CSPs) induced by RNA binding by first considering the interactions of the individual RRMs. The EDEN7 RNA sequence (UGUUUGU) was titrated into a 0.5-mM solution of RRM1 at pH 7.0 and CSPs were detected from a series of 2D 1H/15N-TROSY spectra collected at different molar ratios (Figure 5a). Perturbations to the spectra were manifest as either RNA concentration-dependent fast-exchange movement of TROSY cross-peaks or intermediate-exchange effects which resulted in the movement, broadening and disappearance of peaks at high RNA:protein ratios such that the bound state was difficult to identify. As a consequence, it was possible to track the chemical shift changes between free and bound states for some residues but not others. In Figure 5a–d, measured CSPs are shown as black bars, but resonances which broaden and disappear are shown with arrow heads. In the titration of RRM1 with EDEN7, residues in the β-strands undergo a combination of these effects. In β1 (F19 and G21) and β3 (K59, C61 and C62) the CSPs can be monitored, however, peaks for V20, V23, F63 were rapidly lost. The perturbations readily confirm that RRM1 is interacting strongly with EDEN7 largely through β1 and β3, with additional interactions with the adjacent loops and residues within β4. Perturbations are mapped on to the surface of the structure in Figure 6a. The construction of binding isotherms from cross-peaks in fast exchange suggest tight binding (Kd < 10 µM). Fluorescence analysis at protein concentrations of 5 and 2.5 µM produced steep binding curves saturating at close to 1:1, similarly suggesting binding affinities in the low micromolar range.
Figure 5.
NMR CSP plots for the binding of EDEN 7 (UGUUUGU) and CUGCUG to the two isolated RRMs. In (a) and (b), EDEN7 binding to RRM1 is illustrated, and in (c) and (d), binding of CUGCUG to RRM1 and RRM2. In each case an arbitrary cut-off of 0.1 ppm is shown by the dotted line with residues showing CSPs > 0.1 ppm individually labelled. In addition, proline residues are marked with a black dot and residues which broaden and disappear during the titration are marked with an arrow head. Along the top of the figure the relative position of the protein secondary structure is indicated.
Figure 6.
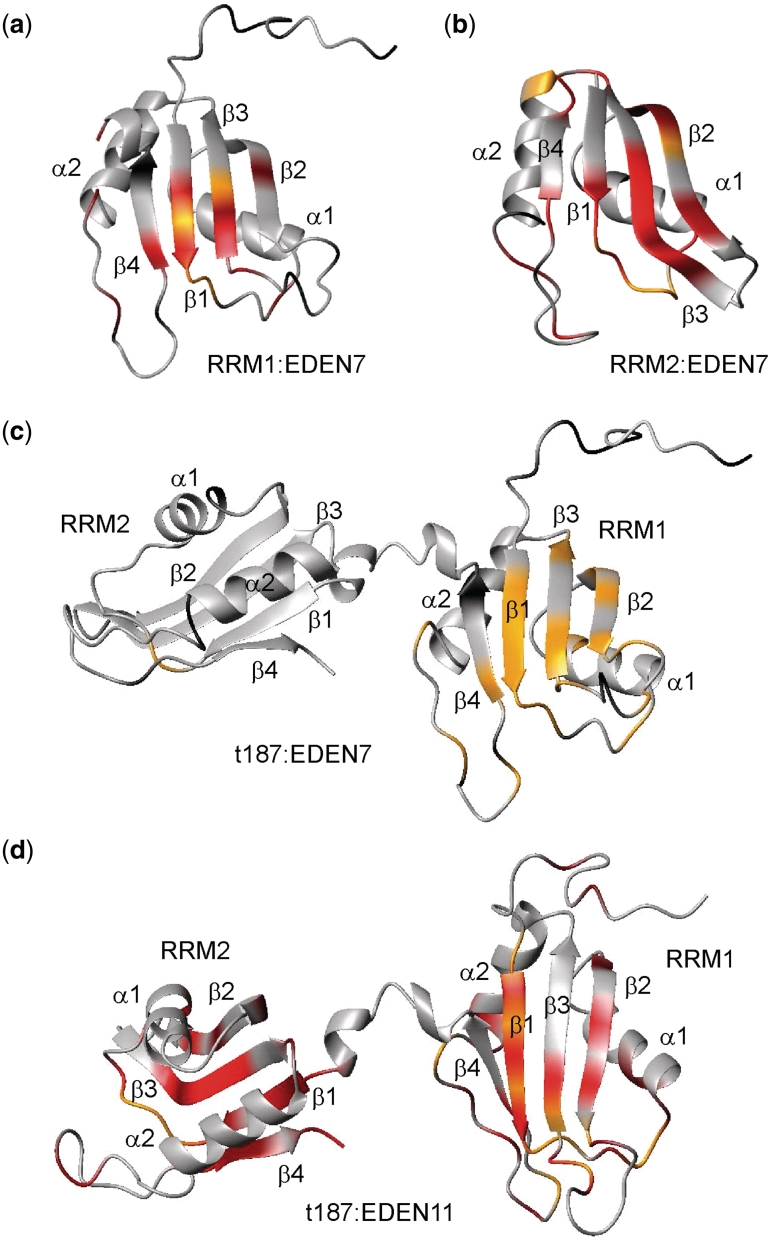
Structural representation of the CSPs mapped to the surface of the RRM1 (a), RRM2 (b) and T187 (c, d) for the RNA indicated. The magnitude of the CSPs is shown on a red to grey scale (largest perturbations shown in brightest red). Residues which broaden and disappear are marked in yellow, as particularly evident for the RRM1 domain of T187 on binding EDEN7.
Similar perturbations were observed for the binding of RRM2 to EDEN7, with significant CSPs in β1 for I112, G113 and V115, and in β3 for C150, A151 and F152 with the peaks for M114, K117, N118 rapidly lost (Figures 5b and 6b). Again, residues in β4 and the preceding loop (V182 and A186) are also affected. The location and magnitude of the shifts suggest that the RNA–protein interactions are quite similar for both RRMs. However, some differences in the binding stoichiometries are readily apparent that are consistent with gel filtration studies (Figure 3). In the NMR titration studies with RRM1 and EDEN7, the CSPs reach a limiting value at a molar ratio close to 1:0.5 (RRM1:EDEN7) which suggests that two RRM1s can be accommodated on the EDEN7 UGU repeat sequence. ESI-MS data confirm that the 2:1 complex is also highly abundant in the gas phase. In contrast, NMR titration studies with RRM2 indicate that saturation of binding occurs much closer to a 1:1 ratio. We explored this further in titration studies with UGUUUAU and a UGU trinucleotide where only a single UGU site was available. Both of these RNA substrates gave similar NMR CSPs to those observed for EDEN7, with limiting shifts at a 1:1 ratio. A control sequence UAUUUAU (ARE) lacking a suitable UGU site did not cause significant CSPs. The data clearly indicate that RRM1 binds to a minimal UGU trinucleotide and that two RRM1s are accommodated on EDEN7 containing a UGU repeat.
We extended the analysis to the binding of RRM1 and RRM2 to CUG repeat sequences by titrating the proteins with CUGCUG. As can be seen (Figure 5c and d), RRM1 gave a similar pattern and magnitude of CSPs to those described for the interaction with EDEN7, however, a few additional peaks (V23) remained visible throughout the titration. RRM1 appears to be promiscuous in binding both the CUGCUG and UGUUUGU sequences using the same β-sheet binding surface, however, there are some subtle differences in the CSPs for the complexes with EDEN7 and CUGCUG. In a few cases, the direction of the movement of cross-peaks and the magnitude of the change suggest sequence specific binding effects. In contrast, RRM2 did not give clear CSPs with the CUGCUG RNA, with few CSPs observed over 0.1 ppm (Figure 5d), indicating a much lower affinity interaction.
Tandem interaction of RRM1 and RRM2 of T187
We examined the tandem interaction of RRM1 and RRM2 with RNA in the context of the longer construct T187 in which the two RRMs are covalently connected through an eight-residue flexible linker. With NMR assignments for >90% of residues we could monitor interactions with both RRMs simultaneously. Titration with EDEN7 resulted in significant perturbations to the resonances of RRM1 but essentially no effects on the cross-peaks from RRM2. The majority of the perturbed resonances in the N-terminal RRM1 broadened and disappeared, making quantitative analysis of CSPs difficult. These are plotted on to the structure shown in Figure 6c. Two conclusions are immediately apparent: first, RRM1 appears to bind with higher affinity to the EDEN7 sequence than RRM2, such that the latter is not able to compete effectively for binding sites. Secondly, EDEN7 appears to be too short to bind both RRMs simultaneously, suggesting a critical minimum spacing between UGU sites that is sterically compatible with both motifs being accommodated at adjacent sites within a UGU repeat.
We examined this hypothesis using the longer EDEN11 (UGUUUGUUUGU) and EDEN15 sequences (UGUUUGUUUGUUUGU). NMR analysis now showed significant CSPs for both RRMs of T187, with the magnitude of the effects comparable to those observed in the titrations of the individual domains (Figure 7a; T187 with EDEN15). Several additional key residues of T187, including C61 in RRM1 and V115 in RRM2 now broaden and disappear, suggesting an enhanced interaction. These RNA sequences demonstrate that a spacing of 5 nt enables both RRMs of T187 to engage in tandem. The perturbations are mapped on to the surface of RRM1, RRM2 and T187 in Figure 6d, with the depth of shading indicating the magnitude of the perturbations.
Figure 7.
NMR CSP plots for the binding of EDEN15 (a) and CUG15 (b) to T187. In each case an arbitrary cut-off of 0.1 ppm is shown by the dotted line with residues showing CSPs > 0.1 ppm individually labelled. In addition, residues which broaden and disappear during the titration are marked with an arrow head. No assignments were obtained for proline residues and these are marked with a black dot. Along the top of the figure the relative position of the protein secondary structure is indicated with the domain boundary between RRM1 and RRM2 shown at residue 110. Representative portions of the 1H/15N TROSY spectra from the two titrations are shown in (c) and (d). In particular, residue C150 in RRM2 is perturbed by the binding of EDEN15 (c), but not by CUG15 (d). Five TROSY spectra are overlayed in each case representing protein:RNA ratios in the range 1:0–1:1. (e) Similar overlayed portions of the 1H/15N TROSY spectrum of T187 showing the limiting CSPs for Cys61 (RRM1) and Cys150 (RRM2) at binding saturation with a series of related RNA sequences containing different U spacers UGU(U)xUGU, where x = 1–7. (f) Histogram plot showing the CSP data from (e) for Cys61 and Cys150 with the CSPs for RRM1 reaching a plateau between x = 2–5; longer U spacers (x = 6 and 7), or a very short spacer (x = 1) result in a reduction in binding affinity for RRM2 but little difference in the shift of Cys61 in RRM1. (g) Binding isotherms at 298 K from ITC studies of T187 with UGU(U)xUGU sequences where x = 1, 2 and 6. The shortest sequence (x = 1) shows a biphasic binding behaviour whereas the two other sequences (x = 2 and 4) fit to a 1:1 binding model.
We examined the importance of the length of the spacer sequence between UGU sites more systematically in NMR titration studies with UGU(U)xUGU, where x = 1–7 (Table 1). The magnitude of the CSP for C150 (RRM2) in 1H/15N TROSY spectra at binding saturation provided an indicator of the relative binding affinities, reaching a plateau between x = 2–5. Residues G113, A151 and F152 within RRM2 showed a similar behaviour but with smaller limiting CSPs. However, longer U spacers (x = 6 and 7) appeared to show a fall-off in binding interaction with RRM2. In contrast, C61 of RRM1 showed the same limiting shift in all cases (Figure 7f), suggesting that RRM1 binds with higher affinity and is anchored to the first UGU site to guide RRM2 into place. As described, the very short spacer (x = 1; EDEN7) is unable to accommodate both RRMs simultaneously and only RRM1 shows evidence of a significant binding interaction. Using the high affinity EDEN4U sequence (x = 4), we also examined whether RNA binding resulted in significant changes in dynamics between the two RRMs of T187 by measuring 1H-15N NOEs (as described above). Where we could resolve the overlapping resonances, we could still detect a smaller heteronuclear NOE for residues 99–101 (NOE ≤ 0.5) (Supplementary Data) which suggests that some of the intrinsic flexibility in the linker is retained in the RNA-bound state of T187 of > 100-fold.
We examined a number of the UGU(U)xUGU sequences by ITC by titrating T187 into RNA to obtain quantitative binding data (Figure 7g). EDEN7 (x = 1) is clearly different from the other sequences showing two distinct sequential binding events. However, for x ≥ 2 the binding isotherms are consistent with a single co-operative binding interaction for T187 with Kd values in the range 0.4–4 µM (Table 3), with similar enthalpies and entropies of interaction (ΔH = –45 ± 2 kJ mol−1 and ΔS = –125 ± 5 J K−1 mol−1). The highest affinity interactions of ~ 0.4 µM are evident for the shorter RNA sequences (x = 2 and 4), with a subsequent 10-fold reduction in binding affinity for the longer sequences (x = 5 and 6), broadly reflecting the results of the NMR titration studies. In contrast, ITC studies of T187 with the trinucleotide UGU demonstrated a weaker interaction with a Kd = 64 ± 2 µM, indicating a substantial enhancement of binding affinity from the co-operative interaction of the two binding motifs of T187.
Table 3.
Isothermal titration calorimetry data for the interaction of CELF1 T187 construct with RNA
| RNA sequence | Kd (mM) | Binding stoichiometry |
|---|---|---|
| UGU | 64 ± 2 | 0.51 |
| UGUUUUGU (x = 2) | 0.43 ± 0.15 | 1.27 |
| UGUUUUUUGU (x = 4) | 0.37 ± 0.15 | 1.35 |
| UGUUUUUUUGU (x = 5) | 3.5 ± 0.5 | 0.91 |
| UGUUUUUUUUGU (x = 6) | 3.4 ± 0.3 | 0.89 |
RRM binding sites underlined.
We also investigated the longer EDEN15 (UGUUUGUUUGUUUGU) sequence by ITC, however, this produced a more complex binding isotherm with at least two sequential binding events with quite different binding affinities (data not shown). The quality of the fit to this simple model was poor suggesting that there could be a number of other degenerate 2:1 binding modes with regard to positioning and orientation on the RNA. However, we can rationalize the overall 2:1 binding stoichiometry on the basis that RRM1 and RRM2 of T187 can bind in tandem with high affinity to two of the UGU sites of EDEN15, with an optimal spacing of 5 nt (underlined), with a second molecule of T187 interacting at the terminal UGU site (bold) only through its RRM1. The observations from NMR titrations and ESI-MS experiments that EDEN7 (UGUUUGU) can accommodate two isolated RRM1s on the two closely spaced UGU sites, suggest that this may also be a plausible model for accommodating two T187 molecules in sequential binding events with EDEN15, with the optimum 5-nt spacer between UGU sites for accommodating in tandem the RRMs of T187. ESI-MS analysis of the T187 interaction with EDEN11 (UGUUUGUUUGU) unambiguously confirms a single high affinity 1:1 complex.
We repeated the NMR titrations of T187 with the CUG-rich sequence CUG15 (Figure 7b) and, surprisingly, found that the CSP effects for RRM1 were reduced compared to shifts of the isolated RRM1 with CUGCUG. The perturbations to RRM2 all fell below the 0.1 ppm CSP cut-off, consistent with the weak interaction apparent in the titration of the isolated domain with CUGCUG. Specific effects in the TROSY spectra are illustrated for residue C150 (RRM2) in which a large CSP is evident in the titration with EDEN15, but no significant effect is observed with CUG15 (Figure 7c and d). The apparent reduction in CSP effects for RRM1 when the two domains bind in tandem suggests that RRM2 may exert some negative allosteric regulation on the binding of RRM1 to CUG sequences.
DISCUSSION
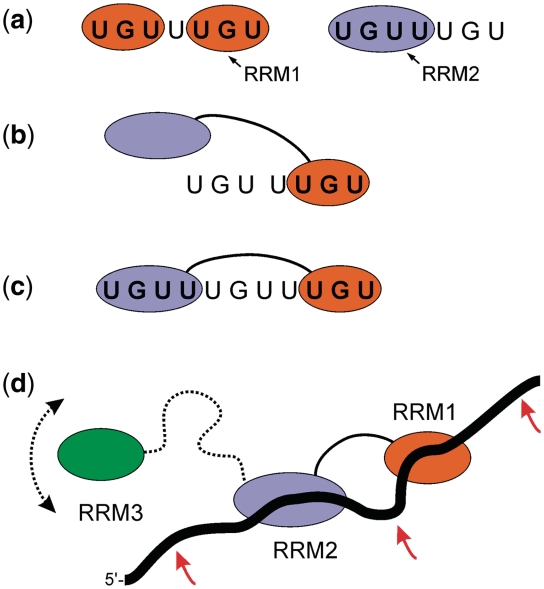
We have demonstrated that the first two RNA recognition motifs (RRM1 and RRM2) of CELF1 are capable of binding independently to a single UGU site. Significantly, RRM1, but not RRM2, is also capable of binding to CUG repeats, which when abnormally extended are linked to a variety of pathological processes including type 1 myotonic dystrophy (14,15). We have shown that the shortest sequence studied with tandem UGU binding sites (UGUUUGU: EDEN7) was able to accommodate two RRM1s, but only a single RRM2 (Figure 8a). In this context, RRM1 appears to recognize each UGU as an independent site, from which we conclude that there is no absolute requirement for a fourth nucleotide for binding. However, the observation that only a single RRM2 can be accommodated on this short RNA suggests that a fourth nucleotide is a prerequisite for RRM2 with binding to a 5′-UGUU site.
Figure 8.
Schematic representation of the binding of RRM1, RRM2 and T187 to RNA substrates. (a) EDEN7 is able to accommodate two RRM1s on adjacent UGU sites, but only a single RRM2 can bind to the same sequence, suggesting specificity for a UGUU binding site. (b) EDEN7 is too short to bind the two RRMs of T187 in tandem, instead we observe an interaction through RRM1 which could be at either of the two UGU sites (one shown). (c) Binding of T187 to both sites of the consensus GRE sequence (EDEN11) with RRM2 bound at the 5′-terminal UGUU site. The spacer sequence of 5 nt between the tandem UGU sites results in high affinity binding. (d) Possible interaction of full-length CELF1 with a GU-rich substrate with RRM1 and RRM2 bound as for the consensus sequence in (c); the long linker between RRM2 and RRM3 permits considerable conformational flexibility in binding a third UGU site up or down stream (position of arrows), or even to the looped-out spacer sequence between the RRM1 and RRM2 binding sites.
We extended our analysis to the tandem recognition of UGU(U) sites by RRM1 and RRM2 in a single protein construct (T187) using a UGU(U)xUGU expansion series of RNA sequences to investigate the effects of spacer length between binding sites. We could find no evidence by NMR for the simultaneous binding of both motifs to the shortest UGUUUGU (x = 1) sequence, only interactions with RRM1 (Figure 8b), suggesting that there are significant steric restraints in accommodating two RRMs at two UGU(U) sites with only a single U spacer. However, tandem high-affinity binding interactions where apparent for longer spacers (x = 2–5 nt) without significant apparent changes in affinity (Figure 8c). The RRM2 interaction, but not RRM1, diminishes for longer sequences (x = 6 and 7), suggesting that RRM2 is transported into position on the back of its stronger binding partner. Quantitative analysis by ITC shows a high affinity interaction (~0.4 µM) for the shorter sequences (x = 2–4) in particular, and a 10-fold reduction in binding affinity for UGU sites that are more dispersed.
Furthermore, we are able to comment on the orientational preference for the binding of T187 to the two UGU(U) sites in tandem on a single nucleotide chain. Within the context of our UGU(U)xUGU expansion series, the apparent preference of RRM2 for a UGUU site suggests that it is accommodated at the 5′-terminus with RRM1 binding to the 3′-end of the sequence. The differences in stoichiometries that we observe for RRM1 and RRM2 binding independently to UGUUUGU are consistent with this analysis. On this basis, we can conclude that the GU-rich GRE consensus regulatory sequence UGUUUGUUUGU (EDEN11), which has been shown to mediate rapid decay of a number of GRE-containing transcripts in primary human T-cells, binds specifically through tandem interactions of RRM1 and RRM2 of CELF1 at a unique site (Figure 9c). The interaction exploits both site preferences for UGU(U) and spacer length (x = 5) for a tandem interaction. In addition, the EDEN15 RNA motif, which is the over-represented 3′-UTR target sequence for Xenopus CELF1 (EDEN-BP) and activates deadenylation and subsequent translational repression of EDEN-containing transcripts, contains four copies of the UGU motif and gives rise to two degenerate high affinity sites each analogous to that of the GRE sequence.
The apparent tolerance to different spacings between UGU(U) binding sites while retaining a high affinity interaction indicates that the two RRMs are flexibly linked with independent dynamics evident in the absence of bound RNA. The two motifs appear to accommodate either a short RNA sequence (x = 2 or 3) in a linear trajectory between RRMs, or form a looped-out conformation with longer RNA spacer sequences (x > 4). We were unable to detect by NMR in titration studies any differences in the interaction with RNAs of different lengths, and concluded that there were no specific RRM1–RRM2 contacts associated with the binding of certain substrates, nor do the flexible RNA spacer sequences between UGU(U) sites appear to be accommodated through specific protein contacts.
The consensus GU-rich binding site derived from the bioinformatics analysis, UGUUUGUUUGU, appears to correspond to the optimum binding site for the tandem interaction of the first two RRMs of full length CELF1 (12,29). As the third RRM of CELF1 has also been reported to bind (UG)3, this indicates that the preferred binding site for full length CELF1 likely requires an additional GU-rich motif, either upstream or downstream of this consensus site (Figure 9d). The poor binding we observe with T187 and the c-Mos EDEN sequence (EDEN19: UGUAUGUGUUGUUUUAUGU) is surprising since it would appear to contain three optimal spacings of 2, 3 and 4 nt between UGU(U) sites for binding to the first two RRMs. It therefore appears that in addition to the spacer length, the sequence context also influences binding, with certain nucleotides not being tolerated adjacent to the UGU sites. Our data indicate that the central G in the 5-nt spacer in EDEN11 can be replaced by a U without a loss of binding, and both natural and synthetic high affinity binding sites have been observed with CUGUC (Figure 2a and e) and GUGUG spacers (12,25,29). In addition, UGUA repeats do bind CELF1, but they appear to have somewhat reduced affinity (7,25). Further study is required to unambiguously identify the sequence context-dependence for the binding of RRM1 and RRM2.
The possibility of multiple binding sites within the CUGCUG RNA sequence precludes an unambiguous determination of which tri-nucleotide is being recognized. However, the similarity of the interaction of RRM1 with both EDEN7 (UGUUUGU) and CUGCUG suggests that it may be UGY (Y = U or C). However, there are a number of CELF1 substrates that are rich in GCC trinucleotides, rather than UGC or CUG, suggesting that the trinucleotide recognition site could be GCY (Y = U or C). How CELF1 binds to combination substrates with GCY and UGU sites would also give information on the orientation of the RNA in the complex and possibly help refine the consensus binding sequence.
There is no available crystal structure of T187 bound to RNA in the cooperative manner we describe, however, during the review process crystal structures of CELF1 RRM1 and tandem RRM1/RRM2 domains (T187) bound to UGU sites in 12 and 13 nt RNAs have also shed light on UGU(U) recognition of the individual RRMs (41). Studies with the T187 construct with GUUGUUUUGUU (equivalent spacing to EDEN2U in our analysis) resulted in RRM2–RNA interactions but not tandem binding of both RRMs to the same nucleotide sequence. The structures revealed that both RRMs target UGU(U/G) sequences and recognize the base and sugar–phosphate backbone of the UG and GU steps of UGU through a similar set of direct and water-mediated hydrogen bonds and stacking interactions. Cys61 in RRM1 and Cys150 in RRM2 are both involved in hydrogen bonding to the ribose 2′-OH of the guanine within the two tandem UGU(U/G) binding sites. Specificity is also achieved by selecting for a G (syn) configuration, with the UG step adopting an unanticipated left-handed z-helix. Although not observed in the crystal structures of T187 complexes, NMR and ITC studies of T187 with the same 12 nt RNA indicate that the tandem interaction of the two RRMs occurs in solution with a binding affinity >150-fold stronger than for the individual RRMs. These structures suggest that both RRMs require a tetra-nucleotide UGU(U/G) motif, however, from our analysis of binding stoichiometries with short RNA fragments this does not appear to be an absolute requirement in the case of RRM1, where UGU appears to be sufficient. Our studies have provided additional insights into the role of the spacer length in accommodating tandem binding interactions.
Both of the reported structural studies confirm that the canonical RRM fold of RRM1 and RRM2 of CELF1 utilizes conserved residues mainly on strands β1 and β3 in UGU(U) recognition, and have shed light on the tandem interaction of these binding motifs in recognizing regulatory RNA sequences. Enhanced binding of the protein to GC rich substrates in muscular dystrophy (31) has been linked to phosphorylation of Ser28, which lies close to residues on the RNA binding face of RRM1. Mutation of Ser28 to Asp enhances the binding of the protein to these substrates. In fact, in the closest relative of CELF1 in Drosophila, Bruno (Genbank; AAB58464), the corresponding Ser has been replaced by Asp, suggesting that this protein has a permanent preference for GC rich substrates. Further experimentation is required to determine the precise molecular basis of these changes in RNA sequence specificities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) (M.S.S. and J.L., grant number BB/F013663/1) and through a BBSRC studentship (to J.E.). The NMR facilities were supported by the University of Nottingham and the School of Chemistry through SRIF funding; BBSRC studentship from the School of Pharmacy (to E.M.); Wellcome Trust (grant number 076179 to C.H.d.M. and A.K.) and the BBSRC (grant number BB/G001847/1 to Cd.M. and A.K.) Funding for open access charge: Predominantly BBSRC but also Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezzeddine N, Paillard L, Capri M, Maniey D, Bassez T, Ait-Ahmed O, Osborne HB. EDEN-dependent translational repression of maternal mRNAs is conserved between Xenopus and Drosophila. Proc. Natl Acad. Sci. USA. 2002;99:257–262. doi: 10.1073/pnas.012555499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paillard L, Legagneux V, Beverley Osborne H. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol. Cell. 2003;95:107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 5.Delaunay J, Le Mee G, Ezzeddine N, Labesse G, Terzian C, Capri M, Ait-Ahmed O. The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 2004;32:3070–3082. doi: 10.1093/nar/gkh627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 7.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barreau C, Watrin T, Beverley Osborne H, Paillard L. Protein expression is increased by a class III AU-rich element and tethered CUG-BP1. Biochem. Biophys. Res. Commun. 2006;347:723–730. doi: 10.1016/j.bbrc.2006.06.177. [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 11.Timchenko LT, Salisbury E, Wang GL, Nguyen H, Albrecht JH, Hershey JW, Timchenko NA. Age-specific CUGBP1-eIF2 complex increases translation of CCAAT/enhancer-binding protein beta in old liver. J. Biol. Chem. 2006;281:32806–32819. doi: 10.1074/jbc.M605701200. [DOI] [PubMed] [Google Scholar]
- 12.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol. Cell. Biol. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huichalaf CH, Sakai K, Wang GL, Timchenko NA, Timchenko L. Regulation of the promoter of CUG triplet repeat binding protein, Cugbp1, during myogenesis. Gene. 2007;396:391–402. doi: 10.1016/j.gene.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Gautier-Courteille C, Le Clainche C, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne HB, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- 18.Legagneux V, Omilli F, Osborne HB. Substrate-specific regulation of RNA deadenylation in Xenopus embryo and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J. Biol. Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldwin BR, Timchenko NA, Zahnow CA. Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPbeta-LIP in mammary epithelial cells. Mol. Cell. Biol. 2004;24:3682–3691. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 23.Cibois M, Gautier-Courteille C, Vallee A, Paillard L. A strategy to analyze the phenotypic consequences of inhibiting the association of an RNA-binding protein with a specific RNA. RNA. 2010;16:10–15. doi: 10.1261/rna.1742610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J. Biochem. 2008;143:377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- 26.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell. Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J. Biol. Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- 31.Salisbury E, Sakai K, Schoser B, Huichalaf C, Schneider-Gold C, Nguyen H, Wang GL, Albrecht JH, Timchenko LT. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp. Cell Res. 2008;314:2266–2278. doi: 10.1016/j.yexcr.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timchenko NA, Wang GL, Timchenko LT. RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein beta by interacting with the alpha and beta subunits of eukaryotic initiation translation factor 2. J. Biol. Chem. 2005;280:20549–20557. doi: 10.1074/jbc.M409563200. [DOI] [PubMed] [Google Scholar]
- 33.Bonnet-Corven S, Audic Y, Omilli F, Osborne HB. An analysis of the sequence requirements of EDEN-BP for specific RNA binding. Nucleic Acids Res. 2002;30:4667–4674. doi: 10.1093/nar/gkf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosson B, Gautier-Courteille C, Maniey D, Ait-Ahmed O, Lesimple M, Osborne HB, Paillard L. Oligomerization of EDEN-BP is required for specific mRNA deadenylation and binding. Biol. Cell. 2006;98:653–665. doi: 10.1042/BC20060054. [DOI] [PubMed] [Google Scholar]
- 36.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyon AM, Reveal BS, Macdonald PM, Hoffman DW. Bruno protein contains an expanded RNA recognition motif. Biochemistry. 2009;48:12202–12212. doi: 10.1021/bi900624j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Moor CH, Richter JD. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Moor CH, Richter JD. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer HA, Radford HE, Wilson LS, Lissenden S, de Moor CH. Translational control of maskin mRNA by its 3' untranslated region. Biol. Cell. 2007;99:239–250. doi: 10.1042/BC20060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.