Abstract
Aberrant DNA methylation is often associated with cancer and the formation of tumors; however, the underlying mechanisms, in particular the recruitment and regulation of DNA methyltransferases remain largely unknown. In this study, we identified USP7 as an interaction partner of Dnmt1 and UHRF1 in vivo. Dnmt1 and USP7 formed a soluble dimer complex that associated with UHRF1 as a trimeric complex on chromatin. Complex interactions were mediated by the C-terminal domain of USP7 with the TS-domain of Dnmt1, whereas the TRAF-domain of USP7 bound to the SRA-domain of UHRF1. USP7 was capable of targeting UHRF1 for deubiquitination and affects UHRF1 protein stability in vivo. Furthermore, Dnmt1, UHRF1 and USP7 co-localized on silenced, methylated genes in vivo. Strikingly, when analyzing the impact of UHRF1 and USP7 on Dnmt1-dependent DNA methylation, we found that USP7 stimulated both the maintenance and de novo DNA methylation activity of Dnmt1 in vitro. Therefore, we propose a dual role of USP7, regulating the protein turnover of UHRF1 and stimulating the enzymatic activity of Dnmt1 in vitro and in vivo.
INTRODUCTION
In mammals, the majority of C-5-cytosine methylation occurs at CpG dinucleotides that are modified to 70–80% in a cell type-specific pattern and generally associated with repressed states of chromatin (1–4). DNA methylation contributes to epigenetic processes such as differentiation and development, transcriptional regulation, preservation of chromosomal stability, silencing of repetitive elements, genomic imprinting, X-chromosome inactivation and DNA repair (2,3). Dnmt3a and Dnmt3b are mainly involved in de novo establishment of DNA methylation marks, whereas the maintenance DNA methyltransferase Dnmt1 maintains methylation patterns on the newly synthesized daughter strand during replication (5,6). Mice depleted for the DNA methyltransferases revealed developmental defects and die early during embryogenesis (7,8). Aberrant DNA methylation patterns are often associated with cancer (9), genome-wide hypo- and hypermethylation correlates with genomic instability, reactivation of retrotransposons, repression of tumor suppressor genes and loss of genomic imprinting (LOI). The DNA methyltransferases were found to interact with many chromatin-associated factors such as methyl-binding proteins (MBD2, MeCP2), histone deacetylases (HDAC), histone methyl transferases (HMT), transcriptional repressors, chromatin remodeling enzymes and Polycomb group proteins (10,11). However, so far the only Dnmt1 complex purified by means of chromatographic fractionation from HeLa nuclear extracts was a transcription repression complex consisting of Dnmt1, pRB, E2F1 and HDAC1 (12).
Recently, UHRF1 (also known as ICBP90 in human or NP95 in mouse) has been shown to be essential in maintaining genomic DNA methylation (13,14). The DNA of UHRF1-deficient ES cells exhibited low DNA methylation levels and methylation defects of the imprinted genes (14), an effect reminiscent of Dnmt1 knockout in ES cells and mice (7). UHRF1 strongly associates with heterochromatin (15,16) and binds preferentially to hemi-methylated DNA via its SRA domain (13,17–19). The latter was also shown to interact with the TS domain of Dnmt1 (20,21). Hence, it has been proposed that UHRF1 would recruit Dnmt1 to heterochromatin to maintain DNA methylation during replication. Furthermore, UHRF1 belongs to the class of Ring finger-type E3-ubiquitin ligases (22) possessing in vitro autoubiquitinylation activity (15,16,23). UHRF1 does also target histones for ubiquitinylation in vitro and in vivo, with a preference for histone H3 (15,16).
USP7 (also known as HAUSP) was initially identified in promyelocytic leukemia nuclear bodies (PML) of herpes simplex virus infected cells (24). It regulates the stability of p53 and MDM2 through its deubiquitination activity (25–28). Furthermore, USP7 has been shown to associate with other factors and substrates including EBNA1 (29), DAXX (30), FOXO4 (31), PTEN (32) and is crucial for development, as mice depleted for USP7 die during embryogenesis (33). USP7 was shown to silence the homeotic genes through Polycomb in Drosophila (34). In particular, the GMP-synthetase interacted with USP7 to enhance the removal of the active ubiquitin mark from histone H2B and to act as a transcriptional corepressor (35).
We biochemically purified human Dnmt1 and show that it is associated primarily with USP7 and co-assembles with UHRF1 on DNA, forming a trimeric complex that associates with silenced genes in vivo. We show a dual role for USP7 in the complex, first stimulating the enzymatic activity of Dnmt1 and second regulating the stability of the UHRF1 protein.
MATERIALS AND METHODS
Cell culture
All cell lines were grown at 37°C, 5% CO2 in medium containing 10% FBS (GIBCO), 100 U/ml penicillin and 100 µg/ml streptomycin (GIBCO). Human cervix carcinoma cells (HeLa S3 cells; ATCC 161) were grown in spinner flasks in RPMI 1640 medium (GIBCO) with a cell density of ~1 × 106 cells/ml. Human colon adenocarcinoma cells HCT-116 (ATCC 581) were cultivated as mono-layers in D-MEM medium (GIBCO) and grown to a confluency of 60–70%. LS174TR1 cells, expressing the Tet-repressor (36,37) were grown in RPMI 1640/HEPES medium (GIBCO) supplemented with 10 µg/ml blasticidin. LS174TR1 cells with plasmids carrying either shRNA (LS88) or N-myc-USP7 (LS89) under the control of a tetracycline/doxycycline inducible promoter were cultivated as LS174TR1 but supplemented with 400 µg/ml Zeocin (28,38). LS174TR1 cells and their derivates were kindly provided by M. Maurice. For doxycycline treatment, cells were seeded on a 10 cm tissue culture plate with a cell number giving rise to 50–60% confluency on the day of harvest. Cells were grown in RPMI1640/HEPES (GIBCO) supplemented with blasticidin (10 µg/ml), Zeocin (200 µg/ml), TET-FBS (Clontech 631106) and 1.0 µg/ml doxycycline. The parental LS174TR1 cells were used as a negative control in all experiments. H1299 (p53−/−; ATCC: CRL-5803) non-small lung cancer cells were cultivated as mono-layers in D-MEM medium (GIBCO). Protein half-life studies were performed by incubating cells with cycloheximide (Sigma, 100 µg/ml) and proteasome inhibitor MG132 (Sigma, 20 µM) for the indicated time points. Transfection reactions with the Fugene6 reagent (Roche) were performed in six-well plates with 3 × 105 cells/well with 2.0 µg total plasmid DNA for 48 h. For protein half-life studies, cells were split 1:4 36 h post-transfection and treated with cycloheximide for the indicated time points.
For knockdown experiments, 2 × 106 HCT-116 cells were electroporated with 80 pmol validated siRNAs (siGENOME non-targeting siRNA #1, Thermo Scientific Dharmacon; UHRF1 Silencer Select siRNA and USP7 Silencer Validated siRNA, both Applied Biosystems) for 10 ms with 350 V and subsequently seeded in six-well plates for 48 h.
Plasmids and constructs
The plasmid pFastBAcHTa-Dnmt1 with an N-terminal 6 × His-tag and TEV cleavage site were a kind gift of F. Lyko. The cDNA of the full-length Dnmt3b2 (kind gift of F. Lyko) was PCR-amplified and cloned into a modified pet11 bacterial expression vector (Novagen) carrying an N-terminal Flag-tag and a C-terminal Thrombin cleavage site followed by a 6 × His-tag (pETM-Dnmt3b2). Full-length human Dnmt3a was PCR-amplified from a cDNA clone (RZPD, IRATp970A0473D) and cloned in-frame into the pENTR3C-vector (Invitrogen) with BamHI/EcoRI. Full-length USP7/HAUSP and the TRAF domain of USP7 (amino acid 1–215, without stop codon) were PCR amplified on pCin4-HAUSP [kindly provided by Yigong Shi, (39)] and cloned into the pENTR SD D TOPO vector (Invitrogen). USP7 C223S mutant was generated by SOE-PCR and transferred into the pDONR221 vector (Invitrogen). The GST fusion constructs of USP7-domains 2 (amino acid 212–561), 3 (amino acid 561–916) and 4 (amino acid 913–1102), cloned as BamHI/XhoI fragments in pGEX-2T (GE Healthcare), were kindly provided by Prof. Dobner. UHRF1 and UHRF1ΔRING (without stop codon) and the SRA domain (TEV protease cleavage site, amino acid 409–635) of UHRF1 were PCR-amplified on pcDNA3.1-ICBP90/UHRF1 [kindly provided by Yusuke Nakamura, (17)] and cloned into the pENTR SD D TOPO vector (Invitrogen). The correct sequence of all plasmids was verified by sequencing (sequences of plasmids and oligonucleotides on request). pENTR3C-Dnmt3a, pENTR-USP7 and pDONR221-USP7 C223S were subsequently transferred via LR-reaction (Invitrogen) into pDEST10 (Invitrogen) and pENTR-SRA into pDEST20 (Invitrogen) creating expression clones for baculovirus generation. pENTR-UHRF1, pENTR-UHRF1ΔRING and pENTR-TRAF-domain were transferred via LR-reaction (Invitrogen) into pDM7 (pET11 based destination vector with C-terminal 6 × His-tag) and pGEX-4T1-DEST (pGEX 4T1 based destination vector), respectively. For transfection studies pIRES EGFP UHRF1-C-Flag and pIRES EGFP N-HA-USP7 were used.
Antibodies
The following antibodies were used for western blot analysis and immunoprecipitation: anti-actin (rabbit polyclonal, Sigma A2066), anti-Dnmt1 DNM-2C1, anti-Dnmt3a D3A2-6A4, anti-Dnmt3b D3B2-2C1 (rat monoclonal antibodies, E. Kremmer, Helmholtz Gesellschaft), anti-Flag (mouse monoclonal, Sigma M2 F1804), anti-Penta-His (mouse monoclonal, Qiagen P-21315), anti-ICBP90/UHRF1 [mouse monoclonal, generously provided by C. Bronner, (40)], anti-Lamin A/C (rabbit polyclonal, Santa Cruz, sc-20681), anti-RNAPII CTD 8WG16 (mouse monoclonal, generously provided by D. Eick) and anti-USP7 (rabbit polyclonal, Bethyl laboratories, A300-033A). Irrelevant IgG control and horseradish peroxidase-coupled secondary antibody antibodies were purchased from Santa Cruz.
DNA methyltransferase assay
Typical DNA methyltransferase reaction (50 µl) contained 20 nM Dnmt1, non-methylated (LP35/37) or hemi-methylated (LP35/36) oligonucleotides at 120–4320 nM (9 CpG), BSA at 0.2 µg/µl and 3H-SAM (GE Healthcare, TRK581-250UCi, 9.25 MBeq with 1.0 mCi/ml 63.0 Ci/mmol) at 480 nM in DNA methyltransferase buffer (20 mM Tris, pH 7.6, 1.0 mM EDTA, 1.0 mM DTT). The reaction was started with the addition of DNA, incubated at 37°C for 10–30 min, and stopped with 10 µl of 10 mM SAM (Sigma). The reaction was spotted on DE81 filter (Whatman), washed three times with 0.2 M NH3HCO3, once with water and ethanol following drying and scintillation counting.
LP35: 5′-GGTACGGATGCGGAATCGTCTAACGCGTGGAATCGTCCCCTTGCGAATTTCGGTGTCGAT-3′, LP36: 5′-CGTAXGGATGXGGAATXGTCTAAXGXGTGGAATXGTCCCCTTGXGAATTTXGGTGTXGAT-3′, LP37: 5′-ATCGACACCGAAATTCGCAAGGGGACGATTCCACGCGTTAGACGATTCCGCATCCGTACC-3′ (X = C5-methyl group).
In vitro ubiquitinylation assay
Standard ubiquitinylation reactions (10 µl) contained 100 ng E1 activating enzyme (Biomol, UW9410), 300 ng E2 conjugating enzyme UBCH5c (BioMol, UW9070), 1.0 µg ubiquitin (Sigma U-6253) or 100 ng Flag-tagged ubiquitin (Boston Biochem, U-120) and 250 ng UHRF1 or UHRF1ΔRING in 1× reaction buffer (50 mM Tris pH 7.5, 50 mM NaCl, 5.0 mM MgCl2, 0.05% NP40, 1.0 mM DTT, 5.0 mM ATP). The reaction was carried out for 2 h at 37°C and stopped with the addition of HU-buffer and boiled for 10 min at 65°C. Samples were resolved by SDS–PAGE and ubiquitination reaction was analyzed by western blot using antibodies directed against the His-tag or Flag-tag. For autoubiquitinylation reactions of UHRF1 in the presence of USP7 or USP7 C223S, contained 5.0 and 50 ng of USP7/USP7 C223S and were supplemented with 10 mM DTT.
Immunoprecipitation
All steps were carried out on ice or at 4°C and buffers supplemented with protease inhibitors PMSF (1.0 mM), Leupeptin (1–10 µg/ml), Aprotinin, Pepstatin (1.0 µg/ml) prior to use. If not stated differently, the following IP buffer was used for IP experiments: 50 mM Tris pH 7.5, 150 mM NaCl, 1.0 mM EDTA, 0.05% NP-40.
Protein mixtures (recombinant proteins, nuclear extracts) were incubated with 50 µl of proteinG sepharose slurry in the presence of 1× IP-buffer, to analyze for unspecific interaction [referred to as ‘preclearing beads’ (PG)].
The precleared sample/lysate was added to 50 µl of proteinG sepharose charged with antibodies and incubated for 2 h at 4°C. The ‘beads’ were washed three times with 1000 µl IP buffer and resuspended in 50 µl Lämmli dye, heated to 95°C and subjected to SDS–PAGE following WB blot analysis.
Protein bands for identification were sent for MALDI analysis.
Preparation of whole-cell extracts
Cells from P15 tissue culture plates were resuspended in 150 µl lysis buffer (20 mM Tris pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 0.5% NP40) following incubation on ice for 30 min, while vigorously vortexing every 10 min. After centrifugation (30 min, 13 000g, 4°C) the supernatant (WCE) was recovered.
Real-time reverse transcription–PCR
Total RNA was extracted from tumor cells in Trizol (Invitrogen), depleted from DNA and subsequently purified using DNase I and RNeasy Mini Kit, respectively (Qiagen). Reverse transcription (RT) of total RNA was performed using random hexamers (Roche Diagnostics) and SuperScriptII reverse transcriptase (Invitrogen). qPCR amplifications were performed in doublets as described for the ChIP analysis. Amplification of the house-keeping gene TATA-Box-binding-Protein (TBP) was performed to standardize the amount of sample RNA. Relative quantitation of gene expression was performed using the ΔΔct method as described earlier (41). We used the following primer pairs (5′ → 3′-orientation): SFRP1-F, CCTGGGACTCAGCACATTGA; SFRP1-R, GATGGCCTCAGATTTCAACTCG; IGFBP3-F, GTCCAAGCGGGAGACAGAATAT; IGFBP3-R, CCTGGGACTCAGCACATTGA; HHIP-F, TGTACATCATTCTTGGTGATGGG, HHIP-R, AGCCGTAGCACTGAGCCTGT; HOXA7-F, TCAGGACCTGACAGGAAGCG; HOXA7-R, TCAGGTAGCGGTTGAAGTGGA and TBP-F, GCCCGAAACGCCGAATAT; TBP-R, CCGTGGTTCGTGGCTCTCT.
Methylation-specific PCR
Genomic DNA of tumor cells was extracted with phenol and chloroform, ethanol precipitated and dissolved in TE buffer following standard procedures (42,43). We used the EpiTect® Bisulfite Kit (Qiagen) for bisulfite-treatment of DNA as recommended by the manufacturer. Methylation status of the promoter region of the SFRP1, IGFBP3, HHIP and HOXA7 genes was analyzed by Methylation-specific PCR (MSP) using the following primer sets (5′ → 3′-orientation): methylated (SFRP1-M-F, TTTGTAGTTTTCGGAGTTAGTGTCGC; SFRP1-M-R, CGACCCTCGACCTACGATCG; IGFBP3-M-F, GCGAGTTTCGAGTTGTACGTTTTC; IGFBP3-M-R, GCCGACCGCTATATAAAAACCG; HHIP-M-F, AGTAGTCGGGTAGTTTCGGAATTTTC; HHIP-M-R, GAACCTTCGAAACCAACCTCG; HOXA7-M-F, GAGTTTAGATAGACGGCGGC; HOXA7-M-R, CCGAAAACGCCTTTATAACG) and unmethylated (SFRP1-U-F, TTTTGTAGTTTTTGGAGTTAGTGTTGTGTG; SFRP1-U-R, CAATAACAACCCTCAACCTACAATCAA; IGFBP3-U-F, TTGGGTGAGTTTTGAGTTGTATGTTTTT; IGFBP3-U-R, AAACACACCAACCACTATATAAAAACCAAA; HHIP-U-F, TTGTAGTAGTTGGGTAGTTTTGGAATTTTT; HHIP-U-R, AAACCTTCAAAACCAACCTCAAAA; HOXA7-U-F, GTTTGAGTTTAGATAGATGGTGGTG; HOXA7-U-R, CATCCAAAAACACCTTTATAACAAA).
MSP primer design was accomplished using Methyl Primer Express (Applied Biosystems) using the following criteria: CpG percentage >55%; observed/expected CpG > 65%; CpG length > 300 bp. The MSP reaction contained 40 ng DNA, 500 nM of forward and reverse primer, 2 mM dNTPs, 1.5 mM MgCl2, 1 U Hot Start Taq DNA polymerase and 1× Hot Start PCR Buffer (MBI Fermentas). Conditions for MSP were 95°C for 4 min, followed by 38 cycles of 94°C for 30 s, 61°C for 30 s and 72°C for 45 s, with a final extension cycle of 72°C for 10 min. The PCR products were resolved by electrophoresis in a 2% agarose gel.
Pyrosequencing
Pyrosequencing was performed using standard procedures (44). In brief, 160 bp of the HHIP promoter region were amplified from bisulfite-treated DNA using Maxima Hot start DNA polymerase (Fermentas) and the following primers: HHIP-Pyro-F, GGGAGGAGAGAGGAGTTT; HHIP-Pyro-R, Biotin-AACCAACCTCCAAAATACTAAACC. Sequencing was performed on a PyroMark24 with PyroMark Gold Q24 reagents (Qiagen) and the sequencing primer HHIP-Pyro-Seq, TTTAGGATTGAGTTTTTGTTTTAAG.
Additionals
A detailed description of the ChIP procedure as well as the preparation of nuclear extracts and proteins is given in the Supplementary Data.
RESULTS
Dnmt1, UHRF1 and USP7 form complexes in solution and on chromatin
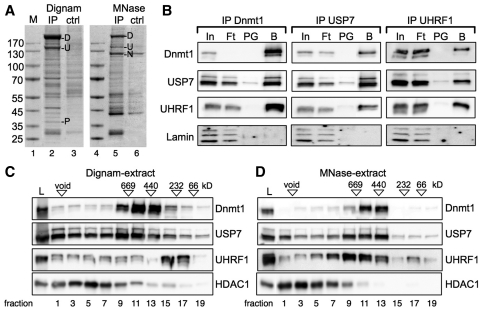
In order to identify Dnmt1 interaction partners, we performed immunoprecipitations with monoclonal antibodies directed against Dnmt1 using nuclear extracts from HeLa S3 cells (Figure 1A) and human placenta (data not shown). Nuclei were isolated and nuclear extracts were either prepared according to the protocol of Dignam (45), or by digestion of chromatin with micrococcal nuclease (MNase) and specifically associated proteins were identified by conventional mass spectrometry of the excised bands, or determined quantitatively by iTraq labeling and mass spectrometry (Supplementary Figure S1). Besides the known interaction partners UHRF1 (N), only present in the MNase-treated extract, and PCNA (P) (13,21,46), we observed a strong enrichment of the protein USP7 (U) in both kind of nuclear extracts (Figure 1A).
Figure 1.
Dnmt1, UHRF1 and USP7 interact with one another in vivo. (A) USP7 is identified as a new interaction partner of Dnmt1. Dnmt1 was immunoprecipitated from nuclear extracts (Dignam-extract and MNase-extract) and subjected to SDS–PAGE and Coomassie blue staining. Protein bands [(U) USP7, (P) PCNA, (N) UHRF1] specific for the Dnmt1-IP were identified by MS analysis (ctrl: proteinG sepharose only). Precipitated Dnmt1 (D) and the molecular weight marker (M) are indicated. (B) Immunoprecipitation of endogenous proteins of the MNase treated extract with the indicated antibodies. Co-precipitated proteins were detected by Western Blot. 2% of the input (In), 10% of the flowthrough (Ft), 10% of the control beads (PG: ProteinG Sepharose) and 10% of the antibody coupled beads (B) were loaded. (C and D) HeLa S3 nuclear extracts [Dignam-extract (C) and MNase-extract (D)] were applied on a supererose6 gelfiltration column (GE Healthcare). Every second fraction was analyzed on SDS–PAGE following western blot analysis with the indicated antibodies. Load (L), void and the migration of the molecular weight reference proteins are indicated.
To verify the specific interaction of the endogenous proteins, immunoprecipitations with antibodies specific for Dnmt1, UHRF1 and USP7 were performed, and the presence of the other proteins was tested by Western Blot analysis (Figure 1B and Supplementary Figure S2). Immunoprecipitation of Dnmt1 showed co-precipitation of UHRF1 and USP7. Vice versa, immunoprecipitation of UHRF1 and USP7 showed interaction with the other proteins of interest revealing an interaction of all three proteins in vivo. Furthermore, digestion of endogenous DNA by DNaseI followed by immunoprecipitation demonstrated that these interactions are direct and not mediated by their independent binding to neighboring DNA elements (Supplementary Figure S2). However, the interaction of Dnmt1 with UHRF1 was significantly weaker after DNaseI digestion, suggesting that the complex with UHRF1 is stabilized on chromatin. To further dissect the interaction of USP7 with Dnmt1 and UHRF1 in vivo, we performed gelfiltration analysis of the nuclear extracts (Dignam-extract, ‘MNase’-extract) analyzing the migration behavior of the three proteins (Figure 1C and D). The major fractions of Dnmt1 and USP7 significantly co-migrated between 440 and 669 kDa in the Dignam-extract, whereas the majority of UHRF1 was not co-fractionating, but migrating at lower molecular weight. However, if the MNase-treated extract was used, we observed co-migration of Dnmt1, UHRF1 and USP7 suggesting the stabilization of the trimeric interaction by its interaction with chromatin (Figure 1D). Analysis of the fractions on SDS–PAGE following silver staining of proteins clearly shows the presence of histones in the fractions of UHRF1 in the ‘MNase’-treated extract but not in the Dignam-extract (Supplementary Figure S1D and E). As UHRF1 was shown to be tightly associated with chromatin (13,15,16), our data suggest a ‘soluble’ Dnmt1/USP7 complex to exist when Dnmt1 is detached from chromatin and that a trimeric complex with UHRF1 forms at the chromatin target sites in vivo. The HDAC1 protein was recently shown to form interactions with USP7, UHRF1 and Dnmt1 (47); however, none of our gelfiltration experiments could reveal a major fraction of HDAC1 co-migrating with the target proteins (Figure 1C and D).
Dnmt1 and UHRF1 interact with USP7 in vitro
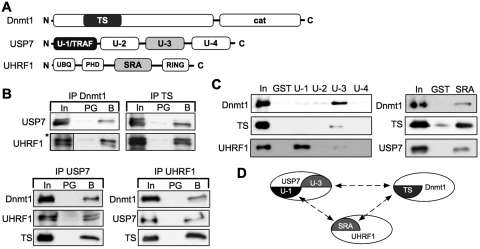
To identify the protein domains mediating the interactions, we purified the recombinant full-length proteins, the TS domain (amino acid 316–601) of Dnmt1, the SRA domain (amino acid 435–586) of UHRF1 and the TRAF domain (amino acid 1–215; U-1), the proteolytic core domain (amino acid 212–561; U-2), the C-terminal domain (amino acid 561–916; U-3) and the C-terminus (amino acid 913–1102; U-4) of USP7. The purified proteins were used in in vitro pull-down experiments to reveal the interaction domains (Figure 2A, Supplementary Figures S3 and S4). Dnmt1 efficiently co-precipitated USP7 in vitro and we were able to map the interaction site of Dnmt1 to the TS domain (Figure 2B). Vice versa USP7 interacted with Dnmt1 and the TS domain as expected. The TS domain does interact at the same time with UHRF1, an interaction that was also shown previously (21). So far the data suggest two dimeric complexes, i.e. Dnmt1/USP7 and Dnmt1/UHRF1. However, we observed as well a direct interaction between UHRF1 and USP7 indicating the potential existence of three different dimeric complexes. Immunoprecipitation experiments using the individual USP7 domains showed that UHRF1 interacts with the TRAF domain (amino acid 1–215, U-1), whereas Dnmt1 and the TS domain were bound by domain 3 of USP7 (amino acid 561–916, U-3) (Figure 2C). In contrast to Dnmt1, the de novo DNA methyltransferases, Dnmt3a and Dnmt3b2 preferentially interacted with the TRAF domain of USP7 (Supplementary Figure S4). Furthermore, the SRA domain of UHRF1 mediated the interaction with both Dnmt1 and USP7 (Figure 2C). Taken together, our in vitro studies allow the discrimination of the following dimeric complexes (Figure 2D): a Dnmt1/UHRF1 interaction between the TS and the SRA domain, an association of UHRF1 and USP7 mediated between the SRA and the TRAF domain, and a Dnmt1/USP7 complex between the U-3 and TS domains, respectively. Moreover, all interactions also suggest the formation of a trimeric complex of USP7, Dnmt1 and UHRF1, arguing for a direct interaction on chromatin in vivo.
Figure 2.
Dnmt1, UHRF1 and USP7 form a trimeric complex. (A) Schematic representation of the domains of Dnmt1, USP7 and UHRF1 (proteins and domains are not in scale). TS (Targeting domain, amino acid 316–601), cat (C-terminal catalytic domain of Dnmt1). U-1 (TRAF domain, amino acid 1–215), U-2 (catalytic domain, amino acid 212–561), U-3 (C-terminal domain, amino acid 561–916), U-4 (C-terminus, amino acid 913–1102). UBQ (ubiquitin-like domain), PHD (plant homeodomain domain), SRA (SET-Ring finger associated domain, amino acid 435–586), RING (Ring finger domain). (B) Co-immunoprecipitation assay to determine the interaction of recombinant proteins with protein-specific antibodies. Detection of co-precipitated proteins via western blot. Input (In, 1.0%), protein G sepharose (PG, 20%), ‘specific beads’ (B, 20%), antibodies used for immunodetection are indicated. The Dnmt1-specific antibody DNM-2C1 recognizes the TS-domain and was used for IP and WB. (Asterisks) Five times shorter exposure of the input signal of UHRF1. (C) GST pull-down assay interaction analysis, using GST, the indicated USP7-domains and the SRA domain fused to GST with Dnmt1, TS-domain, USP7 and UHRF1. The bound proteins were separated and plotted with the respective antibodies. Input (In, 20%); GST or GST-fusion domains (25%). (D) Schematic overview of the protein interactions and protein-domains involved.
USP7 prevents UHRF1 from autoubiquitinylation in vitro and regulates its stability in vivo
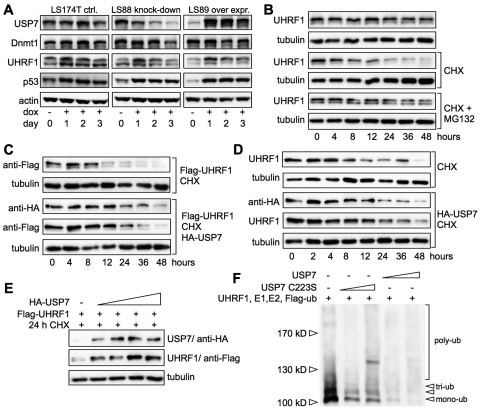
To study the role of the USP7 deubiquitinase activity with respect to the regulation of Dnmt1 and UHRF1 protein stability, the expression of USP7 was either down- or upregulated in doxycycline inducible LS174T cells (Figure 3A). In agreement with previous studies, knockdown (LS88 cells) and overexpression (LS89 cells) of USP7 resulted in stabilization of p53 and thus increase of p53 levels (25–28). Dnmt1 protein levels decreased after 3 days of USP7 knockdown, when USP7 levels were the lowest (Figure 3A), confirming the observation of Wang and colleagues (47). More significant, UHRF1 protein levels decreased or increased after down- or upregulation of USP7, respectively (Figure 3A). This indicates that USP7 affects the stability of Dnmt1 and UHRF1 protein in vivo. Due to the strong and reproducible effects, we further evaluated the stability of the UHRF1 protein. We used the p53 (−/−) non-small lung cancer cell line H1299 to avoid p53-dependent side effects. In order to address whether UHRF1 is turned over in the ubiquitin dependent proteasomal pathway, cells were treated with cycloheximide and the proteasome inhibitor MG132 (48). Incubation of the cells with cycloheximide resulted in a gradual decrease of UHRF1 protein levels (Figure 3B). The addition of MG132 clearly stabilized UHRF1 protein levels in the presence of cycloheximide, indicating that UHRF1 is degraded via the proteasome-dependent degradation pathway.
Figure 3.
USP7 regulates the stability of UHRF1 in vivo and in vitro. (A) LS174T adenocarcinoma cells (LS174TR1, parental) were treated with doxycycline (dox, 1.0 µg/ml) to induce USP7 knockdown (LS88 knockdown) or overexpression of USP7 (LS89 over expr.). Cells were harvested at the indicated days and a WCE was prepared. Samples were subjected to SDS–PAGE and western blot analysis with the indicated antibodies. (B) H1299 cells were treated with cycloheximide (CHX, 100 µg/ml) and proteasome inhibitor MG132 (20 µM) for the indicated time and subsequently treated as described in (A). (C) H1299 cells were transfected with 700 ng Flag-UHRF1 DNA without or with 1.4 µg HA-USP7 DNA. Cells were split 36 h post-transfection, treated with CHX for the indicated time and analyzed as described in (A). (D) Same as for (C) but only transfection of 2.0 µg HA-USP7 DNA. (E) H1299 cells were transfected with 500 ng Flag-UHRF1 DNA and increasing amounts of HA-USP7 DNA (0, 500, 1000, 1500, 2000 ng). Forty-eight hours post-transfection cells were treated with CHX for 24 h and subsequently analyzed as described in (A). (F) In vitro autoubiquitinylation of UHRF1 in the absence or presence of either catalytically active USP7 or inactive USP7 C223S and Flag-tagged ubiquitin was analyzed (substoichiometric USP7 levels were used; 1/5th and 1/50th molar ratio with respect to UHRF1). Proteins were separated on SDS–PAGE and Flag-tagged ubiquitin adducts were detected by western blot with antibodies against the Flag-tag.
USP7 has been described to protect the RING-finger E3-ubiquitin ligases ICP0 (49), Chfr (50) and Mdm2 (51) from autoubiquitinylation, thereby stabilizing their protein levels. We therefore transfected USP7 into H1299 cells to monitor the effect of USP7 on the stability of transfected and endogenous UHRF1 protein (Figure 3C and D). As seen before, cycloheximide treatment led to gradual decrease of UHRF1 levels, but also affected the protein stability of USP7 (timepoints 12–48 h). Notably, the transiently increased USP7 levels were sufficient to stabilize the exogenous and endogenous UHRF1 protein (timepoints: 8–24 h) compared to control cells not transfected with USP7. Transfection of increasing amounts of USP7 following cycloheximide treatment for 24 h (Figure 3E) stabilized UHRF1 protein levels in an USP7-dose-dependent manner. Taken together, these data indicate that USP7 stabilizes UHRF1 protein levels in vivo and suggest that USP7 removes ubiquitin adducts from UHRF1 and therefore prevents proteasomal degradation of UHRF1.
To test whether USP7 directly influences the autoubiquitinylation activity of the RING-finger E3-ubiquitin ligase UHRF1 (15,16,23), we performed in vitro autoubiquitinylation reactions of UHRF1 in the presence of either USP7 or the catalytically inactive mutant USP7 C223S (49) (Figure 3F and Supplementary Figure S5B). The activity of the enzymes and the assay conditions were initially optimized to allow quantification of enzymatic conversion of the substrates, when required (Supplementary Figures S5A and S6). UHRF1 exhibits in vitro autoubiquitinylation activity revealing mono- to tetra-ubiquitin adducts. However, it has to be mentioned that the assay cannot discriminate between the addition of ubiquitin-chains and the addition of multiple mono-ubiquitin molecules on UHRF1. Interaction of USP7 C223S with UHRF1 reduced the autoubiquitinylation activity. However, active USP7 completely abolished the autoubiquitinylation of UHRF1 by removal of ubiquitin adducts. Accordingly, these data clearly indicate that USP7 regulates UHRF1 levels in vivo, and counteracts autoubiquitinylation activity of UHRF1 by removing ubiquitin adducts.
USP7 stimulates the DNA methylation activity of Dnmt1 in vitro and in vivo
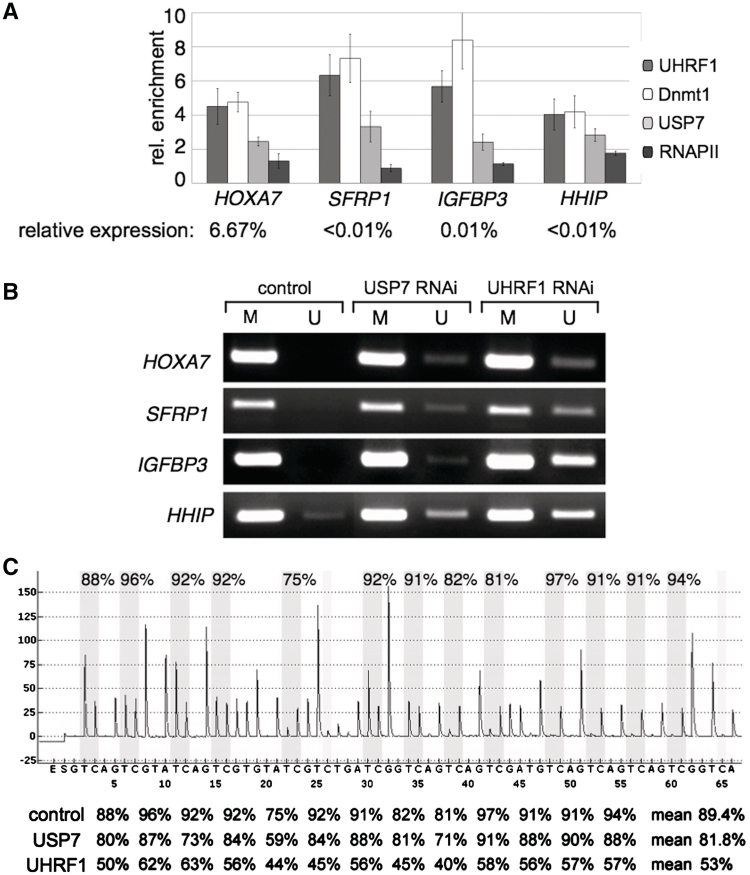
Next, we tested in ChIP assays whether USP7 co-localizes with Dnmt1 and UHRF1 on selected silenced genes in vivo (Figure 4A). Genes were selected according to their reduced expression levels, as determined by quantitative mRNA analysis compared to the expression levels of the TBP gene. The selected genes were either completely silenced SFRP1, IGFBP3, HHIP or active to only 6.6% (HOXA7). Active genes exhibiting high mRNA levels and correspondingly high RNA Polymerase II and low Dnmt1 binding levels were not studied (Supplementary Figure S7). Besides the HOXA7 locus (52), as a previously published control, we show that Dnmt1 localizes as well to the SFRP1, IGFBP3 and HHIP loci together with UHRF1 and in absence of RNA Polymerase II. In agreement with our biochemical characterization we observed an enrichment of USP7 at the SFRP1, IGFBP3, HHIP and HOXA7 loci together with Dnmt1 and UHRF1, suggesting that they form a trimeric and functional complex on DNA.
Figure 4.
USP7 associates with Dnmt1 and UHRF1 on silenced genes in vivo. (A) Chromatin immunoprecipitation (ChIP) was performed with formaldehyde cross-linked HCT-116 cells and the indicated antibodies. The average of three independent ChIP experiments for Dnmt1, UHRF1, USP7 and RNAPII are shown. Standard deviations, target genes of interest and antibodies used for ChIP are indicated. The enrichment of specific IP versus IgG background is plotted. The relative expression levels of these genes compared to the TBP gene are given. (B) Promoter regions of the SFRP1, IGFBP3, HHIP and HOXA7 genes were analyzed for methylated (M) and unmethylated (U) CpG sites by MSP in the presence or absence of USP7 and UHRF1. Proteins were depleted by RNAi-mediated knockdown and the MSP analysis was performed with bisulfite-treated genomic DNA from HCT-116 cells. Representative images of MSP experiments are given. (C) Quantitative analysis of DNA methylation levels after UHRF1 and USP7 knockdown. Pyrogram trace obtained after pyrosequencing analysis of part of the HHIP promoter region containing 13 CpG sites (with potentially methylated cytosines shaded in gray). The y-axis represents the signal intensity in arbitrary units, while the x-axis shows the dispensation order. The percentage of DNA methylation at individual CpG positions of the HHIP promoter of cells transfected with non-targeting control, USP7 and UHRF1 siRNAs are shown below the pyrogram.
To address the effect of the Dnmt1 interacting proteins UHRF1 and USP7 on the DNA methylation activity of Dnmt1, we performed a MSP analysis upon knockdown of UHRF1 and USP7, respectively (Figure 4B). Downregulation of UHRF1 by siRNA resulted in a robust demethylation at the SFRP1, IGFBP3, HHIP and HOXA7 loci. Similarly, knockdown of USP7 resulted in detectable demethylation of all loci analyzed, albeit to a lower degree as UHRF1 (Figure 4B). The quantitative examination of the HHIP promoter region by pyrosequencing revealed a mean decrease in CpG methylation of 7.6 versus 36.4% upon USP7 and UHRF1 knockdown, respectively (Figure 4C).
USP7 stimulates the DNA methylation activity of Dnmt1
USP7 forms a soluble complex with Dnmt1 in solution and a trimeric complex with UHRF1 in chromatin. We addressed the question whether the interaction of USP7, UHRF1 or USP7/UHRF1 with Dnmt1 would affect its DNA methylation activity in vitro. The affinities of USP1, UHRF1 and Dnmt1 toward DNA and the DNA methyltransferase assays were optimized to allow the quantitative and qualitative analysis of these reactions (Figure 5A–C; Supplementary Figures S6, S8 and S9). Although siRNA mediated knockdown of UHRF1 led to reduced DNA methylation in vivo (Figure 4B and C), we did not detect an effect of UHRF1 on the Dnmt1-dependent DNA methylation activity (Figure 5C). In agreement with previous data, this suggests that UHRF1 may solely serve as a recruitment platform determining the location of DNA methylation rather than influencing the activity of Dnmt1 (13,14). Accordingly, we observed a reduction of DNA methylation levels at the SFRP1 and HHIP genes after USP7 knockdown (Figure 4B and C). Strikingly, both the in vitro maintenance and de novo DNA methylation activity of Dnmt1 were stimulated 2-fold in the presence of USP7 (Figure 5D). USP7 and UHRF1 do inhibit Dnmt1-dependent DNA methylation activity when quantitatively bound to DNA, limiting the accessible substrate for Dnmt1 (Supplementary Figure S8). Therefore, the DNA substrate was used in large excess (3-to 24-fold molar ratio), suggesting that USP7 modulates Dnmt1′s enzymatic activity rather than acting as a recruitment factor. Furthermore, USP7 exerts a stimulatory effect on the methylation activity of Dnmt1 even in the presence of UHRF1, which is in agreement to the in vivo data (Figures 4 and 5E). Hence, we identified USP7 as the first factor that has a stimulatory effect on the enzymatic activity of Dnmt1. Furthermore, we describe USP7 as a regulator of UHRF1 protein stability, hence directly affecting Dnmt1-dependent DNA methylation efficiency.
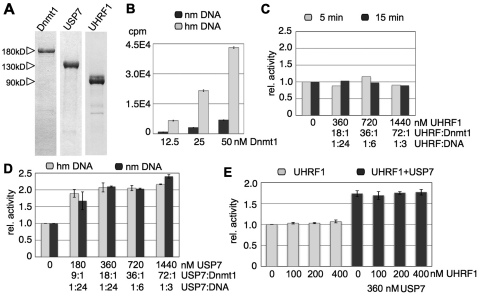
Figure 5.
USP7 activates the DNA methylation activity of Dnmt1 in vitro. (A) Recombinant Dnmt1, USP7 and UHRF1 proteins, purified from baculovirus-infected Sf21 insect cells and E. coli cells via the His-tag. Proteins were separated on a SDS–PAGE and stained with Coomassie blue. (B) Dnmt1 dependent in vitro DNA methylation on non-methylated (nm DNA) and hemi-methylated DNA (hm DNA) was performed with increasing amounts of Dnmt1 and an excess of DNA (4.0 µM) for 15 min. The transfer of the 3H-methyl group onto the DNA was measured and plotted. (C) Dnmt1-dependent DNA methylation reactions in the presence of increasing amounts of UHRF1 were performed in excess of hmDNA for two different time points (5 and 15 min). The relative activity was plotted and UHRF1 ratios to DNA or Dnmt1 are given. (D) Same as for (C) but using USP7 on non-methylated (nm) and hemi-methylated DNA (hm). (E) Same as for (C) but fixed amounts of Dnmt1 and USP7 were incubated with increasing levels of UHRF1 as indicated. The relative methylation activity is plotted and molar ratios of UHRF1:Dnmt1, USP7:Dnmt1, UHRF1:DNA and USP7:DNA are given.
DISCUSSION
Dnmt1 has been extensively described and studied as an epigenetic factor transiently interacting with many other chromatin-associating proteins (10,11). In fact, so far only one Dnmt1-complex containing pRB, HDAC, E2F1 had been biochemically purified (12).
In this study, we identified two novel Dnmt1-containing complexes by means of immunoprecipitation and SEC analysis. First, we observed a strong interaction of Dnmt1 and USP7, forming a soluble dimer complex in vivo. Second, Dnmt1/USP7 could further interact with UHRF1 and strongly associated as a trimeric complex on chromatin (Figures 1, 2 and 4A).
UHRF1 was shown to be tightly associated with chromatin and to bind to hemi-methylated DNA (hmDNA), acting as a recruitment factor for Dnmt1 (13–15). We confirmed the preferential binding of UHRF1 to hmDNA (Supplementary Figure S8A) and demonstrated co-binding of Dnmt1 and UHRF1 to silenced genes in addition to the subunit USP7 (Figure 4A). Upon UHRF1 knockdown, we observed a reduction of the methylation status of the studied genes (Figure 4B and C). As UHRF1 did not stimulate the DNA methylation activity of Dnmt1 in vitro, this clearly indicates that UHRF1′s major function is to ensure Dnmt1 guidance to hmDNA, as previously shown (13,14). USP7 had been described to protect RING-finger E3-ubiquitin ligases from autoubiquitinylation, thereby stabilizing their protein levels (49–51). We were able to show that USP7 levels did directly influence the stability of UHRF1 and was capable to de-ubiquitinylate UHRF1 in vitro (Figure 3F). Transfection of USP7 into H1299 cells stabilized exogenous and endogenous UHRF1 protein upon cycloheximide treatment (Figure 3). Altogether, our data indicate that USP7 removes ubiquitin moieties from UHRF1 and thereby inhibits its proteasomal degradation in the cell. The knockdown of USP7 affected the DNA methylation status of the studied target gene loci, suggesting that it serves as an amplifying module within the trimeric methylating complex, ensuring maintenance of DNA methylation. An effect that apparently resides from UHRF1 stabilization and the upregulation of the Dnmt1 DNA methylation activity, as discussed below. Consistent with the assumption of a close collaboration of those proteins, USP7 knockout mice showed lethal defects in early embryonic development (33), reminiscent of Dnmt1 knockout mice (7).
As the expression of UHRF1 peaks during G1/S-phase transition and is downregulated during G0 and G1 (40,53), USP7 could stabilize UHRF1 during S-phase by constantly removing ubiquitin moieties. After replication, USP7′s activity could be modulated by post-translational modification (54), eventually leading to degradation of UHRF1. Subsequently, the Dnmt1/USP7 complex would dissociate from chromatin consistent with the observation of a diffuse localization of Dnmt1 in the nucleoplasm during G1 and G0 (20).
It was recently suggested that Dnmt1 is stabilized by HDAC1 and USP7 in a cell cycle-dependent manner (47). The authors suggested a stable complex of Dnmt1 with HDAC1 that we did not observe in our immunoprecipitation and gelfiltration assays (Figure 1C and D; Supplementary Figure S2B). This could be related to the fact that we used non-synchronous cells for our purifications and minor amounts of Dnmt1–HDAC1 complexes could have been overlooked, as the published effect is cell cycle dependent. However, in agreement with their data, we see a comparable destabilization of Dnmt1 after 3 days of USP7 knockdown. The mild destabilization of Dnmt1 would be further potentiated by the HDAC1 activity, as described (47).
Besides controlling the ubiquitination status of UHRF1, USP7 plays an additional role in this complex and stimulates both the maintenance and the de novo DNA methylation activity of Dnmt1 in vitro (Figure 5). So far USP7 is the first factor to be shown to exhibit such an activity. Immunoprecipitation and gelfiltration experiments suggest that the majority of Dnmt1 may be associated with USP7 and as shown by Wang and colleagues USP7 does also regulate the stability of Dnmt1 (47). Accordingly, USP7 acts in two ways to stimulate the Dnmt1-dependent DNA methylation activity, i.e. via stimulating its activity and protecting the protein from degradation. The 2-fold activation of the Dnmt1 activity in the Dnmt1/USP7 complex could not be further activated by the addition of UHRF1 confirming the notion that UHRF1 is mainly required to target the complex to hemi-methylated DNA.
From these data, we propose a new role for USP7 as the epigenetic regulator of Dnmt1-dependent DNA methylation activity. First, the enzymatic activity of Dnmt1 in complex with USP7 is directly stimulated by USP7 by factor of two. Second, the Dnmt1-USP7 dimeric complex is recruited to the sites of methylation by UHRF1, forming a trimeric complex on chromatin. As the protein stability of UHRF1 is regulated by USP7, the Dnmt1-USP7 association on chromatin and hence the DNA methylation efficiency is controlled by USP7.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The work was funded by the DFG (LA1331/4-1 to G.L. and KA2274/3-1 to R.K.) and the BayGene program of the state Bavaria (Germany). Funding for open access charge: University of Regensburg.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 4.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freitag M, Selker EU. Controlling DNA methylation: many roads to one modification. Curr. Opin. Genet. Dev. 2005;15:191–199. doi: 10.1016/j.gde.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 8.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 10.Robertson KD. DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 11.Latham T, Gilbert N, Ramsahoye B. DNA methylation in mouse embryonic stem cells and development. Cell Tissue Res. 2008;331:31–55. doi: 10.1007/s00441-007-0537-9. [DOI] [PubMed] [Google Scholar]
- 12.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 13.Bostick K, Estève C, Pradhan, Jacobsen UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:704–715. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 14.Sharif J, Muto M, Takebayashi S-I, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 15.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 18.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 19.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 20.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 21.Achour M, Jacq X, Rondé P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes A, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 22.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell. Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 26.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–692. [PubMed] [Google Scholar]
- 27.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1 p following 486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 28.Meulmeester E, Maurice MM, Boutell C, Teunisse AFAS, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol. Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 29.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 2003;278:47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Qu L-K, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, Yang X. Critical role for Daxx in regulating Mdm2. Nat. Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 31.van der Horst A, de Vries-Smits AMM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BMT. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 32.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Knaap JA, Kumar BRP, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJR, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 35.van der Knaap JA, Kozhevnikova E, Langenberg K, Moshkin YM, Verrijzer CP. Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Mol. Cell. Biol. 2010;30:736–744. doi: 10.1128/MCB.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van De Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 37.Van De Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, Van Leenen D, Holstege FCP, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessler BM, Fortunati E, Melis M, Pals CEGM, Clevers H, Maurice MM. Proteome changes induced by knock-down of the deubiquitylating enzyme HAUSP/USP7. J. Proteome Res. 2007;6:4163–4172. doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- 39.Hu M, Li P, Li M, Li W, Yao T, Wu J-W, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 40.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 41.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eichenmuller M, Gruner I, Hagl B, Haberle B, Muller-Hocker J, von Schweinitz D, Kappler R. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49:482–490. doi: 10.1002/hep.22649. [DOI] [PubMed] [Google Scholar]
- 43.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 45.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 47.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell. Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 49.Canning M, Boutell C, Parkinson J, Everett RD. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 2004;279:38160–38168. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- 50.Oh YM, Yoo SJ, Seol JH. Deubiquitination of Chfr, a checkpoint protein, by USP7/HAUSP regulates its stability and activity. Biochem. Biophys. Res. Commun. 2007;357:615–619. doi: 10.1016/j.bbrc.2007.03.193. [DOI] [PubMed] [Google Scholar]
- 51.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 52.Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36:3590–3599. doi: 10.1093/nar/gkn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujimori A, Matsuda Y, Takemoto Y, Hashimoto Y, Kubo E, Araki R, Fukumura R, Mita K, Tatsumi K, Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome. 1998;9:1032–1035. doi: 10.1007/s003359900920. [DOI] [PubMed] [Google Scholar]
- 54.Fernández-Montalván A, Bouwmeester T, Joberty G, Mader R, Mahnke M, Pierrat B, Schlaeppi J-M, Worpenberg S, Gerhartz B. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 2007;274:4256–4270. doi: 10.1111/j.1742-4658.2007.05952.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.