Abstract
Histone variant H3.3 and heterochromatin protein 1γ (HP1γ) are two functional components of chromatin with role in gene transcription. However, the regulations of their dynamics during transcriptional activation and the molecular mechanisms underlying their actions remain poorly understood. Here, we provide evidence that heat shock-induced transcription of the human HSP70 gene is regulated via the coordinated and interdependent action of H3.3 and HP1γ. H3.3 and HP1γ are rapidly co-enriched at the human HSP70 promoters upon heat shock in a manner that closely parallels the initiation of transcription. Knockdown of H3.3 prevents the stable recruitment of HP1γ, inhibits active histone modifications, and attenuates HSP70 promoter activity. Likewise, knockdown of HP1γ leads to the decreased levels of H3.3 in the promoter regions and the repression of HSP70 genes. HP1γ selectively recognizes particular modification states of H3.3 in the nucleosome for its action. Moreover, HP1γ is overexpressed in three representative cancer cell lines, and its knockdown leads to reduction in HSP70 gene transcription and inhibition of cancer cell proliferation. We conclude that the physical and functional interactions between H3.3 and HP1γ make a unique contribution to acute HSP70 transcription and cancer development related to the misregulation of this transcription event.
INTRODUCTION
The dynamic nature of chromatin is functionally important for the regulation of diverse DNA-dependent processes in the nucleus including transcription, replication and DNA repair. While the mechanisms are not well understood, nucleosome remodeling activities modulate the functional state of chromatin. In addition to ATP-dependent nucleosome remodeling and covalent histone modifications, deposition of histone variants into the nucleosome is thought to be critical in transcriptional regulation (1,2). H3.3 is the predominant form of histone H3 variants, which differs by four amino acids from the replication dependent histones H3.1 and H3.2, (generally referred to as H3) (3,4). The H3.3 variant is expressed throughout the cell cycle and incorporated into chromatin in DNA replication-independent manner, whereas histone H3 is primarily expressed and incorporated during S phase (5). In human cells, replication-independent deposition of H3.3 is primarily mediated by the HIRA chaperone and the ATRX chromatin remodeler through a mechanism distinct from that of replication-coupled deposition of H3 by the CAF-1 (6,7). H3.3 is preferentially distributed over the promoter regions and its accumulation coincides with higher levels of H3-K4 methylation and bound RNA polymerase II (8–11). Further support for such a transcription-coupled H3.3 deposition is provided by studies in Drosophila demonstrating that the heat shock-induced transcription of HSP70 genes coincides with the replacement of H3 with H3.3 (12,13). In addition to its enrichment over active genes, a genome-wide analysis of H3.3 distribution indicated that a fraction of H3.3 also localizes to constitutive heterochromatin regions at telomeres (7,14).
Another group of proteins specifically marking chromatin state consists of heterochromatin protein 1 (HP1). Mammalian cells possess three closely related isoforms of HP1 based on their size and amino acid sequence similarity (15). HP1α primarily localizes to pericentric heterochromatin, HP1β binds to both heterochromatin and euchromatin, and HP1γ exclusively targets euchromatin (16–18). All these isoforms have been originally characterized as regulatory proteins that establish an inactive state of chromatin, but recent studies have challenged this view (19,20). One of the first clear indications that HP1 acts as a positive regulator came from a previous work demonstrating that H3K9me and HP1γ are enriched at the coding regions of actively transcribed genes (21). An activating role of HP1 in transcription was further supported by recent studies showing that HP1γ associates with the Survivin gene in its active state (22). Another notable finding is that Drosophila HP1c (HP1γ homolog) stimulates transcription elongation by bridging the interaction between histone chaperone FACT and phosphorylated RNA polymerase II (23). All these results suggest that, besides its most commonly cited role in heterochromatin formation, HP1 can trigger an active chromatin structure once it is targeted to specific genes within euchromatin.
Of special relevance to the present study, the rapid incorporation of H3.3 and the recruitment of HP1c have been shown to play a role in controlling transcription of the HSP70 genes under heat-shock condition in Drosophila (13,24). These findings raise the question whether H3.3 and HP1γ are also required for higher rates of transcription at induced HSP70 genes in human cells and if so, how their effects are generated. In this study, we addressed these questions under relevant conditions by monitoring independent and cooperative actions of H3.3 and HP1γ in HSP70 gene transcription. We found that H3.3 and HP1γ are co-localized at HSP70 promoters and establish transcriptional competence in response to heat shock. Detailed investigation of the underlying mechanism revealed a selective connection between HP1γ localization and active histone modifications enriched in H3.3 nucleosomes. Furthermore, HP1γ and H3.3 are functionally interdependent as RNA interference (RNAi)-mediated depletion of either HP1γ or H3.3 inhibits HSP70 transcription and cancer cell proliferation.
MATERIALS AND METHODS
Cell culture, antibodies and constructs
HeLa and MCF-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Antibodies used in this study are as follows: H2Aac, H2Bac, H3ac and H4ac from Millipore, H3K9me1/me2/me3, H3K4me1/me2, H3K27me1/me2 and H3.3 antibodies from Abcam, IgG antibody from Santa Cruz Biotechnology, Flag antibody from Sigma, HP1α, HP1β and HP1γ antibodies from Millipore, H3K4me3 antibody from Active Motif and H3K27me3 antibody from Dr J. Rice. For mammalian expression of H3 and H3.3, their cDNAs were PCR-amplified and ligated into pIRES mammalian vector in frame with 5′ Flag and HA sequences.
Nucleosome purification
HeLa cells were transfected with expression vectors for Flag- and HA-tagged versions of human H3 and H3.3. 48 h post-transfection, cells were harvested and lysed with buffer A (20 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothritol and protease inhibitor cocktail) containing 0.2% Triton X-100. Nuclei were pelleted by centrifugation at 1000g, resuspended in buffer A containing 2 mM CaCl2 and digested with 0.6 U microccocal nuclease (MNase, Sigma) at 37°C for 28 min. Digested nuclei were collected and incubated in nuclear extraction buffer (20 mM HEPES, pH 7.4, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EGTA and protease inhibitor cocktail) for 1 h, and centrifuged to remove nuclear debris. After adjusting the salt concentration of the extract to 150 mM NaCl, ectopic H3/H3.3-containing nucleosomes were isolated by two consecutive immunoprecipitation using anti-Flag and anti-HA antibodies (Sigma) in washing buffer (20 mM HEPES, pH 7.8, 300 mM NaCl, 1.5 mM MgCl2, 0.2 mM EGTA, 10% Glycerol, 0.2% Triton X-100 and protease inhibitor cocktail). Bead-bound nucleosomes and proteins were analyzed by western blot analysis.
Immunofluorescence
Heat shock-treated HeLa cells were fixed in freshly prepared 4% formaldehyde solution at room temperature. Cells on cover slips were permeabilized in Solution P (1% BSA and 0.2% Triton X-100 in PBS) for 10 min, washed with PBS and blocked with 10% BSA in PBS. Cells were incubated in stepwise with a primary antibody overnight and a secondary fluorescent antibody for 1 h at 25°C. Images were captured on a Zeiss ApoTome Axiocam MRc microscope equipped with a 60× oil-immersion lens. Image processing was performed with Photoshop.
qRT–PCR, chromatin immunoprecipitation and shRNA
For quantitative reverse transcription PCR (qRT–PCR) analysis of HSP70 gene expression, total RNA was isolated using the Trizol reagent (Invitrogen) following the manufacturer's instructions. RNA was converted to cDNA using iScript cDNA Synthesis Kit (Bio-Rad), and gene expression was assessed by using the IQ SYBR Green Supermix (Bio-Rad) and the MYiQ2 real time cycler (Bio-Rad). Assays were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. The primers used quantitative real-time PCR are listed in the Supplementary data (Supplementary Table S1). ChIP assays were performed essentially as described (25). DNA was recovered from heat/mock-treated HeLa cells by PCR purification column kit. Input DNA (1%) was used for normalization. Primers used in this study are listed in Supplementary data (Supplementary Table S2). The sequences encoding the short hairpin RNAs (shRNA) for this study are listed in Supplementary Data (Supplementary Table S3). The pLKO vectors containing specific shRNA were constructed according to the manufacturer's instructions (Open Biosystems). All the constructs were verified by DNA sequencing. For knockdown experiments, cells (1 × 105) were transfected with shRNA expression constructs using Lipofectamine reagent (Invitrogen) and selected with puromycin (5 μg/ml) for 2 weeks.
MTT and colony formation assays
Cell proliferation was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer's instructions (Calbiochem). Briefly, HP1γ/H3.3-depleted MCF7 cells were incubated with 1 ml of MTT (5 mg/ml) for 3 h at 37°C. The MTT formazan precipitate was dissolved with 1 ml of MTT solvent and cellular proliferation was determined from the conversion of MTT to formazan using a Microplate Reader Model 680 (Bio-Rad) at a wavelength of 570 nm with background subtraction at 650 nm. Experiments were done in triplicate. For soft agar colony formation assay, MCF-7 cells (1 × 105) were suspended in semisolid medium (DMEM 10% FBS plus 0.33% ultra-pure noble agar). The mixture was added over a layer of 0.8% agar in DMEM on 60 mm plate and incubated at 37°C in a humidified 5% CO2 atmosphere. Colonies were counted after 3 weeks. All assays were run in triplicate, and results presented are the average of three individual experiments.
RESULTS
H3.3 and HP1γ are co-enriched at HSP70 promoters
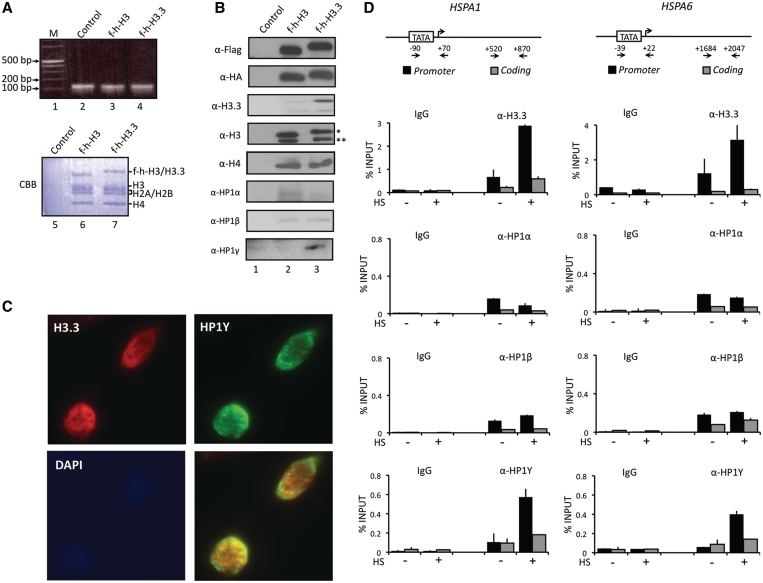
To study the interaction of HP1 isoforms with nucleosomes containing H3.3 or H3, we transiently expressed Flag-HA tagged versions (f-h) of H3.3 and H3 in HeLa cells. Western blot analysis of cell lysates indicated that the expression level of the ectopic histones was <30% of their endogenous counterparts (Supplementary Figure S1B). Soluble chromatin fragments were prepared from cell nuclei of the transfected cells and digested with micrococcal nuclease (MNase) to yield mainly mononucleosomes, as confirmed by gel electrophoresis of nucleosomal DNA (Figure 1A, lanes 2–4). The mononucleosomes were subjected to double immunoprecipitations with anti-Flag and anti-HA antibodies to selectively purify mononucleosomes containing f-h-H3.3 or f-h-H3 (Supplementary Figure S1A). As expected, almost equimolar levels of H2A, H2B, H4 and endogenous/ectopic H3.3/H3 were found in purified nucleosomes (Figure 1A, lanes 6 and 7). Western analyses with H3 antibody recognizing both H3 and H3.3 confirmed that similar levels (~10%) of ectopic H3 and H3.3 were incorporated into nucleosomes (Figure 1B, α-H3). In addition, our analyses using H3.3 specific antibody showed trace amounts of endogenous untagged H3.3 in H3.3-containing nucleosomes (α-H3.3, lane 3), indicating that the lower band detected by western blotting with H3 antibody (α-H3, lane 3) mainly contains endogenous H3. Next, we examined whether any HP1 isoforms are associated with H3.3 nucleosomes by western blotting using antibodies specific to three HP1 isoforms. HP1α and HP1β showed no detectable changes in their interaction with nucleosomes when H3.3 replaced H3 (α-HP1α/HP1β, lane 3). Importantly, however, HP1γ showed a marked preference for H3.3 nucleosomes over H3 nucleosomes (α-HP1γ, lane 3).
Figure 1.
Selective interaction of HP1γ with H3.3 nucleosomes. (A) HeLa cells were transfected with control (lane 2), histone H3 (lane 3) and H3.3 (lane 4) expression vectors for 48 h, and mononucleosomes were prepared as summarized in Supplementary Figure S1A. Total nucleosomal DNA was subjected to 2% agarose gel electrophoresis and visualized with ethidium bromide (lanes 2–4). Mononucleosomes containing H3 or H3.3 were purified by sequential immunoprecipitations using anti-Flag and anti-HA antibodies, and histone compositions of the purified nucleosomes were analyzed by Coomassie staining following 15% SDS–PAGE (lanes 6 and 7). Lane 1, 0.1–12 kb DNA ladder; lane 5, control mock-purified material. (B) Proteins co-purified with H3 and H3.3 nucleosomes were separated by 15% SDS–PAGE, and the presence of indicated proteins were analyzed by Western blotting. Lane 1, mock-purified material; lane 2, H3 mononucleosomes; lane 3, H3.3 mononucleosomes. Asterisk indicates ectopic f-h-H3 or f-h-H3.3 proteins and double asterisks indicate endogenous H3 or H3.3 proteins. (C) HeLa cells were immunostained for HP1γ (green channel), H3.3 (red channel) and DAPI (blue channel). Image overlays show co-localization between HP1γ and H3.3. (D) HeLa cells were mock-treated (−) or heat-treated (+) for 30 min, and quantitative chromatin immunoprecipitation (ChIP) assays of promoter (filled square) and coding (shaded square) regions of HSPA1 and HSPA6 genes were performed using antibodies specifically recognizing H3.3, HP1α, HP1β and HP1γ. IgG antibody was used as a negative control. Input DNA and immunoprecipitated DNA were analyzed by qPCR analyses using primer sets depicted in the top panel. The results are shown as percentage of input. The error bar indicates the means ± SD.
To further support the observed interaction, we conducted immunofluorescence imaging of endogenous H3.3 and HP1γ proteins in HeLa cells. As depicted in Figure 1C and Supplementary Figure S2, double immunostaining of H3.3 and HP1γ proteins showed a significant co-localization of H3.3 with HP1γ, but not with HP1α and HP1β in the cell nucleus. Also of note, the specificity and sensitivity of the antibodies employed in these studies were verified by western blotting using recombinant proteins (Supplementary Figure S1C–E).
Having shown the ability of HP1γ to selectively bind to H3.3 nucleosomes, we sought to determine whether this interaction is related to their cellular functions. Recent studies demonstrated that HP1γ and H3.3 are associated with heat shock-induced transcription of HSP70 genes in Drosophila (12,13). Thus, we first examined the localization of H3.3 and HP1γ at two HSP70 genes, HSPA1 and HSPA6, in HeLa cells after heat shock by chromatin immunoprecipitation (ChIP). Two sets of primers were employed to detect crosslinking of H3.3 and HP1 proteins in the promoter and transcribed regions by qPCR. As shown in Figure 1D, heat-shock treatment resulted in a rapid accumulation of H3.3 protein in the promoter regions of HSPA1 and HSPA6 genes (α-H3.3). On the contrary, H3.3 was minimally localized in the coding region and showed little change in its level after heat-shock treatment (α-H3.3). In exploring the alteration of HP1γ occupancy, we also found that HP1γ level was significantly increased in the promoter region (~10-fold) but only modestly in the coding region during the activation process (α-HP1γ). Remarkably, however, parallel ChIP assays with anti-HP1α and anti-HP1β antibodies repeatedly demonstrated no detectable accumulation of HP1α and HP1β in the promoter and coding regions in response to heat shock (α-HP1α and α-HP1β). To further confirm the above results, the ChIP assays were repeated using cells transiently transfected with Flag-tagged versions of H3.3, HP1α, HP1β and HP1γ; similar results were obtained with these ectopic proteins (Supplementary Figure S3). These results argue strongly against the possibility that H3.3 and HP1γ antibodies cross-react with other proteins, and indicate that the distribution of H3.3 and HP1γ proteins in the cell is indeed accurately established.
Both H3.3 and HP1γ are required for HSP70 transcription
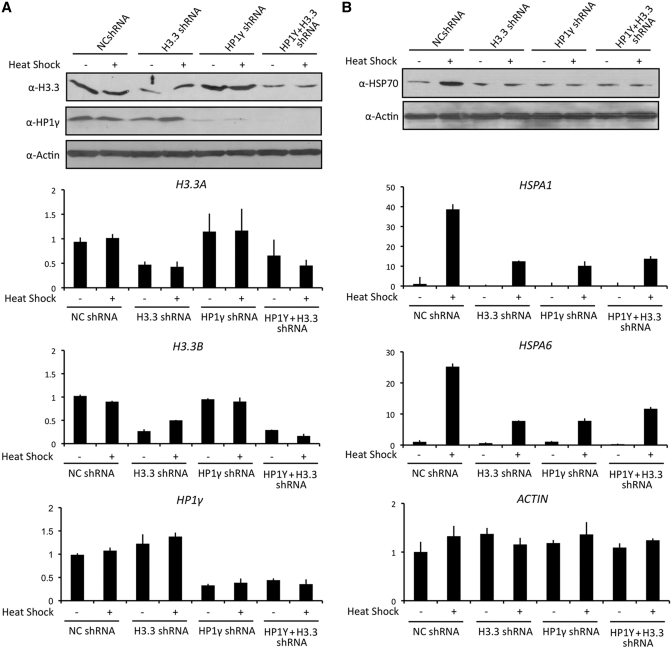
Heat shock-induced enrichments of H3.3 and HP1γ described above raised the possibility that they are required for transcription of HSP70 genes. To check this possibility, we first depleted H3.3 and HP1γ individually by a vector-based RNA interference (RNAi) technique. Western blot and qRT–PCR analyses confirmed that transfection of HeLa cells with shRNAs against H3.3 and HP1γ efficiently depleted their responsive target proteins (Figure 2A, H3.3 shRNA and HP1γ shRNA). When we measured the level of heat shock-induced transcription of HSPA1 and HSPA6 genes by qRT–PCR, we detected a distinct reduction in transcription of HSPA1 and HSPA6 genes upon shRNA-mediated silencing of HP1γ (Figure 2B, HP1γ shRNA). Similarly, upon the knockdown of H3.3, there was a substantial decrease in transcription under heat shock conditions (Figure 2B, H3.3 shRNA). On the other hand, the knockdown of H3.3 or HP1γ minimally changed transcription of HSPA1 and HSPA6 genes under non-heat shock conditions. Considering the possibility that H3.3 and HP1γ could act together to regulate HSP70 genes, we also checked whether double knockdown of H3.3 and HP1γ exhibits more severe defect in transcription. Interestingly, however, the simultaneous knockdown of H3.3 and HP1γ attenuated HSP70 transcription in similar level as that observed in individual knockdown of H3.3 or HP1γ (Figure 2B, H3.3+ HP1γ shRNA). These data constitute a powerful argument that both H3.3 and HP1γ are indeed essential for the transcriptional activation of HSP70 genes upon heat shock.
Figure 2.
Requirements for H3.3 and HP1γ in HSP70 transcription. (A) HeLa cells were stably transfected with shRNAs targeting to non-specific control, H3.3 or HP1γ. The efficiency and selectivity of knockdown were determined under non-heat shock and heat shock conditions by western blotting (upper panel) and qRT–PCR (lower panel). All transcription levels were normalized to that of GAPDH. Note that H3.3 protein is encoded by two different genes, H3.3 A and H3.3B. β-Actin was used as an internal control for equal loading in all western blot analyses. (B) Whole-cell extracts were prepared for analyses of HSP70 expression in these cells by western blotting analysis. Total mRNA was purified and subjected to qRT–PCR to examine transcription levels of HSPA1 and HSPA6 genes under non-heat shock and heat shock conditions. Average and SD of three independent experiments are shown.
H3.3 and HP1γ are interdependent for their localization at HSP70 promoters
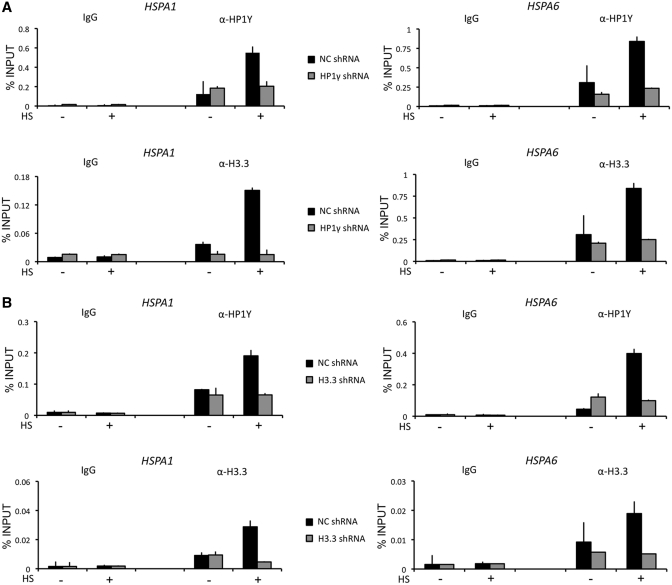
To gain support for the knockdown results above, we next checked whether the depletion of H3.3 or HP1γ is directly attributable to their promoter occupancy. As expected, the mock-depleted cells showed an apparent increase of HP1γ and H3.3 at HSPA1 and HSPA6 promoters after heat-shock treatment (Figure 3A and B, NC shRNA). When ChIP experiments were performed using HP1γ-depleted cells, significant loss of HP1γ was detected at both promoters (Figure 3A, upper panel, α-HP1γ, HP1γ shRNA). Unexpectedly, however, HP1γ-depleted cells also showed a substantial reduction of H3.3 occupancy in promoter (Figure 3A, lower panel, α-H3.3, HP1γ shRNA). This observation indicates that the heat shock-stimulated incorporation of H3.3 into the promoter nucleosomes is dependent on the presence of HP1γ. Analogously, when ChIP experiments were repeated using H3.3-depleted cells (Figure 3B, lower panel, α-H3.3, H3.3 shRNA), a decreased level of H3.3 in the promoter nucleosomes coincided with a base line level of promoter-bound HP1γ (Figure 3B, upper panel, α-HP1γ, H3.3 shRNA), suggesting that H3.3 is necessary component for the promoter localization of HP1γ. Since HSPA1 and HSPA6 promoters are minimally occupied by H3.3 and HP1γ in the absence of heat shock, knockdown of H3.3 or HP1γ failed to show any apparent alteration and interdependency in their promoter occupancy under this normal condition. We also repeated the entire experiments in cells transiently transfected with Flag-tagged H3.3 and HP1γ; similar results were obtained from these experiments (Supplementary Figure S4).
Figure 3.
Interdependent localization of H3.3 and HP1γ at HSP70 promoters. (A) Mock-depleted (filled square) and H3.3-depleted (shaded square) cells were heat-treated as in Figure 1D, and ChIP assays of HSP70 promoter regions were performed using antibodies against H3.3 and HP1γ. (B) ChIP assays were essentially identical to Figure 3A, but in H3.3-depleted cells.
Active histone marks are enriched at promoter-positioned nucleosomes
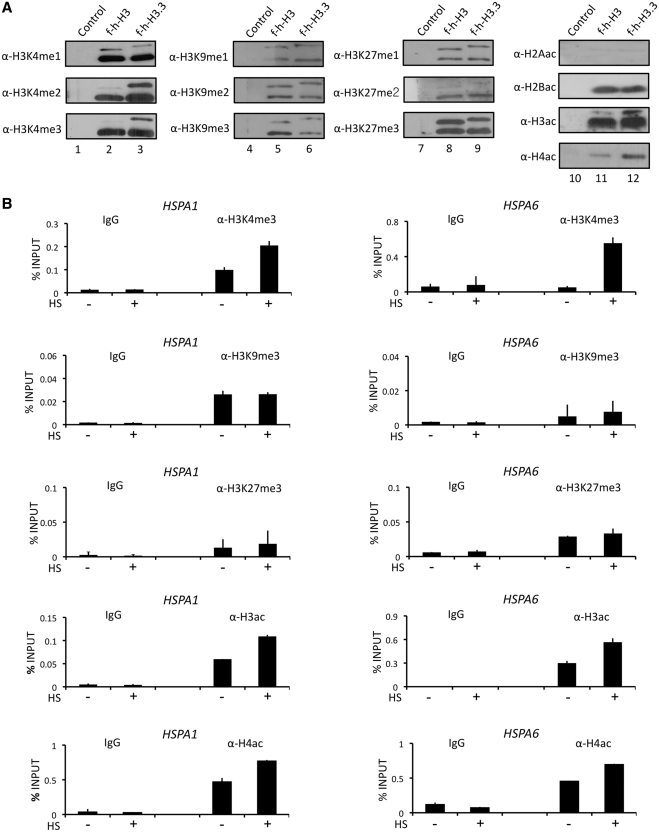
One possible explanation for the observed co-localization of HP1γ and H3.3 is that H3.3 nucleosomes are enriched with specific histone modifications to promote a stable binding of HP1γ. To investigate this possibility, we prepared nucleosomes containing f-h-H3 or f-h-H3.3 as described in Supplementary Figure S1A and determined the levels of histone methylation and acetylation by western blotting. Since H3 and H3.3 are identical across the N-terminal domain, all antibodies specific for H3 modifications should recognize H3.3 modifications with the same affinity. Given that only trace amount of endogenous H3.3 was detected in purified nucleosomes (Figure 1B, α-H3.3), our western blot analysis mainly evaluated the modifications of ectopic H3.3. In determining the degree of methylation, we found much higher levels of di- and tri-methylation of ectopic H3.3 lysine 4, compared to those of ectopic H3 lysine 4 (Figure 4A, lanes 2 and 3, α-H3K4me2/H3K4me3). This was not due to a general preference of H3.3 for HMTs present in the nucleus, as the levels of K9 and K27 methylations of ectopic H3.3 were similar to those of ectopic H3 (lanes 5, 6, 8 and 9). When we checked the acetylation status of H3 and H3.3 in purified nucleosomes, we found that ectopic H3.3 was acetylated to a higher extent than ectopic H3 (lanes 11 and 12, α-H3ac). Interestingly, a higher level of H4 acetylation (H4ac) was also detected in the H3.3 nucleosomes (lanes 11 and 12).
Figure 4.
Enrichment of active modifications in H3.3 nucleosomes. (A) Mononucleosomes containing ectopic H3 or H3.3 were prepared as in Figures 1A and S1A, and analyzed by western blotting using antibodies that recognize H3–K4 methylation (α-H3K4me1/me2/me3) H3-K9 methylation (α-H3K9me1/me2/me3), H3–K27 methylation (α-H3K27me1/me2/me3), H2A acetylation (α-H2Aac), H2B acetylation (α-H2Bac), H3 acetylation (α-H3ac) and H4 acetylation (α-H4ac). Lanes 1, 4, 7 and 10, mock-purified materials; lanes 2, 5, 8 and 11, H3 mononucleosomes; lanes 3, 6, 9 and 12, H3.3 mononucleosomes. (B) Cells were mock-treated (−) or heat-treated (+) for 30 min, and modification status of promoter nucleosomes was determined. ChIP assays were essentially as described in Figure 3D.
We next determined whether similar modifications are critical for the transcriptional activation of HSP70 genes by employing the same ChIP protocol as for examination of HP1γ and H3.3 localization. Of note, antibodies specific for H3 tail modifications are unable to distinguish H3 modifications from H3.3 modifications in our ChIP assays. In agreement with our purification results (Figure 4A), we found that the level of H3K4me3 distinctly increased at HSPA6 promoter in response to heat shock (Figure 4B, HSPA6, α-H3K4me3). Similar results were observed at transcriptionally active HSPA1 promoter, although the increase was not as high due to the fact that high signal was detected before heat shock treatment (HSPA1, α-H3K4me3). In marked contrast, two repressive modifications, H3K9me3 and H3K27me3, failed to show any changes after heat shock treatment (α-H3K9me3 and α-H3K4me27). In parallel ChIPs with antibodies against acetylated H3 (H3ac) and H4 (H4ac), it was observed that both promoters are preloaded with basal levels ofH3ac and H4ac under non-heat shock conditions (α-H3ac and α-H4ac, −). However, an apparent increase in H3ac and H4ac at the promoters was seen after heat-induced stimulation of HSP70 transcription (α-H3ac and α-H4ac, +). Taken together, these data imply that cooperative functions of H3.3 and HP1γ in HSP70 transcription are accompanied by the acquisition of active histone marks at the promoter nucleosomes.
HP1γ interacts preferentially with H3.3 nucleosomes carrying active histone marks
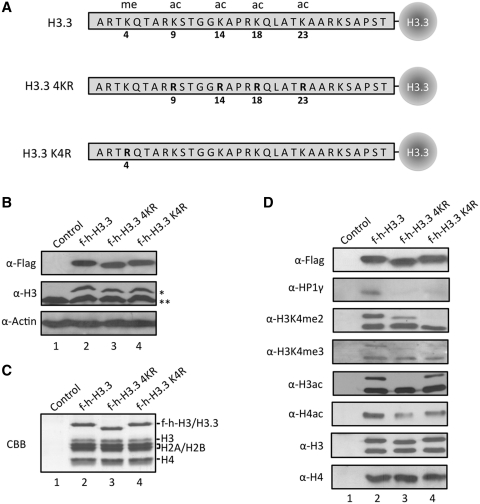
The observed enrichments of H3K4me2/3, H3ac and H4ac in H3.3 nucleosomes of HSP70 promoters encourage the possibility that these modifications may play a role in the interplay between H3.3 and HP1γ by serving as recognition sites for HP1γ. In an attempt to test this possibility, we mutated target sites for those active modifications in H3.3, expressed mutant versions of H3.3 histones in HeLa cells, and prepared mononucleosomes containing the ectopic H3.3 following the protocol described in Figure 1 (Figure 5A–C). When we analyzed the acetylation status of ectopic H3.3 in purified nucleosomes, we detected a high level of acetylation of wild type H3.3, but no acetylation of H3.3 mutated at four major acetylation sites (K9, K14, K18 and K23) (Figure 5D, lane 3). We also found that K4 methylation was highly selective for wild-type H3.3 relative to K4-mutated H3.3 present in nucleosomes (lane 4). Interestingly, the level of acetylation of endogenous H4 in nucleosomes carrying mutant H3.3 was found to be relatively lower than those in wild type H3.3 nucleosomes (compare lanes 3 and 4 with lane 2). Similarly, western blot analysis of K4 methylation of endogenous H3 reproducibly showed a weaker signal in mutant H3.3-containing nucleosomes than it did for wild type H3.3 nucleosomes. These results point to the importance of acetylation and K4 methylation of H3.3 for the active modifications of H3 and H4. In checking the association of HP1 protein with purified nucleosomes, we found that HP1γ bound avidly to wild type H3.3 nucleosomes, but bound minimally to nucleosomes containing acetylation sites-mutated H3.3. Under the same purification conditions, K4-mutated H3.3 nucleosomes also diminished HP1γ binding (α-HP1γ, lane 3 and 4). These results argue that the active modifications of H3 and H4 may depend on the prior modification of H3.3 and again support earlier indications (Figure 4) that active histone marks in H3.3 nucleosomes are critical for HP1γ-H3.3 cooperation. Thus, our results bring up several interesting questions: What is the temporal order of these modifications upon heat shock? Which factors are responsible for the observed modifications? What are the sequence and motif of HP1γ to allow the recognition of active histone marks present in HSP70 promoters?
Figure 5.
Preferential binding of HP1γ to H3.3 containing mononucleosomes with active modifications. (A) Schematic diagrams of wild type (H3.3), acetylation site-mutated (H3.3 4KR) and K4-mutated (H3.3 K4R) H3.3. Four acetylatable lysine residues (K9, K14, K18 and K23) and one methylatable lysine residue (K4) are mutated to arginine. (B) HeLa cells were transfected with control (lane 1), wild type (lane 2) and mutant H3.3 (lane 3 and 4) expression plasmids for 48 h, and the expression levels of ectopic H3.3 were confirmed by western blotting. (C) Mononucleosomes containing wild type and mutant H3.3 were immunoprecipitated from total mononucleosomes, essentially following the procedure employed in Figure 1. Histone compositions of the purified nucleosomes were analyzed by 15% SDS–PAGE followed by Coomassie staining. (D) H3.3 mononucleosomes were purified as in Supplementary Figure S1A, and analyzed by western blotting using antibodies that recognize H3 acetylation (α-H3ac), H4 acetylation (α-H4ac), H3K4 dimethylation (α-H3K4me2), H3K4 trimethylation (α-H3K4me3), H3 (α-H3), H4 (α-H4) and HP1γ (α-HP1γ). Asterisk indicates ectopic f-h-H3 or f-h-H3.3 proteins and double asterisks indicate endogenous H3 or H3.3 proteins.
Cooperative roles of H3.3 and HP1γ are deregulated in cancer cells
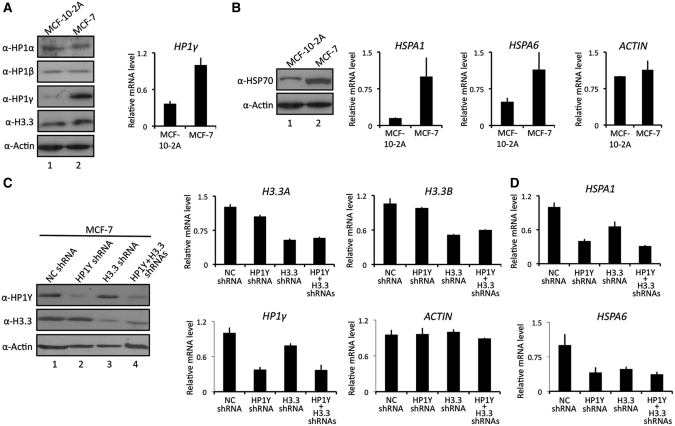
HSP70 has been positioned as a cancer-relevant survival protein and misregulation of HSP70 expression is directly linked to cell proliferation and dedifferentiation in human cancer (26–29). Intrigued by the demonstrated action of H3.3 and HP1γ in HSP70 transcription, we next assessed the expression levels of H3.3 and HP1γ in three different human cancer cell lines; breast cancer cell line (MCF-7), prostate cancer cell line (LNCaP) and bladder cancer cell line (LD611). From our western blot and qRT–PCR analyses of these cancer cells, a much higher level of HP1γ expression was evident in comparison with their normal counterparts (MCF-10-2 A, MLC and Urotsa) (Figure 6A and Supplementary Figure S5). Additionally, similar western blot analyses detected a slightly lower level of HP1α in prostate (LNCaP) and bladder (LD611) carcinoma cells (Supplementary Figure S5), but no difference in the levels of HP1α, HP1β and H3.3 in MCF-10-2 A and MCF-7 (Figure 6A). Furthermore, the expression levels of HSP70 as well as HSPA1 and HSPA6 genes were much higher in MCF-7 breast cancer cell lines compared to the normal cells (Figure 6B).
Figure 6.
Affects of H3.3/HP1γ knockdown on HSP70 transcription. (A) MCF-7 breast cancer cells and untransformed MCF-10-2 A breast epithelial cells were subjected to western blot analysis using antibodies specific for HP1α, HP1β, HP1γ and H3.3 (left panel). β-Actin served as a control for equal protein loading. qRT–PCR was performed as in Figure 2A (right panel). (B) HSP70 expression was assessed by western blot with anti-HSP70 antibody (left panel). Transcription of HSPA1 and HSPA6 genes in MCF-10-2 A and MCF-7 cells was quantified by qRT–PCR and corrected for expression of the control gene (GAPDH) (right panel). qRT–PCR was also performed to measure β-Actin mRNA expression (ACTIN). (C) MCF-7 cells were transfected with shRNAs targeting H3.3 and/or HP1γ, and individual and simultaneous depletions of H3.3 and HP1γ were confirmed by western blot analysis (left panel) and qRT–PCR (center and right panels). (D) Transcription levels of HSPA1 and HSPA6 genes of H3.3/HP1γ-depleted MCF-7 cells were analyzed as in Figure 6B.
In line with the cooperative action of H3.3 and HP1γ during HSP70 gene induction, their requirement on HSPA1 and HSPA6 transcription was examined in MCF7 breast cancer cells. Accordingly, we depleted H3.3 and HP1γ either individually or simultaneously, as confirmed by qRT–PCR and western blot analyses (Figure 6C). Congruent with our results from HeLa cells, the selective knockdown of HP1γ resulted in a distinct reduction in HSPA1 and HSPA6 mRNA levels (Figure 6D). Interestingly, although a similar level of H3.3 expression was found in normal MCF-10-2 A and cancer MCF7 cells (Figure 6A), H3.3 depletion also repressed heat shock-induced transcription of HSPA1 and HSPA6 genes (Figure 6D). Importantly, when both H3.3 and HP1γ were depleted, HSPA1 and HSPA6 transcription was repressed, but the level of repression was similar to that detected in single knockdown of H3.3 or HP1γ (Figure 6D), again supporting the idea of the dual requirement of H3.3 and HP1γ for HSP70 regulation.
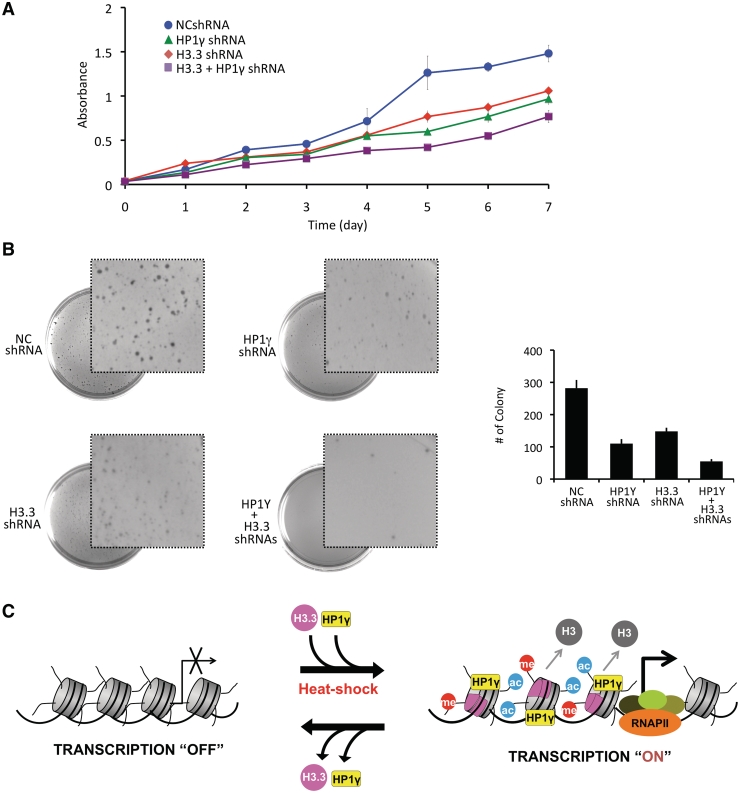
We next examined if knockdowns of H3.3, HP1γ, or both proteins in MCF7 cells have effects on cell proliferation by employing MTT and clonogenic assays. As summarized in Figure 7A, MCF7 cells depleted of HP1γ or H3.3 displayed a significantly reduced cell growth, showing ~60% lower than mock depleted cells. Interestingly, simultaneous depletion of HP1γ and H3.3 resulted in a somewhat greater reduction in colony formation compared to depletion of HP1γ or H3.3. One possible explanation for these results is that other cellular reactions beside HSP70 transcription might also have been affected after depletion of both genes. Similarly, MCF7 cells were depleted of HP1γ and H3.3, and the percentage of surviving colonies was assessed after two weeks of growth in soft agar. Cell survival decreased to about 40% after individual depletion of HP1γ and H3.3 as determined by the average percentage of colony numbers (Figure 7B). Our results also showed the colonies from MCF7 cells depleted of HP1γ or H3.3 was much smaller in size than those from the control cells. The rate of cell proliferation and colony formation were more obviously decreased after double knockdown of H3.3 and HP1γ (Figure 7B), again reflecting other functions of H3.3 and HP1γ that are not required for HSP70 transcription.
Figure 7.
H3.3/HP1γ knockdown-induced alterations in cancer cell growth. (A) MCF-7 cells were depleted of H3.3 and/or HP1γ as in Figure 6C and cell proliferation was measured by MTT assay. Results represent the mean ± SD of three experiments performed in triplicate. (B) H3.3/HP1γ-depleted MCF-7 cells were subjected to soft agar colony formation assay. The graph illustrates the total number of colonies present on the plate after three weeks of culture. Error bars on the graph indicate the standard deviation from triplicate experiments. (C) Models showing the combinatorial role of H3.3 and HP1γ in heat shock-induced HSP70 transcription. Upon heat shock, H3.3 is incorporated at HSP70 promoters and contributes to rapid changes in active histone modifications to stimulate the stable localization of HP1γ at the promoter nucleosomes. The co-enrichment of H3.3 and HP1γ converts HSP70 gene from a repressor state to an active state, leading to a great increase in transcription of HSP70 genes. Lack of H3.3 nucleosomes disrupts the recruitment of HP1γ, which drives the equilibrium further toward H3.3 dissociation, resulting in blockage of transcription initiation. See the ‘Discussion’ section for more details.
DISCUSSION
It has been known for years that H3.3 incorporation and HP1γ binding to nucleosomes are related to changes in transcriptional competency of chromatin (9,11,12,27), but it is only recently that biochemical studies have merged to elucidate the dynamic actions of these chromatin regulators (30,31). In this study, we combined in vitro and in vivo experiments to investigate the H3.3 exchange and HP1γ recruitment occurring at human HSP70 genes induced by heat shock stimulus. Our results indicate that H3.3 and HP1γ exert a critical role in heat shock response network through the regulated transcription of HSP70, which is upstream component of the heat shock signal transduction pathway. By using knockdown and ChIP techniques, we were able to confirm that H3.3 exchange is essential for transcription of two HSP70 genes, HSPA1 and HSPA6, in response to heat shock. The heat shock-induced enrichment of H3.3 appears to be mainly localized at the promoter, as only modest changes in their levels were detected within the coding region. This finding is in agreement with previous results demonstrating that H3.3-containing nucleosomes mark promoters of transcriptionally active genes, e.g. in the MyoD promoter (32). Somewhat surprisingly, our data show that HP1γ is brought into HSP70 promoters when HSP70 genes are activated by heat shock, in the same manner that heat shock stabilizes H3.3 localization at HSP70 genes. A detectable increase in H3.3 and HP1γ occupancy in coding regions was also observed after heat shock, but the change was relatively modest. It is therefore likely that alterations in promoter nucleosomes by H3.3 exchange and HP1γ binding are responsible for establishing active state of HSP70 genes.
Another intriguing finding of our study is that the recruitments of H3.3 and HP1γ to HSP70 promoters are mutually dependent, which underscores a complex relationship between H3.3 and HP1γ in heat shock response. The mechanistic basis for the H3.3 dependency in HP1γ recruitment is not fully elucidated in our study. However, given that H3.3 incorporation promotes activating modifications in the nucleosome, one obvious possibility is that these H3.3-induced modifications stimulate HP1γ binding to the promoter regions (Figure 7C). To support this model, our study confirmed that HP1γ overrides with HP1α and HP1β in interacting with H3.3 nucleosomes enriched by H3/H4 acetylation and H3K4 methylation. Such a selective interaction could enable HP1γ to interpret a set of combinatorial modifications and to cooperate with H3.3 at HSP70 promoters. The data presented here thus provide a glimpse into the positive interplay of active histone modifications in HP1γ recruitment and afford a conceptual framework on which to build similar models for other HP1γ-dependent transcription processes. However, this model does not provide the explanation for HP1γ-dependent function of H3.3 at the HSP70 promoter. Along with indications that most histone variants are in dynamic equilibrium between deposition and dissociation (2), one obvious possibility is that HP1γ binding to H3.3 nucleosome would shift the equilibrium toward deposition. Hence, although HP1γ is not required to facilitate the initial deposition of H3.3, we believe that HP1γ-based decorations of H3.3 nucleosomes might be critical for the intrinsic stability of H3.3 in the nucleosome. Due to technical limitations, this possibility has not been examined in the current study, but should be considered to shed light on the regulatory mechanism for HP1γ-H3.3 cooperation. To further support the functional significance of HP1γ and H3.3, our work demonstrates a much higher level of HP1γ expression in several human cancer cell lines in comparison to the corresponding normal cells. These data constitute a powerful argument that overexpression of HP1γ in cancer cells results in the misregulation of the cooperative action between H3.3 and HP1γ during HSP70 gene induction. In fact, single or double knockdown of H3.3 and HP1γ in these cancer cell lines leads to an apparent reduction in HSP70 transcription rate. That cell proliferation and colony formation were obviously decreased after the same knockdown also points to the interplay of H3.3 and HP1γ in these cancer cells. Therefore, further characterization of cooperative activities of H3.3 and HP1γ has potential implications in terms of cancer treatment.
It is currently unknown whether the ability of H3.3 and HP1γ to occupy other chromatin regions is attributed by the same mechanism with which they are co-localized in HSP70 genes. We note that a similar, but unidirectional, mechanism has been proposed for pericentrometic heterochromatin in which H3.3 deposition and K9 methylation contribute to the recruitment of HP1γ (30). These data hint at the possibility that H3.3 and HP1γ are also involved in condensing inactive chromatin harboring high levels of the repressive mark H3-K9 methylation. Therefore, a better understanding of cooperative actions of H3.3 and HP1γ will require further studies on how specific histone marks are initially established and how they contribute to H3.3 exchange and HP1γ localization at distinct chromatin regions. Also of note, Drosophila HP1c, which is human HP1γ homolog, has been linked to transcription elongation by acting as a bridge between FACT and elongating RNA Pol II (23). The observed effects of HP1c are intriguing, as HP1c might perform an analogous cooperative action with H3.3 in the post-initiation control of transcription. Hence, whether HP1γ is capable of recognizing H3.3 nucleosomes in both the initiation and elongation phases of transcription is an intriguing question that needs to be addressed in future studies. Overall, our findings could be incorporated in a model that highlights the combinatorial roles of H3.3 and HP1γ in governing signal integration, competence and specificity in heat-induced stimulation of HSP70 transcription (27) (Figure 7C). Such cooperative actions should provide additional levels of specificity and efficiency to the signaling pathways involved in transcriptional regulation of heat shock proteins. What emerges from this model is a cell's response to heat shock to be precisely regulated through highly restricted communication between HP1γ and H3.3. The challenge for us in the future is to determine whether other genes are also controlled by the HP1γ-H3.3 interplay and how this process is properly regulated in human cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes Health (Grant R01GM84209); ACS Research Scholar Grant DMC-1005001 (awarded to W.A.). Funding for open access charge: National Institutes of Health (R02GM84209).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G.D. Crump for H3.3 cDNA and A.S. Lee for HSP70 antibody. We also thank Chenyin Ou from M.R. Stallcup laboratory for providing technical advises on soft agar colony formation assay, Kwangho Lee for constructive discussion of this article, and members of the An laboratory for instructive discussions and insightful suggestions.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 3.Wells D, Hoffman D, Kedes L. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 1987;15:2871–2889. doi: 10.1093/nar/15.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albig W, Bramlage B, Gruber K, Klobeck HG, Kunz J, Doenecke D. The human replacement histone H3.3B gene (H3F3B) Genomics. 1995;30:264–272. doi: 10.1006/geno.1995.9878. [DOI] [PubMed] [Google Scholar]
- 5.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 6.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 9.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions' of active promoters and other regulatory regions. Nat. Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh PB, Miller JR, Pearce J, Kothary R, Burton RD, Paro R, James TC, Gaunt SJ. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue A, Hyle J, Lechner MS, Lahti JM. Perturbation of HP1 localization and chromatin binding ability causes defects in sister-chromatid cohesion. Mutat. Res. 2008;657:48–55. doi: 10.1016/j.mrgentox.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Dialynas GK, Terjung S, Brown JP, Aucott RL, Baron-Luhr B, Singh PB, Georgatos SD. Plasticity of HP1 proteins in mammalian cells. J. Cell Sci. 2007;120:3415–3424. doi: 10.1242/jcs.012914. [DOI] [PubMed] [Google Scholar]
- 18.Serrano A, Rodriguez-Corsino M, Losada A. Heterochromatin protein 1 (HP1) proteins do not drive pericentromeric cohesin enrichment in human cells. PLoS One. 2009;4:e5118. doi: 10.1371/journal.pone.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hediger F, Gasser SM. Heterochromatin protein 1: don't judge the book by its cover! Curr. Opin. Genet. Dev. 2006;16:143–150. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Hiragami K, Festenstein R. Heterochromatin protein 1: a pervasive controlling influence. Cell. Mol. Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 2010;24:2133–2145. doi: 10.1101/gad.1959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H, Heo K, Kim JH, Kim K, Choi J, An W. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J. Biol. Chem. 2009;284:19867–19877. doi: 10.1074/jbc.M109.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes JA, Dix DJ, Collins BW, Luft C, Allen JW. Expression of inducible Hsp70 enhances the proliferation of MCF-7 breast cancer cells and protects against the cytotoxic effects of hyperthermia. Cell Stress Chaperones. 2001;6:316–325. doi: 10.1379/1466-1268(2001)006<0316:eoihet>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galluzzi L, Giordanetto F, Kroemer G. Targeting HSP70 for cancer therapy. Mol. Cell. 2009;36:176–177. doi: 10.1016/j.molcel.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhang R, Liu ST, Chen W, Bonner M, Pehrson J, Yen TJ, Adams PD. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol. Cell. Biol. 2007;27:949–962. doi: 10.1128/MCB.01639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Font-Burgada J, Rossell D, Auer H, Azorin F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 2008;22:3007–3023. doi: 10.1101/gad.481408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.