Abstract
There is a close relationship between histone acetylation and ATP-dependent chromatin remodeling that is not fully understood. We show that acetylation of histone H3 tails affects SWI/SNF (mating type switching/ sucrose non fermenting) and RSC (remodels structure of chromatin) remodeling in several distinct ways. Acetylation of the histone H3 N-terminal tail facilitated recruitment and nucleosome mobilization by the ATP-dependent chromatin remodelers SWI/SNF and RSC. Tetra-acetylated H3, but not tetra-acetylated H4 tails, increased the affinity of RSC and SWI/SNF for nucleosomes while also changing the subunits of SWI/SNF that interact with the H3 tail. The enhanced recruitment of SWI/SNF due to H3 acetylation is bromodomain dependent, but is not further enhanced by additional bromodomains found in RSC. The combined effect of H3 acetylation and transcription activators is greater than either separately which suggests they act in parallel to recruit SWI/SNF. Besides enhancing recruitment, H3 acetylation increased nucleosome mobilization and H2A/H2B displacement by RSC and SWI/SNF in a bromodomain dependent manner and to a lesser extent enhanced ATP hydrolysis independent of bromodomains. H3 and H4 acetylation did not stimulate disassembly of adjacent nucleosomes in short arrays by SWI/SNF or RSC. These data illustrate how histone acetylation modulates RSC and SWI/SNF function, and provide a mechanistic insight into their collaborative efforts to remodel chromatin.
INTRODUCTION
SWI/SNF and the related RSC complex in Saccharomyces cerevisiae use DNA-dependent ATP hydrolysis and DNA translocation to remodel chromatin with a variety of different outcomes, such as translational repositioning of histone octamers along DNA (1), H2A/H2B dimer eviction, or complete nucleosome disassembly (2–4). Despite their similar biochemical properties, RSC and SWI/SNF exhibit significant differences inside the cell (5). RSC is ~10 times more abundant than SWI/SNF with two distinct forms containing either Rsc1 or Rsc2 (6) and most of the subunits in RSC are essential for viability (7,8). While RSC controls transcription by RNA polymerases (RNA pol) I, II and III (9), SWI/SNF regulates only RNA pol II transcription. Both these remodelers control non-overlapping sets of mRNA genes (10–12). SWI/SNF has no apparent role in cell-cycle control, but RSC is involved in kinetochore function and sister chromatid cohesion (13,14). RSC contributes to both types of double-strand break (DSB) repair, but SWI/SNF plays a role in only homologous recombination (HR) based DSB repair and not in non-homologous end joining (NHEJ) (15,16). Even in HR based DSB repair SWI/SNF and RSC are associated with distinct steps (17,18). RSC plays an early role in DSB sensing and is also required later during donor DNA and recipient synapsis. SWI/SNF has an early role in DSB repair by HR and is required at the strand invasion step.
All the SWI/SNF homologs contain a ~70 amino acid domain called the bromodomain that recognizes acetylated lysines. SWI/SNF contains a single bromodomain in the Swi2/Snf2 catalytic subunit and 8 out of the 15 bromodomains found in yeast are in RSC. The presence of bromodomains in RSC and SWI/SNF suggests a functional connection between acetylation and these complexes. Consistent with this idea, histone acetylation precedes SWI/SNF recruitment and/or remodeling in the activation of the human β-interferon gene (19), PHO8 (20), and genes regulated by the RAR/RXR heterodimers (21). At the human α1 antitrypsin promoter, hBrm, homolog of Snf2, and two histone acetyl transferase (HAT) complexes, CBP and P/CAF, are recruited simultaneously after preinitiation complex assembly to stimulate transcription (22). There is also genetic data supporting interactions between SWI/SNF and histone acetylation. Combining mutations in the subunits of SWI/SNF and the HAT complex SAGA resulted in strong synthetic phenotypes (23,24). Similar synthetic phenotypes were obtained when combining mutations in RSC subunits Rsc1 or Rsc2 with SAGA subunit Gcn5, demonstrating an important relationship between these complexes (6).
Although SWI/SNF and HATs functionally interact, how this interaction facilitates chromatin remodeling remains poorly understood. There have been some attempts to study how histone acetylation directly affects ATP-dependent chromatin remodeling. However the interpretation of these studies, in general, has been complicated by the lack of substrate specificity of HATs, variable extents of acetylation, and with the HAT complex potentially interacting with the ATP-dependent chromatin remodeler (25–28). In this study we examined if H3 and H4 histone tail acetylation only impacts remodeler targeting or does it also affect nucleosome movement and/or disassembly independent of the remodeler's binding ability. We have included three different SWI/SNF complexes containing six, one or no bromodomains to determine if RSC has any strategic advantage over SWI/SNF when it comes to recruitment or remodeling of acetylated chromatin. We have used native chemical ligation for the synthesis of histones H3 and H4 with uniformly acetylated N-terminal tails to avoid the drawbacks of enzymatic modification (29). Using these chemically acetylated nucleosomes our studies have revealed that H3 and not H4 tail acetylation modulates the activity of SWI/SNF and RSC at multiple steps in the remodeling process. Furthermore, H3 tail acetylation has a greater impact on nucleosome movement and H2A/H2B displacement by RSC than SWI/SNF. The higher responsiveness of RSC to histone acetylation than SWI/SNF can provide additional regulatory mechanisms for RSC which might ultimately account for their different functional roles inside the cell.
MATERIALS AND METHODS
Purification of RSC, SWI/SNF, ΔBr SWI/SNF, Piccolo and Ada2 subcomplexes, Gal4-VP16 and histones
RSC was purified from C-terminally TAP-tagged Rsc2 expressing S. cerevisiae strain YLR357W (Open Biosystems) by tandem affinity purification (TAP) (30). SWI/SNF was purified by FLAG affinity purification from the S. cerevisiae strain YBB001 expressing C-terminally FLAG tagged Snf6 and HA-V5-6× His tagged Swi3 as described (31). ΔBr SWI/SNF was analogously purified from the S. cerevisiae strain YBB002 containing Snf6 tagged at the C-terminus with FLAG epitope. The C-terminus of Swi2/Snf2 was truncated from residue 1547–1703 and tagged with V5 and 6× His. The Piccolo NuA4 and Gcn5/Ada2/Ada3 subcomplexes used for enzymatic acetylation of nucleosomes were prepared as described previously (32). Gal4-VP16 was overexpressed in Escherichia coli Xa-90 and purified as described (31). Wild type Xenopus laevis core histone proteins and mutant histones with cysteine 110 replaced with alanine in H3 and serine 53 replaced with cysteine in H2B were overexpressed, purified and refolded into octamers as described (33).
Native peptide ligation and acetylated octamer refolding
Tetra-acetylated H3 (K9, 14, 18, 23Ac) and H4 (K5, 8, 12, 16Ac) histones were prepared using a native peptide ligation method as described (34,35). Histone H3 or H4 N-terminal peptides (1–24 or −22 amino acid) containing covalently modified acetylated lysine residues at desired locations and a C-terminal thioester moiety were generated by solid phase peptide synthesis. The rest of the histone H3 or H4 protein encompassing the globular core and the C-terminal tail was prepared with an N-terminal cysteine residue by recombinant protein expression in E. coli. Native ligation chemistry was used to fuse the two protein fragments followed by purification of the full-length modified histones by ion-exchange and reverse phase chromatography and refolding them with other histones to generate the tetra-acetylated H3 and H4 octamers and purified by gel filtration chromatography.
DNA synthesis and nucleosome reconstitution
Mononucleosomes (29N59 or 69N59) were assembled with 235 or 276 bp DNA containing the 601 positioning sequence (36) flanked by 29 and 59 bp or 69 and 59 bp of extranucleosomal DNA respectively. Dinucleosomes [40N(601)-31-N(603)6] were assembled with a 371 bp DNA containing 601 and 603 positioning sequences separated by a 31-bp intervening sequence with 40 bp of linker DNA at the 601 end and 6 bp of linker DNA at the 603 end (36). All three carrier DNAs contained a single Gal4 site in the longer linker or in the 59 bp linker 23 bp away from the nucleosomal edge. The carrier DNA for mono and dinucleosomes were prepared by PCR from p159-1Gal4-27 and p159-1Gal4-27-601-603 plasmid DNA templates respectively (31). The 5′-end of the top strand of the 371-bp carrier DNA was biotinylated. Nucleosomes were assembled at room temperature by stepwise salt dialysis from 2 M to 100 mM NaCl (37) and analyzed by 4% (35.36:1 acrylamide to bisacrylamide) native PAGE.
Nucleosome binding assays with competitor DNA and histone crosslinking
Non-acetylated (H3 or H4) or acetylated (H3 Ac or H4 Ac) 29N59 mononucleosomes at 10 nM final concentration were titrated with remodeler to determine conditions for full binding in the absence of competitor DNA. The intrinsic nucleosome binding activity of TAP-RSC was relatively poor and ~5–8 times more RSC was used for complete binding of H3 or H4 nucleosomes as compared to FLAG purified SWI/SNF and ΔBr SWI/SNF (Supplementary Figures S2 and S3; Supplementary Table S1). Binding reactions of 6.3 µl were incubated at 30°C for 30 min with saturating amounts of RSC, SWI/SNF or ΔBr SWI/SNF and 1.8–50 ng of sheared salmon sperm DNA competitor. The differences observed in the amount of remodeler bound to acetylated versus non-acetylated nucleosomes with increasing amounts of competitor DNA directly correlates to the ratio of the KD of the remodeler for acetylated versus non-acetylated nucleosomes. This ratio is best observed at the highest amount of competitor DNA in which the amount of bound nucleosomes can still be accurately measured with both types of nucleosomes. Reaction conditions were 20 mM HEPES-NaOH (pH 7.8), 3 mM MgCl2, 6% (v/v) glycerol, 70 mM NaCl and 0.1 μg/μl BSA. Gal4-VP16 dependent SWI/SNF binding to nucleosomes was achieved by including sheared salmon sperm DNA in the binding reaction. Gal4-VP16 was pre-bound to nucleosomes at 30°C for 30 min followed by the addition of SWI/SNF or ΔBr SWI/SNF and incubating for another 30 min. The binding reactions were analyzed on 4% (79:1 acrylamide to bis-acrylamide) polyacrylamide native gel.
PEAS-I125 modified Biotin-69N59 nucleosomes (16 nM) were immobilized on Dynabeads and incubated with 64 nM Piccolo NuA4 or Gcn5/Ada2/Ada3 and 42 µM acetyl CoA (SIGMA) at 30°C for 30 min. Reaction conditions were 50 mM Tris–HCl (pH 8), 50 mM KCl, 0.1 mM EDTA (pH 8), 10 mM sodium butyrate, 5% glycerol, 1 mM DTT and 1 mM PMSF. The HAT complexes were washed off and the acetylated or non-acetylated nucleosomes were bound to SWI/SNF, UV irradiated to crosslink followed by radio-label transfer by disulfide reduction and finally analyzed on 4–12% Bis–Tris SDS–PAGE (31).
Rate of ATP hydrolysis
The rate of ATP hydrolysis was determined either under conditions of saturating enzymes or nucleosomes. ATPase assays with saturating amounts of nucleosomes was carried out with 3 nM RSC or SWI/SNF or ΔBr SWI/SNF, and 15 nM H3 or H3 Ac dinucleosomes. Background signal was determined from samples with no remodeler added and any intrinsic ATPase activity was observed with remodeler in the absence of nucleosomes. Pre-binding was performed at 30°C for 15 min followed by lowering to 18°C. The [γ-32P] ATP was added and stopped with SDS (1.5%) and EDTA (50 mM). Inorganic phosphate and ATP were separated by thin layer chromatography on polyethyleneimine cellulose plate (J. T. Baker, Germany) and developed with 0.5 M LiCl and 0.5 M formic acid. The mean and standard deviation were calculated from three independent experiments.
Nucleosome mobilization assays
H3 or H3 Ac mono- or dinucleosomes (10 nM) were bound for 15 min at 30°C with RSC, SWI/SNF or ΔBr SWI/SNF with 6 ng of competitor DNA in a volume of 6.3 µl. After addition of ATP (1 mM) reactions were incubated for 20 min at 30°C and stopped with 10 μg of sonicated salmon sperm DNA and γ-thio ATP (1.2 mM). Samples were analyzed on either 5% high resolution (60:1 acrylamide to bis-acrylamide) or 4% (35.36:1 acrylamide to bis-acrylamide) polyacrylamide native gels in 0.2× TBE with pump recirculation or 0.5× TBE. Remodeling was measured by quantifying the disappearance of the original nucleosomal band or the formation of remodeled species I and II during the course of the reaction. The rate of remodeling was estimated with saturating amounts of RSC, SWI/SNF or ΔBr SWI/SNF as determined by gel shift assay (Supplementary Figure S4 and data not shown). After binding the enzyme with nucleosomes at 30°C for 15 min the reaction temperature was lowered to 18 or 25°C and remodeling was initiated by adding ATP (4 or 55 μM). At regular time intervals samples were removed and stopped by adding 6.3 μl of sample to 2 μl of a 1:1 mixture of 10 μg/μl sonicated salmon sperm DNA and 10 mM γ-thio ATP. Samples were analyzed on native polyacrylamide gels as mentioned in the figure legends. Disappearance of the original nucleosomal band or appearance of remodeled species I and II over time was plotted using GraphPad (PRISM). The initial rate of remodeling was estimated by fitting the data to a hyperbolic curve (non-linear fitting) using the Solver add-in function in Microsoft Excel over 1000 iterative cycles followed by differentiation of the curve and solving for time equals zero (38). The mean and standard deviation were estimated from two independent repeats.
RESULTS
Acetylated H3, but not H4, tails increase RSC and SWI/SNF affinity for nucleosomes
SWI/SNF and RSC share many identical, or highly homologous, protein subunits that are evolutionarily conserved across higher eukaryotes (39). A common domain within several subunits is the bromodomain, depicted in Supplementary Figure S1B. The catalytic subunit (Sth1 or Swi2/Snf2) of both complexes has a single bromodomain near the C-terminus. Unlike the Swi2/Snf2 bromodomain, the one in Sth1 when removed renders cells temperature sensitive and arrests the cell cycle resulting in cells with highly elongated buds (40–42). As shown in Supplementary Figure S1B, the auxiliary subunits Rsc1, Rsc2 and Rsc4 of RSC each have two bromodomains (BD1 and BD2) (6,43). While, only one of the two bromodomains (BD2) in Rsc1 and Rsc2 are critical for RSC function, the tandem bromodomains in Rsc4 are essential for cell survival (6,43). It is not clear how RSC remodeling activity might vary from that of SWI/SNF due to the former having a higher number of bromodomains, but we suspected these differences might only be observed when using acetylated nucleosomes.
Histone N-terminal tails are acetylated at lysines 9, 14, 18 and 23 in H3 by Gcn5/SAGA (44,45) and at lysines 5, 8, 12 and 16 in H4 by Esa1/NuA4 (46). The contribution of these acetylated histone tails on the recruitment and remodeling activities of RSC and SWI/SNF was analyzed using tetra-acetylated H3 and H4 nucleosomes.
The purity of the tetra-acetylated histone octamers and absence of any unligated globular core of the histone proteins H3 and H4 are evident by analysis on a 17% SDS-PAGE (Supplementary Figure S1A, lanes 1 and 2). Although the chemically acetylated recombinant histones are of the same length as the non-acetylated histones and have the expected mass (data not shown), their mobility on SDS–PAGE is altered due to their acetylation (Supplementary Figure S1A). These octamers were subsequently assembled with 601 DNA to generate nucleosomes acetylated at H3 or H4 histone tails.
We used competitor DNA in our binding assays to more effectively detect differences in affinity based on our previous experience with Gal4-VP16 recruitment of SWI/SNF (31). A fixed amount of RSC or SWI/SNF and acetylated nucleosomes with increasing amounts of sonicated salmon sperm DNA was used in the binding assays (Figure 1). Besides RSC and SWI/SNF, a version of SWI/SNF was used which has no bromodomain (ΔBr), maintains complex integrity (Supplementary Figure S1C) and has comparable nucleosome mobilization activity with unmodified nucleosomes as wild type SWI/SNF (data not shown). Both RSC and SWI/SNF exhibited significant preferential binding to nucleosomes with acetylated H3 tail (H3 Ac) over non-acetylated (H3) (Figure 1A–D) in presence of competitor DNA (1.8–50 ng). Unlike RSC and SWI/SNF, the ΔBr SWI/SNF mutant showed only small differences in its affinity for H3 Ac and H3 nucleosomes (Figure 1E and F). In contrast to the 7- and 9-fold enhancement in RSC and SWI/SNF binding, ΔBr SWI/SNF binding affinity was enhanced only 2-fold with 6 ng of competitor DNA (compare Figure 1D and F) suggesting that H3 tail acetylation mostly exerts its effect on RSC and SWI/SNF binding through their bromodomains. RSC and SWI/SNF binding was increased equally by H3 acetylation which suggests that the additional bromodomains of RSC do not help in this regard. The difference in RSC or SWI/SNF affinity for H3 Ac versus H3 nucleosomes measured in this way is significantly greater than that observed in the absence of competitor DNA (Supplementary Figure S2 and Supplementary Table S1). Thus, we have found it important to conduct these binding experiments in the presence of competitor DNA to more accurately observe the preference for acetylated nucleosomal substrates which is otherwise not seen due to the high affinity of RSC and SWI/SNF for nucleosomes.
Figure 1.
H3 tail acetylation enhances the affinity of RSC and SWI/SNF for nucleosomes. Gel shift assays were performed with 80 nM RSC (A), 16 nM SWI/SNF (C) or 13 nM ΔBr SWI/SNF (E) and 10 nM mononucleosomes with (H3 Ac) or without (H3) tetra-acetylated H3 tails. Samples in (A and C) contained increasing amounts of competitor DNA (0.25, 0.45, 0.9, 1.8, 3.6 and 7.2 ng/µl in lanes 3–8 and 11–16, respectively) or no competitor DNA (lanes 2 and 10). Lanes 1 and 9 are nucleosomes only. In (E) the competitor DNA used was 0.45, 0.9, 1.8, 3.6 ng/µl, respectively, in lanes 3–6 and 9–12. Quantification of the gel shift assays in (A, C and E) are shown in (B, D and F), respectively. Numbers above the bars indicate the ratio of H3 Ac versus H3 nucleosomes binding for that particular concentration of competitor DNA. The binding ratios are included for only those gel shift lanes in which we have high confidence in the quantification of each species.
A side-by-side comparison of RSC and SWI/SNF binding under identical conditions was carried out to delineate the effect of acetylated H4 tails. There was no preference for acetylated H4 over non-acetylated nucleosomes for RSC, SWI/SNF or ΔBr SWI/SNF (Supplementary Figure S3) and even increasing the amount of competitor DNA did not make a difference in this case (Figure 2A–D). Acetylation of H3 tails and not of H4 tails enhanced the binding of SWI/SNF and RSC to nucleosomes.
Figure 2.
H4 tail acetylation does not increase the affinity of RSC or SWI/SNF for nucleosomes. (A–F) The sections in this figure are the same as shown in Figure 1, except that 10 nM of tetra-acetylated H4 mononucleosomes (H4 Ac) are used instead of tetra-acetylated H3 mononucleosomes. Un-acetylated nucleosomes are represented as H4.
Changes in H3 tail interactions with SWI/SNF are mediated by acetylation
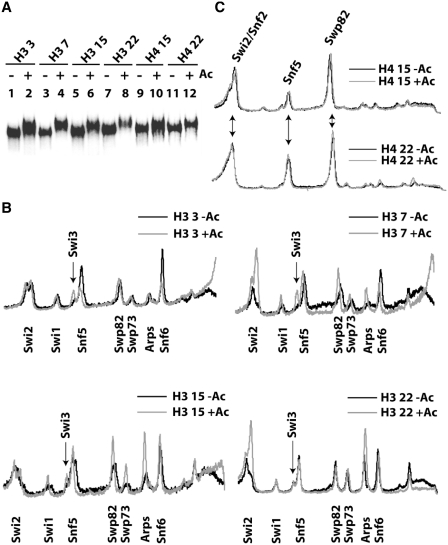
To understand how H3 tail acetylation enhances the nucleosome binding affinity of SWI/SNF, the effects of acetylation on the interactions of SWI/SNF with the H3 tail was examined in a nucleosomal context by site-directed histone crosslinking and label transfer (31). Cysteine was incorporated into different positions in the histone H3 and H4 tails and coupled to a photoreactive reporter called I125-PEAS for probing the interactions of SWI/SNF. Specific amino acid residues that are not very highly conserved were selected for changing to cysteine. Four positions, residues 3, 7, 15 and 22, in the H3 tail were examined. Incorporation of cysteine at these sites did not alter the Gcn5 recognition motif GKXXP (47) or RKT/SXGX(Kac)XPR/K (48) in the H3 N-terminus and hence was expected not to interfere with acetylation. The histone tails of the photoreactive nucleosomes were enzymatically acetylated using yGcn5/Ada2/Ada3 [SAGA sub-complex, primarily H3, (49)] and Piccolo [NuA4 sub-complex, primarily H4 and to a lesser extent H2A, (50,51)] to map the effects of histone acetylation on the interactions between SWI/SNF and H3 and H4 histone tails (Figure 3A). The Gcn5 or Esa1 catalytic subunit alone acetylates only free histones, but the sub-complexes and holo complexes can efficiently acetylate both histones and nucleosomes (32). The yGcn5/Ada3/Ada2 complex preferentially acetylates lysines 18, 14 and 9 in H3 and to a lesser extent lysine 23 (49). The PEAS modified nucleosomes upon acetylation by the HAT complexes exhibited reduced mobility in comparison to a non-acetylated control (Figure 3A).
Figure 3.
Acetylation changes the interactions of SWI/SNF with the H3 histone tail. (A) Mononucleosomes coupled to photoreactive I125-PEAS at residues 3, 7, 15 or 22 in the H3 tail were acetylated by yGcn5/Ada2/Ada3 SAGA subcomplex (lanes 1–8). Other nucleosomes had I125-PEAS coupled to residues 15 or 22 in the H4 tail and were acetylated by Piccolo NuA4 (lanes 9–14). Nucleosomes were analyzed before (odd lanes) and after acetylation (even lanes) on a 4% native PAGE. (B) SWI/SNF subunits labeled by crosslinking at positions 3, 7, 15 and 22 of H3 were separated on 4–12% Bis–Tris SDS–PAGE and visualized by phosphorimaging. The quantification of these profiles are overlaid using Image Quant software (Molecular Dynamics) for non-acetylated and yGcn5/Ada2.Ada3 acetylated nucleosomes as shown along with the positions of the Swi2/Snf2, Snf5, Swp82, SWP73, Arp7/9 and Snf6 subunits of SWI/SNF. (C) The same approach as in (B) was used to determine which SWI/SNF subunits were crosslinked at positions 15 and 22 of H4 in non-acetylated and Piccolo NuA4 acetylated nucleosomes.
Acetylation of H3 by the yGcn5/Ada3/Ada2 SAGA sub-complex changed the pattern and efficiency of SWI/SNF subunits crosslinked to the histone H3 tail (Figure 3B). H3 tail acetylation enhanced crosslinking of the Swi2/Snf2 subunit at H3 histone residues 7 and 22, but not at residues 3 or 15. While crosslinking of Arps was also enhanced at residues 7 and 22, the striking difference is that they were greatly enhanced at residue 15 upon acetylation. It appears that binding of the core SWI/SNF complex consisting of Swi2/Snf2, Arp7 and 9 are generally enhanced across the region acetylated by yGcn5/Ada3/Ada2 (2). Binding of the Swp82 subunit was also increased at residues 7 and 15 of H3 upon acetylation. Although Swp82 does not appear to be essential for SWI/SNF remodeling activity, it has been shown previously to be one of only a few subunits of SWI/SNF that comes into close proximity to the nucleosome surface (31). The lower amount of Swi3 crosslinking observed at positions 3 and 7 was dependent on acetylation. Snf5 and Snf6 showed reduced crosslinking to the H3 tail at residue 3 upon acetylation. Together, these crosslinking data show that acetylation by yGcn5/Ada3/Ada2 enhances the binding of several key core subunits of the SWI/SNF complex to the H3 histone tail.
Parallel experiments were conducted with I125-PEAS conjugated in the H4 histone tail at two positions, residues 15 and 22. Swi2/Snf2, Snf5 and Swp82 were found to be associated with the H4 tail at amino acid residues 15 and 22 (Figure 3C). No significant change in the SWI/SNF subunit crosslinked to the H4 tail at residues 15 and 22 was observed after acetylation with Piccolo (Figure 3C) consistent with no changes in the binding affinity of SWI/SNF seen in Figure 2. Together these crosslinking data suggest that acetylation of H3 tails alters the interactions of SWI/SNF with nucleosomes in a way specific to the H3 tail and could be the basis for the increased affinity of SWI/SNF for H3 Ac nucleosomes observed in Figure 1.
Acetylated H3 tails promote SWI/SNF recruitment in synergy with transcription activators
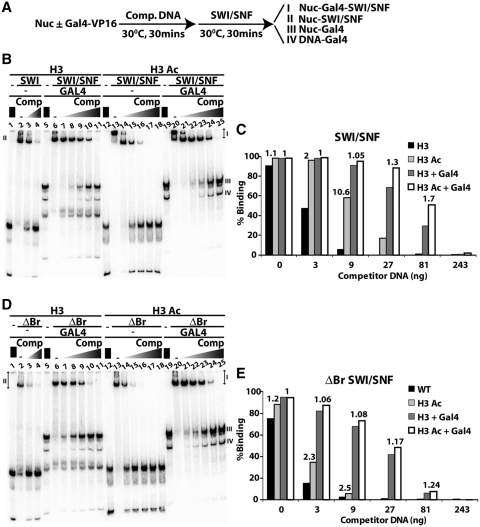
SWI/SNF may be targeted to specific genomic sites by sequence specific DNA binding transcription activators or H3 acetylation (52,53). It might be that both can function together, thereby further enhancing the affinity of SWI/SNF for that particular target site. SWI/SNF binding to H3 and H3 Ac nucleosomes was studied under conditions in which SWI/SNF was recruited by the Gal4-VP16 transcription activator to investigate how these two modes of SWI/SNF recruitment compare to each other and whether they act in parallel (Figure 4). Both types of nucleosomes had one Gal4-VP16 transcription activator binding site in the extranucleosomal DNA for recruitment of SWI/SNF and ΔBr SWI/SNF. H3 and H3 Ac nucleosomes were pre-bound with or without Gal4-VP16 and different amounts of competitor DNA before addition of SWI/SNF (Figure 4A). Consistent with the results in Figure 1, SWI/SNF bound ~10 times more efficiently to H3 Ac nucleosomes as compared to H3 nucleosomes with 9 ng of competitor DNA in a bromodomain dependent manner (Figure 4B and C; Figure 4D and E compare lanes 4–15).
Figure 4.
SWI/SNF recruitment by the transcription activator Gal4-VP16 and H3 acetylation. (A) The order of addition of nucleosomes, competitor DNA and SWI/SNF is shown for the recruitment assays in (B and D). Nucleosomes contain one Gal4 site in extranucleosomal DNA, 27 bp from the entry site. Gel shift assays are shown with 15 nM SWI/SNF (B) or 10 nm ΔBr SWI/SNF (D) that had 10 nM nucleosomes and 25 nM Gal4-VP16 where indicated. H3 acetylated nucleosomes (H3 Ac) are in lanes 12–25 and nucleosomes without acetylation (H3) are in lanes 1–11. Increasing amounts of competitor DNA were added ranging from 0.45 to 35 ng/µl for lanes 7–11 and 21–25 or from 0.45 to 1.3 ng/µl for lanes 3–4 and 14–18. Species I, II, III and IV refer to nucleosome-Gal4-SWI/SNF, nucleosome-SWI/SNF, nucleosome-Gal4 and DNA-Gal4, respectively. Quantification of (B and D) are shown in (C and E), respectively, for SWI/SNF and ΔBr SWI/SNF binding to H3 Ac versus H3 nucleosomes with and without Gal4-VP16. The numbers above the bars is the ratio of H3 Ac versus H3 nucleosomes binding with or without Gal4-VP16 added. The binding ratios are included for those particular gel shift lanes in which each species could be accurately quantified.
Similar to H3 acetylated nucleosomes, the addition of competitor DNA was necessary to distinguish direct binding versus SWI/SNF being recruited to H3 nucleosomes by Gal4-VP16 (Figure 4B lanes 2–11 and Figure 4C). From these experiments it seems that the addition of competitor DNA is imperative in order to see the selective recruitment of SWI/SNF by either a transcription activator or H3 acetylation. The bromodomain of SWI/SNF was not required for recruitment by Gal4-VP16 (compare lanes 4 and 8 or lanes 15 and 22 in Figure 4D and E), but was more effective than H3 acetylation under higher competitor DNA concentrations (Figure 4B compare lanes 7–11 with lanes 14–18 and Figure 4C). Four times more SWI/SNF was bound to nucleosomes with Gal4-VP16 than with H3 acetylation in the presence of 27 ng of competitor DNA (Figure 4B compare lanes 16 and 23, and Figure 4C). The combination of H3 acetylation and Gal4-VP16 had an additional 1.3 to 1.7-fold increase in SWI/SNF binding relative to that of Gal4-VP16 alone with 27–81 ng of competitor DNA (Figure 4C). These data indicate that histone H3 acetylation and the transcription activator recruit SWI/SNF in parallel pathways that together have an overall combined effect greater than either individually. This additional stimulation in SWI/SNF binding due to H3 tail acetylation and Gal4-VP16 was not observed when the bromodomain of SWI/SNF was missing (Figure 4D). These results are consistent with the additive effect being due to the Swi2/Snf2 bromodomain binding to the acetylated histone H3 tail.
H3 tail acetylation increases the rates of nucleosome movement and dimer displacement by RSC and SWI/SNF without much alteration in the rates of ATP hydrolysis or nucleosome disassembly
The effect of H3 tail acetylation on nucleosome stimulated ATP hydrolysis by RSC and SWI/SNF was determined by measuring the initial rate of ATP hydrolysis. RSC, SWI/SNF and ΔBr SWI/SNF, all hydrolyzed ATP at similar initial rates with H3 dinucleosomes, but H3 acetylation caused a ~1.5–1.8 times increase for RSC and SWI/SNF (Supplementary Figure S5A–C and Supplementary Table S2). This increase in the rate of ATP hydrolysis was not much more than that observed for ΔBr SWI/SNF of 1.4 times with H3 tail acetylation. These assays used saturating amounts of enzyme as determined on the basis of the binding titration in Supplementary Figure S4, but the results were not significantly different when nucleosomes were at saturating amounts rather than enzymes (Supplementary Figure S6 A–C and Supplementary Table S3). Acetylation of H3 tails only modestly stimulates the ATPase activity of RSC and SWI/SNF in a bromodomain independent manner.
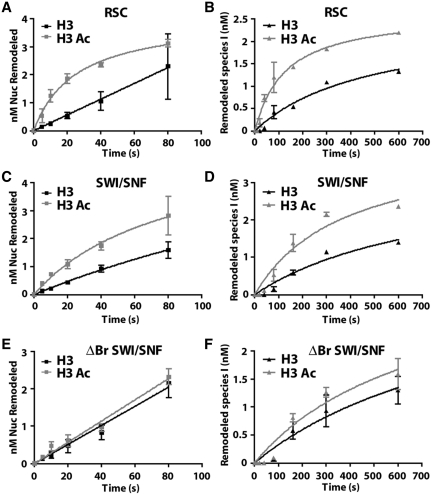
Two types of nucleosome remodeling assays were performed to find if H3 acetylation affected nucleosome mobilization, H2A/H2B dimer displacement, or nucleosome disassembly by RSC and SWI/SNF in a manner distinct from that of the ATPase activity. The first one had competitor DNA and nucleosomes in the reaction with limiting amounts of enzyme; whereas, the other had only nucleosomes present and enzyme was in excess. Under these two conditions the effects of H3 acetylation were examined either under recruitment limiting conditions or independent of recruitment, respectively. Besides acetylated and non-acetylated nucleosomes, mono- and dinucleosomes were used to assess both nucleosome mobilization and disassembly activities of these remodelers (4).
For conditions when competitor DNA was present, acetylation of H3 tails stimulated RSC and SWI/SNF movement of mononucleosomes by eight times or more at lower concentrations of remodeler (Supplementary Figure S7). In addition to nucleosome movement, displacement of H2A/H2B dimer and complete disassembly of nucleosomes by RSC and SWI/SNF was stimulated by H3 acetylation as shown with dinucleosomes in the presence of competitor DNA (Supplementary Figures S8 and S9). SWI/SNF lacking the bromodomain was not stimulated as much by H3 acetylation and indicates that this effect is bromodomain specific (Supplementary Figures S7–S9). Since these experiments were performed under competitive conditions where binding is limiting, these effects could be due to acetylation increasing the affinity of the remodeler.
In order to find if histone acetylation influences the remodeling activities of RSC and SWI/SNF in a manner other than recruitment to nucleosomes, remodeling was carried out with saturating amounts of SWI/SNF and RSC, and the initial rates of nucleosome mobilization, H2A/H2B displacement and nucleosome disassembly measured. These conditions are the same as those used for the first set of ATPase assays shown in Supplementary Figure S5 and as such can be used to track the initial movement in dinucleosomes along with the loss of one H2A/H2B dimer. Previously, the two remodeled dinucleosome species observed by native gel electrophoresis, referred to as species I and II, were shown to have lost one H2A/H2B dimer (species I) or completely lost one nucleosome (species II) (4). The histone dimer is lost first followed by a much slower step in which the entire histone octamer is lost (4). The same type of gel shift assay with H3 acetylated nucleosomes showed that acetylation enhanced the movement of dinucleosomes by five times for RSC and 2.7 times for SWI/SNF (Figure 5, Supplementary Figure S10 and Table 1). No significant change in the rate of nucleosome movement was observed with ΔBr SWI/SNF and suggests that for SWI/SNF the bromodomain is required for acetylation enhanced nucleosome movement. Acetylation of H3 increased the rate of nucleosome movement more for RSC than for SWI/SNF which correlates with RSC having multiple bromodomains and SWI/SNF having only one. H2A/H2B dimer displacement by RSC and SWI/SNF occurred in a slower step, but was stimulated similarly as nucleosome movement by H3 tail acetylation (Figure 5, Supplementary Figure S10 and Table 1). The increase in H2A/H2B dimer displacement when nucleosomes are acetylated could be due to the faster rate of nucleosome movement observed with the dinucleosomes.
Figure 5.
H3 tail acetylation stimulates nucleosome mobilization in a bromodomain dependent manner. The rate of nucleosome movement by 80 nm RSC (A and B), 20 nM SWI/SNF (C and D), and 20 nm ΔBr SWI/SNF (E and F) was determined under the same conditions as in Supplementary Figure S5 by gel shift assay using 10 nM 601–603 dinucleosomes. (A, C and E) The concentration of mobilized dinucleosomes moved versus time was plotted and fitted non-linearly to the Michaelis–Menten equation using Graph Pad Prism. (B, D and F) The rate at which the first H2A/H2B dimer is displaced from dinucleosomes was estimated by tracking the appearance of the remodeled species I in two independent experiments. This rate was determined in the same way as the rate of nucleosome movement.
Table 1.
The rates of nucleosome movement and H2A/H2B displacement of SWI/SNF and RSC with non-acetylated and acetylated H3 nucleosomes
| Initial Rate (nM NCP/s) |
RH3 Ac:RH3 | Initial Rate (nM species I/s) |
RH3 Ac:RH3 | RateNuc Slid : RateDimer Loss |
||||
|---|---|---|---|---|---|---|---|---|
| H3 | H3 Ac | H3 | H3 Ac | H3 | H3 Ac | |||
| RSC | 2.8 × 10−2 | 1.4 × 10−1 | 5 | 5.1 × 10−3 | 2 × 10−2 | 4 | 5.5 | 7 |
| SWI/SNF | 2.5 × 10−2 | 6.7 × 10−2 | 2.7 | 4.9 × 10−3 | 1.3 × 10−2 | 2.7 | 5 | 5 |
| ΔBr SWI/SNF | 2.6 × 10−2 | 2.8 × 10−2 | 1.1 | 4.5 × 10−3 | 5.8 × 10−3 | 1.3 | 5.7 | 4.9 |
The initial rates of nucleosome movement and H2A/H2B displacement are shown for RSC, SWI/SNF and ΔBr SWI/SNF with excess remodeler to dinucleosomes. The rates of nucleosome movement and dimer displacement were determined by native gel electrophoresis as shown in Figure 5 and Supplementary Figure S10. The ratio of the rates with H3 tail acetylated versus non-acetylated dinucleosomes is shown for nucleosome movement and dimer displacement.
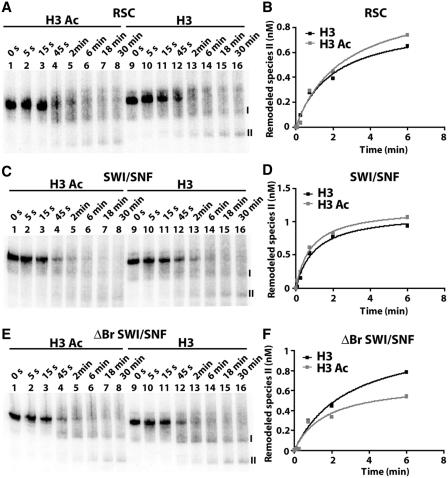
The remodeled species II formation, however, was much less under the conditions of 4 µM ATP and 18°C temperature (Supplementary Figure S10). Hence, the initial rate of formation of species II could not be accurately measured from these experiments. The rate of remodeling was increased by changing ATP to 55 µM and the reaction temperature to 25°C to better observe the formation of remodeled species II and therefore nucleosome disassembly. H3 acetylation did not significantly increase the rate of nucleosome disassembly by RSC or SWI/SNF and there was no noticeable difference between RSC and SWI/SNF as had been observed before with the rate of nucleosome movement and H2A/H2B displacement (Figures 5, 6 and Table 2). Similar assays were also done with tetra-acetylated H4 containing nucleosomes and H4 acetylation was not found to increase the rate of nucleosome disassembly by ΔBr SWI/SNF, SWI/SNF or RSC (Supplementary Figure S11A–F and Supplementary Table S4). Nucleosome disassembly by RSC and SWI/SNF is a much slower step than nucleosome movement or H2A/H2B displacement and cannot be stimulated by H3 or H4 tail acetylation as seen in these experiments.
Figure 6.
Nucleosome disassembly by RSC and SWI/SNF is not enhanced by H3 tail acetylation. The rate at which one nucleosome is removed from the 601–603 dinucleosome was determined using conditions similar to that in Figure 5, except that the ATP concentration was increased to 55 µM and incubated at 25°C. The appearance of the second remodeled species (II) was followed by gel shift on a 4% native polyacrylamide gel for RSC (A), SWI/SNF (C), and ΔBr SWI/SNF (E). The concentration of remodeled species II formed by RSC (B), SWI/SNF (D), and ΔBr SWI/SNF (F) was plotted against time and fitted non-linearly to the Michaelis–Menten equation when using non-acetylated (H3) or H3 acetylated (H3Ac) dinucleosomes.
Table 2.
The rate of RSC and SWI/SNF disassembly of nucleosomes with H3 tail acetylated or non-acetylated dinucleosomes
| Initial Rate (nM species II/s) |
RH3 Ac:RH3 | ||
|---|---|---|---|
| H3 | H3 Ac | ||
| RSC | 5.5 × 10−3 | 6.8 × 10−3 | 1.2 |
| SWI/SNF | 1.3 × 10−2 | 2.0 × 10−2 | 1.5 |
| ΔBr SWI/SNF | 5.1 × 10−3 | 3.7 × 10−3 | 0.7 |
The initial rate of nucleosome disassembly was estimated in terms of appearance of the remodeled species II by gel shift assay for RSC, SWI/SNF and ΔBr SWI/SNF as shown in Figure 6. The ratio of the rates for nucleosome disassembly is shown for H3 tail acetylated versus non-acetylated dinucleosomes.
DISCUSSION
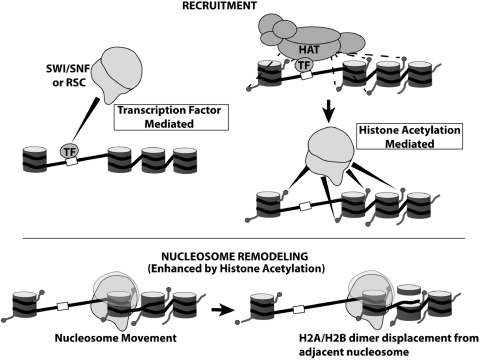
Histone acetylation regulates chromatin remodeling by SWI/SNF and RSC in two distinct ways. The first is that SWI/SNF and RSC are recruited to genes that are poised for transcription due to acetylation of the neighboring chromatin. We have found using competitive binding conditions that SWI/SNF and RSC is selectively recruited to H3 tail acetylated nucleosomes in a manner similar to recruitment by transcription activators such as Gal4-VP16 (Figure 4). The KD of RSC and SWI/SNF for nucleosomes is about seven to nine times greater when H3 tails are acetylated (Figure 1B and D, with 6 ng of competitor DNA). In comparison the ratio of the KD of SWI/SNF and ΔBr SWI/SNF for nucleosomes with and without Gal4-VP16 is 17 and 31, respectively with non-acetylated nucleosomes (Figure 4C and E, with 9 ng of competitor DNA). The affinity of SWI/SNF for Gal4-VP16 is therefore approximately two times greater than for acetylated nucleosomes (Figure 4C and E, and 9 ng competitor DNA). There is evidence for two inter-related pathways for recruiting SWI/SNF and RSC to particular genomic DNA sites that involve DNA sequence-specific transcription factors (Figure 7). Particular genomic regions can be marked for RSC or SWI/SNF recruitment by hyperacetylation of these regions mediated through DNA sequence-specific transcription factors and histone acetyltransferases. The other pathway is the sequence-specific DNA binding factor directly recruiting SWI/SNF or RSC to the correct target site. Even in the case of the strong transcription activator Gal4-VP16 there is a mild synergistic effect when SWI/SNF is recruited through the combination of histone acetylation and Gal4-VP16 (Figure 4B and C). It is likely that this synergistic effect may become even more significant when the transcription activator is not as strong as Gal4-VP16. The recruitment of RSC by H3 acetylation seems to be through just one of six bromodomains since the affinity of RSC for acetylated nucleosomes was equivalent to that of SWI/SNF with its one bromodomain (Figure 1).
Figure 7.
H3 tail acetylation modulates SWI/SNF and RSC function by distinct mechanisms. RSC and SWI/SNF recruitment occurs by two independent pathways mediated by some DNA sequence specific transcription factors (TF). Transcription activators recruit via direct interactions with SWI/SNF. Alternatively, transcription factors mediate recruitment of histone acetyl transferases (HATs) that catalyzes site-specific histone H3 tail acetylation and leads to SWI/SNF and RSC recruitment via bromodomain-acetyl lysine interaction. Recognition of acetyl marks on H3 tails via their bromodomains also modulates the nucleosome remodeling function of these complexes apart from recruitment. Specifically, H3 acetylation facilitates nucleosome movement in nucleosomal arrays and enhances H2A/H2B displacement from neighboring nucleosomes.
It is interesting that H3 tail acetylation caused an increase in the affinity of ΔBr SWI/SNF, although to a much lesser extent than when the bromodomain is present (Figure 1). These data suggest that there may be another part of SWI/SNF other than the bromodomain that might bind less well to acetylated H3 and yet contribute to its affinity for acetylated nucleosomes. On the other hand it could be that acetylation of H3 tails modestly increases the affinity of SWI/SNF for nucleosomes by disrupting the tail interactions with DNA. The reduced binding of tails to DNA could make particular domains of the nucleosome more open and accessible for binding by SWI/SNF thereby indirectly increasing its affinity. The overall higher affinity of SWI/SNF for H3 acetylated nucleosomes observed here is likely connected to the previous observation that H3 acetylation stabilizes the interactions of SWI/SNF with nucleosomes (25). This earlier study found that acetylation of H4 tails by NuA4 stabilizes SWI/SNF interactions, but in our more defined system we find no evidence for H4 acetylation increasing the affinity of SWI/SNF for nucleosomes (Figure 2). Our site-directed crosslinking studies also showed no change in the interactions of SWI/SNF with the H4 tail upon acetylation consistent with no increase in affinity (Figure 3). Earlier reports had shown that H3 tail acetylation only slightly increased the affinity of RSC for nucleosomes, the same as shown in our study when no competitor DNA is used [Supplementary Figure S2, Supplementary Table S1 and (54)]. We have found in this study that it is imperative to examine the binding affinities under competitive conditions due to the high affinity of SWI/SNF and RSC for nucleosomes and the inability to dilute nucleosomes further without them being destablized.
Histone acetylation also enhances the intrinsic nucleosome mobilizing activities of SWI/SNF and RSC independent of recruitment and their ATPase activities. RSC and SWI/SNF remodeling was performed under full binding conditions so that recruitment was not a factor in the efficiency of nucleosome movement. Under these conditions acetylation of H3 and not H4 histone tails increases the rate of nucleosome movement much more than the ATP hydrolysis rate. The increase in the rate of nucleosome movement with acetylated H3 tails is not due to histone acetylation making nucleosomes more prone to move as shown by SWI/SNF without its bromodomain (ΔBr SWI/SNF) moving acetylated and non-acetylated nucleosomes at the same rate (Figure 5E and Table 1). The increase in the rate of nucleosome movement associated with H3 acetylation required the complex to contain a bromodomain and was further accentuated when there were more bromodomains (Figure 5A and C). The 2-fold increase in the rate of nucleosome movement by RSC over SWI/SNF suggests that more than one of the six bromodomains in RSC are likely to be involved in recognizing acetylated H3 and promoting nucleosome movement, but not all otherwise the increase might be larger. One of the two bromodomains in Rsc4 has been shown to interact with acetylated lysine 14 in H3 and is a likely candidate along with the bromodomain of Sth1 to be involved in recognizing acetylated H3 tails and stimulating nucleosome movement (55). It seems that RSC may be more responsive to histone acetylation than SWI/SNF and likely reflects a key difference between the two classes of mammalian SWI/SNF that are distinguished by the presence or absence of the polybromo subunit BAF180 (PBAF and BAF).
While histone acetylation increases the efficiency of nucleosome movement it had no effect on the nucleosome disassembling activities of SWI/SNF and RSC (Figure 6 and Supplementary Figure S11). H3 acetylation only stimulates H2A/H2B dimer displacement which may be a direct consequence of enhanced nucleosome movement (Figure 7). Consistent with our current model of stepwise nucleosome disassembly (4) the rate of nucleosome disassembly with both RSC and SWI/SNF is much slower than H2A/H2B dimer displacement (Figures 5, 6, Supplementary Figure S11 and Table 1). H3 as well as H4 tail acetylation does not affect the rate limiting step of disrupting the H3/H4 tetramer interactions with DNA (Figure 6, Supplementary Figure S11, Supplementary Tables S2 and S4). The tetra-acetylated H4 experiments were prompted by the prior observation that H4 acetylation increased the rate of octamer transfer by RSC (54). Octamer transfer however appears to respond differently as it is stimulated by H4 tail tetra-acetylation (54) but nucleosome disassembly is not (Supplementary Figure S11 and Supplementary Table S4).
Given that histone acetylation and bromodomains have direct roles not only in recruiting remodeling complexes to nucleosomes, but also in helping to mobilize nucleosomes the next step will be to find out how acetylation and bromodomains facilitate nucleosome movement. We feel it is important to point out that H3 acetylation did not merely increase the binding of the catalytic subunit to the histone H3 tail, but rather we saw, by site-directed crosslinking, that several of the core subunits of SWI/SNF interacted more with the H3 tail after acetylation (Figure 3). These results suggest that acetylated H3 tail may cause some kind of switch in SWI/SNF and RSC conformation that enhances their ability to mobilize nucleosomes. When acetylated H3 tail peptide is added to RSC, the complex transitions from a mix of open and closed conformations to primarily a closed one as observed by electron microscopy (56). This switch in RSC conformation occurred only with the acetylated and not the non-acetylated H3 tail and may represent a more active state.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM 48413 to B.B., GM 79663 to M.S.K. and GM060489 to S.T.). Funding for open access charge: National Institutes of Health (grant GM 48413).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Tom Owen-Hughes for help with the data analysis.
REFERENCES
- 1.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat. Struct. Mol. Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 3.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee N, Sen P, Bartholomew B. In: Handbook of cell signalling. Bradshaw RA, Dennis EA, editors. Vol. 3. Oxford: Academic Press; 2009. pp. 2345–2356. [Google Scholar]
- 6.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 7.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 8.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 9.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- 11.Monahan BJ, Villen J, Marguerat S, Bahler J, Gygi SP, Winston F. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat. Struct. Mol. Biol. 2008;15:873–880. doi: 10.1038/nsmb.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Hsu JM, Laurent BC. The RSC nucleosome-remodeling complex is required for Cohesin's association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Laurent BC. A Role for the RSC chromatin remodeler in regulating cohesion of sister chromatid arms. Cell Cycle. 2004;3:973–975. [PubMed] [Google Scholar]
- 15.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong LY, Recht J, Laurent BC. Chromatin remodeling and repair of DNA double-strand breaks. J Mol. Histol. 2006;37:261–269. doi: 10.1007/s10735-006-9047-4. [DOI] [PubMed] [Google Scholar]
- 17.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell's response to DNA damage. Curr. Biol. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 20.Reinke H, Gregory PD, Horz W. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell. 2001;7:529–538. doi: 10.1016/s1097-2765(01)00200-3. [DOI] [PubMed] [Google Scholar]
- 21.Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol. Cell. 2000;6:1049–1058. doi: 10.1016/s1097-2765(00)00103-9. [DOI] [PubMed] [Google Scholar]
- 22.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 23.Pollard KJ, Peterson CL. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 26.Hassan AH, Awad S, Prochasson P. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J. Biol. Chem. 2006;281:18126–18134. doi: 10.1074/jbc.M602851200. [DOI] [PubMed] [Google Scholar]
- 27.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 28.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Shogren-Knaak MA. Cross-talk between histone H3 tails produces cooperative nucleosome acetylation. Proc. Natl Acad. Sci. USA. 2008;105:18243–18248. doi: 10.1073/pnas.0804530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittmeyer J, Saha A, Cairns B. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 2004;377:322–343. doi: 10.1016/S0076-6879(03)77020-7. [DOI] [PubMed] [Google Scholar]
- 31.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrios A, Selleck W, Hnatkovich B, Kramer R, Sermwittayawong D, Tan S. Expression and purification of recombinant yeast Ada2/Ada3/Gcn5 and Piccolo NuA4 histone acetyltransferase complexes. Methods. 2007;41:271–277. doi: 10.1016/j.ymeth.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 34.Shogren-Knaak MA, Fry CJ, Peterson CL. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J. Biol. Chem. 2003;278:15744–15748. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 35.Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- 36.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 38.Chang EY, Ferreira H, Somers J, Nusinow DA, Owen-Hughes T, Narlikar GJ. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry. 2008;47:13726–13732. doi: 10.1021/bi8016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 42.Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl Acad. Sci. USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 45.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 46.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 48.Ferreira R, Eberharter A, Bonaldi T, Chioda M, Imhof A, Becker PB. Site-specific acetylation of ISWI by GCN5. BMC Mol. Biol. 2007;8:73. doi: 10.1186/1471-2199-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 50.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selleck W, Fortin I, Sermwittayawong D, Cote J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 53.Cote J, Peterson CL, Workman JL. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl Acad. Sci. USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol. Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skiniotis G, Moazed D, Walz T. Acetylated histone tail peptides induce structural rearrangements in the RSC chromatin remodeling complex. J. Biol. Chem. 2007;282:20804–20808. doi: 10.1074/jbc.C700081200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.