Abstract
Friedreich’s ataxia (FRDA) is caused by biallelic expansion of GAA repeats leading to the transcriptional silencing of the frataxin (FXN) gene. The exact molecular mechanism of inhibition of FXN expression is unclear. Herein, we analyze the effects of hyperexpanded GAA repeats on transcription status and chromatin modifications proximal and distal to the GAA repeats. Using chromatin immunoprecipitation and quantitative PCR we detected significant changes in the chromatin landscape in FRDA cells relative to control cells downstream of the promoter, especially in the vicinity of the GAA tract. In this region, hyperexpanded GAAs induced a particular constellation of histone modifications typically associated with heterochromatin-like structures. Similar epigenetic changes were observed in GFP reporter construct containing 560 GAA repeats. Furthermore, we observed similar levels of FXN pre-mRNA at a region upstream of hyperexpanded GAA repeats in FRDA and control cells, indicating similar efficiency of transcription initiation. We also demonstrated that histone modifications associated with hyperexpanded GAA repeats are independent of initiation and progression of transcription. Our data provide strong evidence that FXN deficiency in FRDA patients results from a block of transition from initiation to a productive elongation of FXN transcription due to heterochromatin-like structures formed in the proximity of the hyperexpanded GAAs.
INTRODUCTION
Friedreich’s ataxia (FRDA) is an inherited degenerative disease that is characterized by progressive ataxia, including uncoordinated gait and limb movements, weakened muscle strength, and diminished senses of position and vibration. FRDA is caused by an insufficient level of Frataxin (FXN) (1,2). FXN is an evolutionarily conserved mitochondrial protein that is involved in iron homeostasis in cells (3). Reduced levels of the FXN gene expression in FRDA patients are caused by a hyperexpanded tract of repeated GAA triplets in intron 1 of the FXN gene (4,5). In FRDA patients, the GAA tract frequently consists of >1000 triplets, whereas unaffected individuals have 66 or fewer repeats at the FXN gene (4). Pathological expansion of the GAA repeats is associated with localized chromatin changes and transcriptional silencing at the gene; however, the underlying molecular mechanisms of hyperexpanded GAA-induced transcriptional defects are not yet clear.
The hyperexpanded GAA repeats at the FXN gene have been reported to adopt a heterochromatin-like structure that is characterized by high levels of di- and tri-methylated lysine 9 of histone H3 (H3K9me2/3) and underacetylated H3 and H4 (6–9). Inhibitors of histone deacetylases increase levels of FXN expression in FRDA primary lymphocytes and in a murine model (8,10). Additionally, altering histone modifications, especially levels of acetylation, can partially reactivate expression of the FXN gene. Thus, the results of these studies suggest that changes in chromatin structure upstream of the hyperexpanded GAA repeats induce FXN silencing. However, it is not clear whether the heterochromatin-like structure induced by the hyperexpanded tract of GAA repeats impacts initiation and/or elongation of FXN transcription. Some studies indicate that the heterochromatin-like conformation induced by the hyperexpanded GAA repeats extend to the promoter region and affect initiation of FXN transcription (11,12). Repressive marks such as H3K27me3 and H3K9me3, as well as heterochromatin protein (HP1) are enriched at the transcription start site (TSS) of the FXN gene in FRDA fibroblast lines, which results in the failure of FXN transcription initiation. These heterochromatin marks may also affect expression of antisense transcripts at the region upstream of the FXN TSS, thereby interfering with initiation of FXN sense transcripts in FRDA patients (11). Other studies suggest that FXN deficiency results not only from defective initiation, but also transcript elongation (12). Levels of both H3K4me3 at the TSS (represents active transcription initiation) of the FXN gene and H3K36me3 (an indicator of transcription elongation) are decreased at the FXN gene in FRDA cell lines.
Epigenetic changes induced by the hyperexpanded GAA repeats are one of the primary therapeutic targets in FRDA. A number of studies have demonstrated that specific histone deacetylase inhibitors (HDACi) are capable of enhancing histone acetylation and FXN expression in FRDA cells (8–10). On the contrary, a repressive mark, H3K9me3, observed in the proximity of the long GAA repeats is sustained during HDACi treatment (8,10). Additionally, inhibition of H3K9 methylation with BIX-01294 has no effect on FXN expression in FRDA cells (17). These results suggest that simultaneous targeting of two or more epigenetic silencing pathways may be required to restore full activity of the FXN gene. Therefore, a detailed definition of the landscape of histone modifications associated with hyperexpanded GAA repeats is necessary in order to further understand the molecular mechanisms underlying chromatin changes in FRDA cells and their relationship to FXN deficiency.
While previous studies have examined selected histone modifications at the FXN gene, each study was limited in either the scope of the modifications examined or the number of cell lines analyzed. Here we report a comprehensive analysis of histone modification patterns in multiple lymphoid cell lines derived from FRDA patients and unaffected controls. We found that the hyperexpanded GAA repeats affect chromatin structure in the proximity of the GAA repeats, but do not extend to the promoter. In contrast, the distribution of RNA pol II and histone methylation marks associated with transcription elongation were underrepresented in the presence of the hyperexpanded GAA repeats at the FXN gene in FRDA cell lines. These results strongly support the hypothesis that the transcription machinery correctly initiates synthesis of the FXN mRNA. Interestingly, RNA pol II is enriched and appears to be paused at the FXN promoter-proximal region in unaffected cell lines, but not FRDA cells. We also found that altered enrichment of H3K4me3 and H3K79me2 at the FXN gene reflects defective transcription at a post-initiation step. Taken together, our study provides strong evidence that FXN deficiency in FRDA patients results from a block in the transition between initiation and elongation of FXN transcription. This deficiency is possibly attributed to heterochromatin-like structures formed in the proximity of the hyperexpanded GAAs that act as an obstacle to the elongation of FXN pre-mRNA.
MATERIALS AND METHODS
Cell lines
Lymphoid cell lines were purchased from NIGMS Human Genetic Cell Repository at The Coriell Institute for Medical Research, Camden, NJ, USA. We selected three cell lines derived from FRDA patients: GM15850 harboring two expanded alleles of approximately 650 and 1030 GAA repeats; GM16798 harboring two expanded alleles of approximately 750 and 1000 GAA repeats and GM16209 harboring approximately 800 GAA repeats on both alleles of the FXN gene. As controls we also used three lymphoid cell lines (GM15851, GM03928 and GM05152) derived from healthy, unaffected individuals containing short GAA repeat tracts within a normal range. Lymphoid cell lines were grown in RPMI 1640 medium supplemented with 15% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2. The GFP_GAA systems [GFP_(GAA·TTC)0 and GFP_(GAA·TTC)560] were generated by integration of a tract of GAA repeats derived from the GM16210 affected cells as described previously (9). A DNA fragment harboring approximately 560 GAA repeats was PCR amplified from genomic DNA isolated from the GM16210 cell line (NIGMS Human Genetic Cell Repository at The Coriell Institute for Medical Research, Camden, NJ, USA) as described in (9). PCR product was cleaved by Bsu36I and BssHII endonucleases and the GAA repeat-containing fragment was cloned into intron 1 (1.2 kb from the exon1/intron1 junction) of the GFP gene of the pGFP_Int plasmid (13,14). Plasmids containing 0 and 560 GAA repeats were integrated by site-specific recombination into the genome of the HEK293Flp-InT-Rex cell line (Invitrogen). The use of identical sites of integration for both GFP_(GAA·TTC)0 and GFP_(GAA·TTC)560 constructs allows direct comparison between cell lines and eliminates any potential bias resulting from random integration events in different chromosomal contexts. Integrants were selected using hygromycin (200 μg/ml); individual hygromycin-resistant colonies were isolated and analyzed for repeat size and GFP expression level. Correct splicing of the GFP mRNA was determined as described earlier (4). All constructs were sequenced prior to as well as after establishing the stable cell lines. The GFP_GAA lines were selectively maintained in Dulbecco’s Modified Eagle’s Medium with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2 supplemented with hygromycin (200 μg/ml) and blasticidin (5 μg/ml). To induce transcription of the GFP minigene in the GFP_GAA system, cells were treated with 0.1 μg/ml tetracycline for 24 h.
Polymerase chain reaction
The size of the hyperexpanded tract of GAA repeats in lymphoid cell lines were determined by conventional PCR using primers 2500F and 629 under previously described conditions (4). The insertion of the GAA repeats at an intron of the GFP gene in the GFP_GAA system was detected by the conventional PCR using the primers listed in Table 1. The PCR products were analyzed on 0.8% agarose gels.
Table 1.
Primers used for analyses in the GFP_GAA system
| Primer (5′–3′) |
||
|---|---|---|
| Forward | Reverse | |
| Repeat | CTTCCCTTTACACAACGTTTGGGTT | GTACTGTTTGGATTCAGTGAGGGACT |
| Ex1/Ex2 | GCGACGTAAACGGCCACAAGTT | ATGCCCTTCAGCTCGATGCGGT |
| Ex1/Int | GACGACGGCAACTACAAGACC | CTAGGACAAAGGTGCCTAAGACC |
| Up | AATAGCCTCCTGACCACAGATCCTT | CCATGTGACATCTAGCCCCGCA |
| Down | CCCACAGGCCTGAAACACT | TTCATGCGTGCTAGGGTAAA |
| Int/Ex2 | CCCTAGCACGCATGAACC | ATGCCCTTCAGCTCGATGCGGTa |
aThis primer is the same as the GFP Ex1/Ex2 reverse primer.
To perform quantitative reverse transcriptase–PCR (qRT–PCR), total RNAs from lymphoid cell lines and the GFP_GAA system were isolated using the RNeasy Mini Kit (Qiagen). DNase I (TURBO DNA-free; Ambion) was added to remove genomic DNA contamination from isolated RNAs. The qRT–PCR was conducted using the Power SYBR Green RNA-to-CT 1-Step Kit (7500 Fast Real Time–PCR System, Applied Biosystems). As a control, reactions were also performed without reverse transcriptase to confirm a removal of genomic DNA. To calculate levels of FXN expression, the delta Ct value was generated by subtraction of the Ct value of GAPDH from the Ct value of either FXN mRNA or four different region transcripts of each cell line. Levels of FXN mRNA from each cell line were normalized to the delta Ct of GM15851. For levels of FXN pre-mRNA expression, each delta Ct value of four different regions at the FXN gene was normalized to the delta Ct value of the region upstream of the GAA repeat from GM15851. All primers used in this study are listed in Tables 1 and 2.
Table 2.
Primers used for analyses in lymphoid cell lines
| Primer (5′–3′) |
||
|---|---|---|
| Forward | Reverse | |
| Ex3/Ex4 | CCTTGCAGACAAGCCATACA | GGTCCACTGGATGGAGAAGA |
| In1Ex2 | AGCACTCGGTTACAGGCACT | GCCCAAAGTTCCAGATTTCC |
| −242 | CGCATTTTATAAACAAGGCACA | GTATGTGGGGCCAGGAGAC |
| Pro (or −133) | CCCCACATACCCAACTGCTG | GCCCGCCGCTTCTAAAATTC |
| +48 | AAGCAGGCTCTCCATTTTTG | CCGCAGGCACTCTTCTGT |
| Up (or +1231) | GAAACCCAAAGAATGGCTGTG | TTCCCTCCTCGTGAAACACC |
| +1394 | GGTACGCCGCATGTATTAGG | GCAACCAATCCCAAAGTTTC |
| Down | CTGGAAAAATAGGCAAGTGTGG | CAGGGGTGGAAGCCCAATAC |
| In2Ex3 | GGTAATCATGTTTTGGGTTTTGTGC | AGTCCTCAAACGTGTATGGCTTGTC |
Number on the name of primers denotes the first base of forward primer on the target region with respect to the TSS, +1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the EZ ChIP instructions (Upstate Biotechnology Inc.). Briefly, proteins and DNA we re cross-linked with 1% formaldehyde for 10 min (15 min for RNA pol II ChIP) at room temperature. The cross-linking reaction was quenched with 125 mM glycine for 5 min (10 min in the case of RNA pol II ChIP). Whole lysates were prepared using a cell lysis buffer (50 mM Tris–HCl at pH8.0, 10 mM EDTA and 1% SDS) supplemented with protease inhibitor (Mini EDTA-free protease inhibitor cocktail, Roche) and sonicated to obtain 100–300 bp DNA fragments using a Bioruptor Sonicator (Diagenode). The fragmented chromatin was diluted 10 times with dilution buffer (16.7 mM Tris–HCl at pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100 and 0.01% SDS). The chromatin fragments were pre-cleared for 1 h using Protein A agarose (Millipore) pre-blocked with BSA and salmon sperm DNA. The equivalent of 5 × 106 (1 × 107 cells in the case of RNA pol II ChIP) cells was immunoprecipitated overnight with 5 μg of antibodies recognizing histones and histone modifications (15 μg of RNA pol II antibody in the case of RNA pol II ChIP). The immunoprecipitates were immobilized on the pre-blocked protein A agarose beads for 1 h. Subsequently, the beads were washed with buffers containing low salt, high salt, and LiCl. Chromatin was eluted from the beads with elution buffer (100 mM NaHCO3 and 1% SDS) at room temperature for 15 min twice and subsequently subjected to the reverse cross-linking reaction using 5 M NaCl at 65°C for at least 5 h. The DNAs from chromatin complexes were isolated with Tris–EDTA buffer (80 mM Tris–HCl at pH 6.5 and 20 mM EDTA) supplemented with Proteinase K and RNase A at 37°C for 30 min followed by 42°C for 1 h. DNA fragments were purified using phenol/chloroform extraction before quantitative real-time PCR (qPCR). The qPCR was conducted using the Power SYBR Green-CT Kit (7500 Fast Real Time-PCR System, Applied Biosystems). The qPCR was carried out as follows: 10 min at 94°C, 50 cycles of 30 s at 94°C followed by 60 s at 60°C. As changes in nucleosome occupancy occur upon gene activation and gene silencing (15), it is noteworthy that the relative abundance of histone modifications (referred to as the ‘ratio’) determined by qPCR were analyzed by normalizing the quantity of the immunoprecipitated sample to the quantity of total histone H3 (total H4 in the case of H4K20me3) after normalization with inputs.
Antibodies
The antibodies used in this study were: anti-rabbit IgG as a negative control (Cell Signaling), anti-total H3 (Cell Signaling or Active Motif), anti-total H4 (Abcam), anti-H3K9/14ac (Upstate), anti-H3K4me2 (Active Motif), anti-H3K4me3 (Abcam or Active Motif), anti-H3K9me3 (Upstate or Active Motif), anti-H3K27me3 (Abcam), anti-H3K36me3 (Upstate), anti-H3K79me2 (Upstate or Active Motif), anti-H4K20me3 (Active Motif) and an antibody against the large subunit of RNA polymerase II (N20; Santa Cruz Biotechnology).
Statistics
A two-way analysis of variance followed by the Bonferroni post hoc test were performed to determine the statistical significance of the results of qRT–PCR and ChIP analysis between unaffected and FRDA cell lines.
RESULTS
Reduced level of the FXN pre-mRNA downstream of the hyperexpanded GAA tract in FRDA cell lines
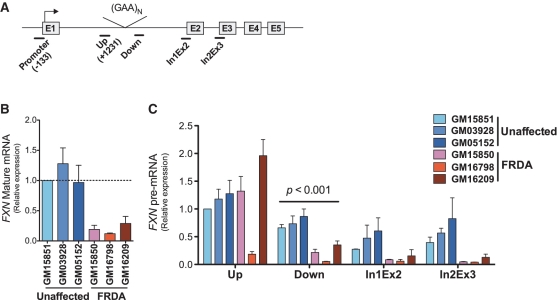
To confirm FXN insufficiency in FRDA cell lines (GM15850, GM16798 and GM16209), FXN mRNA expression was examined by qRT–PCR using a primer set recognizing exon 3 as a forward primer and exon 4 as a reverse primer (Figure 1B). The level of the FXN mRNA expression varies significantly among FRDA patients. Affected individuals can express as little as 5% and as much as 35% of FXN relative to a control individual (16). In order to establish a common epigenetic profile of the FRDA individuals, we selected a representative cohort of FRDA cell lines based on the expression of the FXN mRNA. We selected cell lines expressing low (GM16798) medium (GM15850) and high (GM16209) levels of the FXN mRNA corresponding to ~8, 17 and 29% of the amount of the FXN transcript found in the GM15851 control cell line, respectively (Figure 1B).
Figure 1.
Decreased level of FXN pre-mRNA downstream of the hyperexpanded GAA repeats. (A) Schematic diagram of the FXN gene. Black lines under the schematic diagram indicate regions amplified by designated primers used in this study. (B) FXN mRNA was analyzed in three control and three FRDA cell lines by qRT–PCR with primers complementary to the exons 3 and 4. Blue-series bars designate control cell lines, while Red-series bars designate FRDA cell lines. FXN expression was normalized to the expression level determined by control GM15851 cells. The experiment was conducted in triplicate. All data are expressed as mean ± SD. (C) Relative level of the FXN pre-mRNA was determined in various regions of the transcript. ‘Up’ and ‘Down’ primers amplify regions in the immediate vicinity of the GAA repeats. In1Ex2 and In2Ex3 anneal to the junctions between the corresponding introns and exons of the FXN pre-mRNA. The FXN pre-mRNA expression was normalized to the level of the fragment amplified upstream of the GAA region in the control GM15851 cells. The experiment was performed in triplicate.
In order to further determine the effect of the hyperexpanded GAA repeats on the progression of transcription throughout the FXN gene, we examined levels of FXN pre-mRNA upstream (from +1231 to +1344 within the intron 1) and in three regions downstream (variable positions, depending on repeat tract length) of the GAA repeats as schematized in Figure 1A. Importantly, all primer pairs used in this experiment mapped within the pre-mRNA to avoid a bias from amplification of mature FXN transcript.
The qRT–PCR analyses revealed similar levels of the FXN pre-mRNA at the region upstream of the hyperexpanded GAA repeats in all three unaffected control cell lines and two of the three FRDA lymphoid cell lines (Figure 1C). The GM16798 FRDA cell line that exhibited a lower level of the FXN pre-mRNA upstream of the GAA repeats, as compared to the remaining five cell lines, also exhibited the lowest level of mature FXN RNA. In contrast to the similar levels of FXN pre-mRNA upstream of the GAA triplet repeats, levels of FXN pre-mRNA were reduced ~4-fold at regions downstream of the repeats in all three FRDA lymphoid cell lines relative to unaffected cell lines.
We also conducted analyses of antisense transcription in the FXN locus using strand-specific RT–PCR to evaluate the relative contribution of sense and antisense transcripts from the FXN locus. In agreement with previous data (11), we were unable to detect antisense transcripts at the regions specified in Figure 1A, either upstream or downstream of the GAA repeats (data not shown). Thus, all FXN transcripts measured in our analysis reflect sense RNA transcribed in the vicinity of the GAA tract. These results strongly suggest that the GAA repeats affect later steps of the transcription process that are downstream of initiation.
RNA pol II distribution is influenced in the region upstream of the GAA repeats but not at the promoter in FRDA cell lines
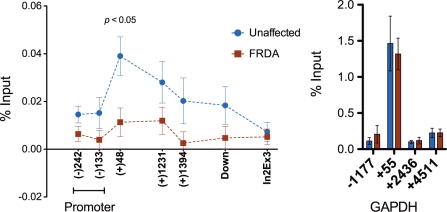
In an effort to further understand how the hyperexpanded tract of GAA repeats affects progression of FXN transcription, we analyzed the distribution of total RNA pol II at the FXN gene by ChIP assay using an antibody that recognizes both phosphorylated and unphosphorylated forms of the large subunit of RNA pol II (Figure 2). We used seven primer sets to assure the fine mapping of the distribution of RNA Pol II in the FXN gene. Based on prior studies demonstrating transcriptional pausing at the long GAA tracts in vitro, as well as in cell-culture systems (17,18), we expected that RNA pol II progression would be impeded in the region upstream of the hyperexpanded GAA repeats in FRDA cell lines, which would result in the accumulation of transcriptional machinery at the region upstream of the GAA repeats. However, we observed decreased levels of RNA pol II across the entire coding region of the FXN gene in FRDA cell lines compared to unaffected cell lines. Importantly, levels of RNA pol II were not changed in the FXN promoter region (from −242 to −12) in FRDA cell lines compared to control cell lines, suggesting that FXN promoter activity is not affected in FRDA cell lines. These results are in agreement with previous findings that similar levels of FXN pre-mRNA at the region upstream of the GAA repeats are observed in both FRDA and unaffected control cell lines (Figure 1C).
Figure 2.
Distribution of RNA polymerase II is affected in the region upstream of GAA repeats in FRDA cells. Level of total RNA pol II was determined by immunoprecipitating the large subunit of RNA pol II in FRDA and control cells. Average occupancy of RNA pol II across the FXN gene in all three FRDA and all three control cells is shown using red and blue dots, respectively. The position of the first nucleotide for the forward primer relative to TSS is indicated below the X-axis. The specificity of RNA pol II antibody for ChIP was verified by RNA pol II pausing at the +55 bp of the GAPDH gene. Error bars represent standard error of the mean. The experiment was conducted in triplicate using three FRDA and three control cell lines. The P-value was generated by comparing the average percentage of input of total RNA pol II between unaffected and FRDA lymphoid cell lines.
Interestingly, RNA pol II accumulated at a promoter-proximal region (from +48 to +150) in unaffected cell lines. This was an unexpected finding as the length of the GAA repeats in these cells are short, ranging from 7 to 20 repeats. The accumulation of total RNA pol II at the FXN promoter-proximal region is similar to what is observed in several other genes (19), where RNA pol II is known to pause. Pausing of RNA pol II was not detected in the FXN promoter-proximal region of the FRDA cell lines.
Collectively, these results suggest that transcriptional defects in FRDA cells begin at a post-initiation step of FXN transcription, which leads to decreased occupancy of the transcriptional machinery at the promoter-proximal region, as well as throughout the FXN gene.
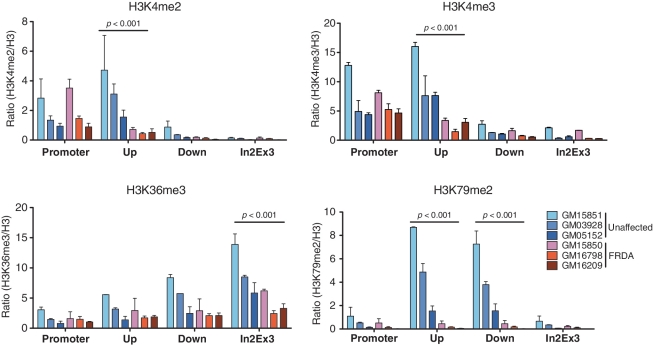
Decreased levels of H3K4me2 and H3K4me3 at the region immediately upstream of the GAA repeats in FRDA cell lines
We next examined distributions of H3K4me2 and H3K4me3 at the FXN gene, as these marks at the promoter are reflective of the transcriptional activity of a gene. Although FXN mRNA levels are known to vary considerably among FRDA individuals and unaffected controls (20), the majority of studies to date have used a single unaffected individual as a control to compare chromatin modification changes at the FXN gene in FRDA patients (10,21). Here, we analyzed cells from three unaffected controls and three FRDA patients to delineate consistent and significant changes in chromatin structure associated with the hyperexpanded GAA repeats in the lymphoid cell lines.
Although variations in H3K4me2 and H3K4me3 levels were observed within compared groups of cell lines, levels of these histone marks were not different at the promoter region (from −133 to −12) of the FXN gene in FRDA cell lines and unaffected controls (Figure 3, upper panels). These results are consistent with our quantitative pre-mRNA analyses showing that the region upstream of the GAA repeats is transcribed at the same level in both unaffected and FRDA cell lines. Interestingly, in all three FRDA cell lines levels of H3K4me2 and H3K4me3 were significantly lower at a region immediately upstream of the GAA repeats (from +1231 to +1344). In contrast, enrichment of H3K4me2 and H3K4me3 levels were the highest at this region in the unaffected cells, which correlated with the distribution of total RNA pol II at the FXN gene (Figure 2). Levels of H3K4me2 have been correlated with non-methylated CpG residues more tightly than with transcriptional activity (22,23). Indeed, high levels of DNA methylation have been reported in the region upstream of the GAA repeats in FRDA cell lines (24,25). Therefore, the decreased levels of H3K4me2 in FRDA cell lines could reflect increased DNA methylation specifically at this region. Previous reports have shown that levels of H3K4me3 in coding regions are associated with the efficiency of the post-initiation processes during active transcription (26). The low levels of H3K4me3 in FRDA cell imply that the hyperexpanded GAA repeats affect the transition between initiation and elongation of FXN transcription. Taken together, these results suggest that the tract of hyperexpanded GAA repeats do not affect chromatin structures at the FXN promoter region, but do affect levels of H3K4me2 and H3K4me3 in the vicinity of the GAA repeats, which indicates a defective transition between initiation and elongation of FXN transcription in FRDA.
Figure 3.
Transcription associated histone modifications are reduced in FRDA cell lines compared to unaffected controls. Transcription status at the FXN gene was determined by the distribution of histone modifications using the ChIP assay. DNA from chromatin immunoprecipitated using antibodies specific for the indicated histone modifications was subjected to qPCR using primers amplifying promoter region, fragments upstream (Up) and downstream (Down) of the GAA repeats and junction between intron 2 and exon 3 (In2Ex3). The experiment was conducted in triplicate using three FRDA and three control cell lines. Data are expressed as mean ± SEM. P-values were calculated by comparing the averages of the ratios between unaffected and FRDA lines. For all histone modification analyses shown in Figures 3–6, ChIP data are presented relative to input DNA and normalized to the total H3 (or H4 in the case of H4K20me3) in each region.
Decreased methylation levels of H3K36 and H3K79 in FRDA cell lines
To further understand how the hyperexpanded GAA repeats affect the transcription process throughout the FXN gene, we measured the levels of H3K36me3 and H3K79me2, which are characteristic markers of transcription elongation. In unaffected controls, the level of H3K36me3 gradually increased toward the 3′-end of the FXN gene, consistent with normal RNA pol II progression as shown in Figure 2. However, the level of H3K36me3 was reduced across the entire FXN gene in FRDA cell lines, indicating defective transcription elongation in FRDA cell lines.
Recently, a genome-wide study revealed that full-length transcription is often characterized by co-enrichment of both H3K4me3 and H3K79me2 at regions downstream of the TSS (27). In contrast, enrichment of H3K4me3 alone is characteristic of genes that undergo transcription initiation, but not elongation (28). We found high levels of H3K79me2 in regions upstream and downstream of the GAA repeats in the unaffected control cells, whereas levels of H3K79me2 were significantly decreased throughout the entire FXN gene in FRDA cells (Figure 3, bottom panel). Because levels of H3K4me3 were unchanged at the FXN promoter in FRDA cell lines (Figure 3, upper panel), these results support the initial observation (Figure 1) that FXN transcription is hampered at an elongation step after successful initiation.
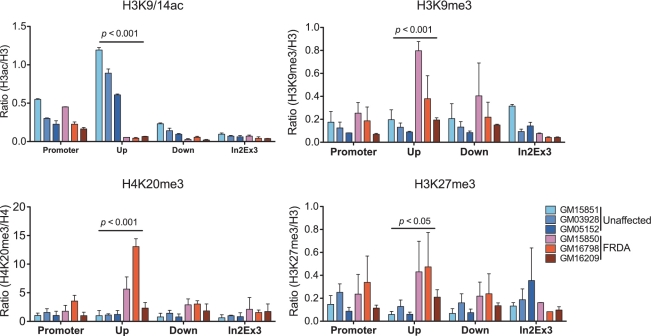
Enriched H3K9 methylation is accompanied by increase of H3K27 and H4K20 methylation at the FXN gene harboring the hyperexpanded GAA repeats
Other groups have reported decreased histone acetylation and increased methylation of H3K9 in the regions flanking the hyperexpanded tract of GAA repeats in FRDA cells, patient autopsy tissues and mouse FRDA models (7,10,11,21,24). We confirmed that the levels of H3K9/14ac were significantly decreased at the FXN gene, specifically at the region upstream of the GAA repeats, in FRDA cells when compared with unaffected cells (Figure 4, upper panel). In contrast to previous findings, in the promoter region of the FXN gene, the levels of H3K9/14ac were similar in both groups. Consistent with the results of H3K4me3, RNA Pol II and FXN pre-mRNA expression analyses, these data indicate that the hyperexpanded GAA repeats do not influence promoter activity.
Figure 4.
The region upstream of the GAA repeats in FRDA cells is enriched with histone modifications associated with repressive chromatin structure. Chromatin structure at the FXN gene was defined by ChIP using antibodies specific to the histone modifications indicated in each graph. The experiment was conducted in triplicate using three FRDA and three control cell lines. Data are expressed as mean ± SEM.
We also confirmed that H3K9me3 was greatly enriched in the region upstream of the GAA repeats in FRDA cell lines (Figure 4, upper panels). Although the majority of H3K9me3 is associated with repressed and silenced regions throughout the genome, this histone modification is also found in the coding regions of transcriptionally active genes (29). In centromeric heterochromatin, where gene expression is repressed, enrichment of H4K20me3 is accompanied by enrichment of H3K9me3 (30,31). To ascertain whether high levels of H3K9me at the FXN gene represent a heterochromatin-like structure adopted by regions surrounding the hyperexpanded GAA repeats, we determined the status of H4K20me3 at the FXN gene in FRDA and control cell lines (Figure 4, bottom panel). ChIP experiments showed that enrichment of H3K9me3 was also associated with significant enrichment of H4K20me3 at the region upstream of the hyperexpanded GAA repeats in FRDA cells. These data further demonstrate that the region immediately upstream of the hyperexpanded GAA repeats adopts a heterochromatin-like conformation in FRDA cell lines.
Repressive chromatin structures, such as the inactive X chromosome and silenced inducible genes, are frequently associated with enrichment of H3K27me3 (32,33). Mapping of H3K27me3 in the FXN gene revealed an over representation in the region upstream of the GAA repeats in FRDA cell lines, co-localizing with H3K9me3 and H4K20me3 enrichments. Taken together, our data indicate that the hyperexpanded tract of GAA repeats in the FXN gene is associated with a unique constellation of repressive histone modifications that lead to the formation of a heterochromatin-like conformation.
Heterochromatin-like structure associated with the hyperexpanded GAA repeats is formed in the absence of transcription
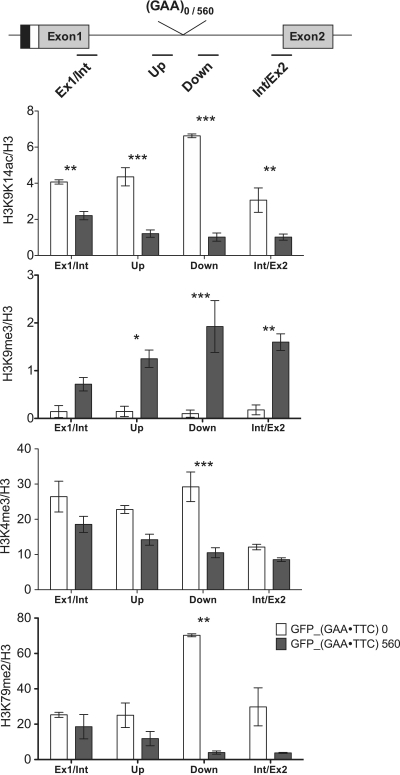
To investigate whether the alteration of histone modifications at the FXN gene result from inherent characteristics of the FXN gene or from the hyperexpanded GAA repeats, we employed a GFP_GAA reporter system that harbors a single copy of the GFP reporter gene containing 560 or 0 GAA repeats in the intron as previously described (9). The GFP_GAA system was established by inserting a hyperexpanded tract of GAA repeats, derived from a FRDA cell line, into the intron of a GFP reporter gene. GFP transcription, driven by a CMV promoter, is controlled by tetracycline repressor/operator. The GFP_(GAA·TTC)560 and GFP_(GAA·TTC)0 cells mimic FRDA and unaffected control cells, respectively. Increased length of the GAA tract up to 560 repeats leads to 2-fold decrease in the expression of the GFP reporter gene (9). To assess changes in chromatin caused by the insertion of long GAA repeats in the context of the GFP gene, we analyzed the status of histone modifications in the vicinity of the GAA repeats in the GFP_GAA system (Figure 5). We confirmed lower levels of H3K9/14ac and high levels of H3K9me3 in the proximity of the GAA repeats in GFP_(GAA·TTC)560 cells as previously demonstrated (9). We found that both H3K9/14ac and H3K9me3 enrichment extended up to the junction between exon 1 and the intron as well as the intron and exon 2 of the GFP gene in the GFP_(GAA·TTC)560 cells. Next, we examined levels of H3K4me3 and H3K79me2 at the GFP gene. No significant changes in the levels of H3K4me3 were detected at the junction between exon 1 and intron as well as in the region upstream of the repeats in the GFP_(GAA·TTC)560 cells relative to the construct lacking the GAA repeats. Levels of H3K4me3 were reduced in the region downstream of the GAA repeats in the GFP_(GAA·TTC)560 cells. This distribution of H3K4me3 is similar to that found at the FXN gene in the FRDA cell lines. Additionally, we found that H3K79me2 was dramatically reduced at the region downstream of the GAA repeats in the GFP_(GAA·TTC)560 cells when compared to the corresponding region in the GFP_(GAA·TTC)0 cells, indicating an impediment of transcription elongation. These results clearly demonstrate that the hyperexpanded GAA repeats not only induce epigenetic changes in the surrounding chromatin independent of the DNA context, but also are sufficient to inhibit transcription elongation. Moreover, the landscape of histone modifications in the GFP_(GAA·TTC)560 cells is similar to that observed in FRDA cell lines. These facts strongly suggest that elongation, but not initiation, is the major step affected by the hyperexpanded GAA repeats in FRDA patients.
Figure 5.
Long GAA repeats induce repressive chromatin conformation in the GFP reporter gene. The levels of histone modifications in the GFP_GAA reporter system, schematically shown in the top panel, were determined by ChIP assay. The black box represents CMV promoter; the white box designates tetracycline operator that allows regulating transcription initiation by tetracycline. The black lines correspond to the location of the PCR products amplified following ChIP. White bars represent the GFP_(GAA·TTC)0 cells. Gray bars represent the GFP_(GAA·TTC)560 cells. Ex1/Int, junction between exon 1 and intron; Up, region upstream of the GAA repeats; Down, region downstream of the GAA repeats; Int1/Ex2, junction between intron and exon2. Data are expressed as mean ± SEM. Asterisk indicates statistical significance (one: P < 0.05, two: P < 0.01, three: P < 0.001). The abundance of histone modifications is shown relative to input DNA in the PCR and normalized to a total H3 for each region.
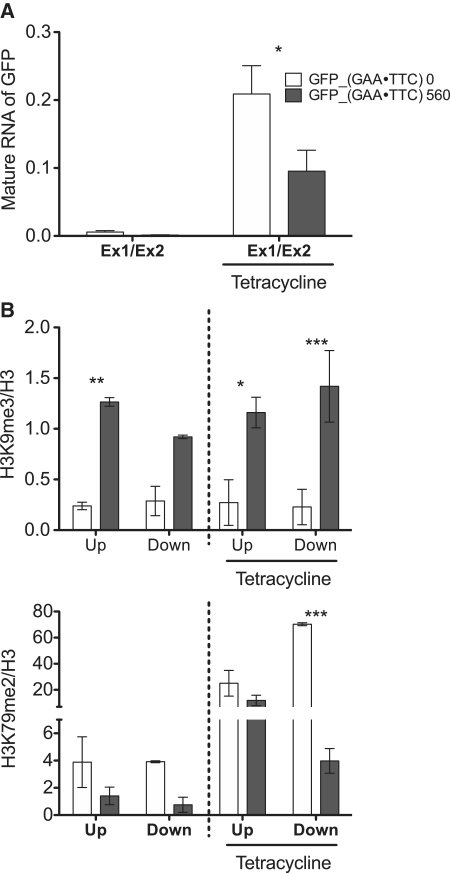
Hyperexpanded GAA repeats are capable of inhibiting the progression of RNA polymerases in vitro and in vivo (17,18). Moreover, transcription arrest is associated with changes in posttranslational histone modifications (34–36). To determine if RNA pol II arrest can trigger the cascade of silencing events, we used the GFP_GAA system to precisely control initiation of transcription using a tetracycline-regulated operator/repressor (Figure 6). We used GFP_(GAA·TTC)560 and GFP_(GAA·TTC)0 cells that were sub-cultured for less than 10 passages prior to the experiment and had never been maintained in the presence of tetracycline. Prior to ChIP analyses the cells were cultured in parallel in the presence or absence of the tetracycline. Upon the inhibition of transcription, the levels of H3K9me3 were enriched in the regions flanking the GAA repeats in the GFP_(GAA·TTC)560 cells. After induction of transcription by the tetracycline, levels of H3K9me3 were unchanged in the vicinity of the GAA repeats in the GFP_(GAA·TTC)560 cells, as compared to the non-tetracycline treated cells. However, levels of H3K79me2 were significantly decreased at the region downstream of the GAA repeats in the GFP_(GAA·TTC)560 cells upon tetracycline treatment when compared to the cells cultured in the absence of the tetracycline. These results strongly suggest that the hyperexpanded GAA repeats are the underlying cause for the formation of the heterochromatin-like structure. The arrest of RNA pol II is unlikely to serve as a signal instigating epigenetic changes associated with long GAA repeats. Taken together, our results demonstrate that FXN deficiency in FRDA patients is a consequence of defective transcription elongation through the heterochromatin-like structure induced by the hyperexpanded GAA repeats.
Figure 6.
The tract of GAA repeats is sufficient to form the heterochromatin-like structure in the absence of transcription. (A) The expression of GFP mRNA was analyzed in the absence/presence of tetracycline using qRT–PCR. White bars represent results obtained from cell line lacking the GAA repeats in intron of the GFP gene while gray bars representing cells harboring intronic 560 GAAs. Data are expressed as mean ± SEM. The experiment was conducted in triplicate and P-values were calculated from the average of all three determinations for the GFP_(GAA·TTC)560 and the GFP_(GAA·TTC)0 cells. (B) Effect of transcription on the formation of the heterochromatin-like structure was analyzed using ChIP assay in the GFP_GAA system. Enrichment of histone modification at the region upstream (Up) and the region downstream (Down) were determined in the absence and presence of tetracycline (0.1 μg/ml for 24 h).
DISCUSSION
In this study we demonstrated that the heterochromatin-like structure induced by the hyperexpanded GAA repeats leads to the inhibition of transcription elongation. Moreover, this phenomenon is independent of the sequence context of the hyperexpanded GAA repeats. The shift in histone modification patterns induced by GAA repeats is locally restricted to the regions upstream of the hyperexpanded GAA repeats at the FXN gene in FRDA cells and does not extend to the promoter region. Consequently, similar levels of the FXN pre-mRNA were detected upstream of the GAA repeats in FRDA and unaffected cell lines. On the contrary, a dramatic decrease of the FXN pre-mRNA was observed at the region downstream of the hyperexpanded GAA repeats, indicating an impediment of FXN transcription elongation in FRDA cell lines. These results are corroborated by differences in the histone modifications as well as RNA pol II occupancy throughout the FXN gene in FRDA cell lines. Similar to the mutated FXN gene in FRDA cells, introduction of the longer GAA repeats into a reporter gene results in formation of heterochromatin-like conformation and silencing of the transgene. The changes in the chromatin conformation triggered by the hyperexpanded GAA repeats are independent of transcription, indicating that the hyperexpanded GAA repeat tract is the primary inducer of the heterochromatin-like structures that subsequently inhibit the transcription process.
Comparison of histone modifications and RNA pol II distribution at the FXN gene between control and FRDA cell lines provides a strong indication that FXN deficiency in FRDA cell lines originates at a post-initiation step of transcription. Several reports using genome-wide approaches have suggested that post-initiation events are rate-limiting in the regulation of transcription (19,37). This phenomenon was first described for a heat shock—inducible gene, Hsp70, in Drosophila (38). High levels of RNA pol II were detected at the promoter of the uninduced Hsp70 gene. The paused RNA pol II was released by heat shock stimuli, which resulted in activation of Hsp70 transcription (38). The increased level of RNA pol II in the promoter-proximal region correlates with the enrichment of active chromatin marks such as H3K9/14ac and H3K4me3 at pause sites in the human genome (19). In our analyses, pausing of RNA pol II at the promoter-proximal region could be readily detected in control cell lines, suggesting that this is part of physiological regulation of the FXN gene expression. Although the occupancy of RNA pol II at the promoter region is similar in unaffected and FRDA cell lines, pausing of RNA pol II is absent at the promoter-proximal region in the FXN gene containing the hyperexpanded GAAs. Pausing of RNA pol II may facilitate remodeling of the chromatin, as shown in our results, to stimulate further transcription progression, as well as stabilizing the binding of transcription machinery facilitating subsequent rounds of transcription initiation. Indeed, new FXN mRNA is slowly synthesized in FRDA cell lines compared to unaffected cell lines (21). The depleted RNA pol II in the coding region of the genes is associated with low levels of H3K4me2, H3K4me3, hyper-methylated DNAs at CpG residues, and hyper-methylated histone H3 at lysine 9 (22,23). As expected, we detected low levels of total RNA pol II in the region upstream of the GAA repeats in FRDA cells. Moreover, the region immediately upstream of the GAA repeats exhibits low levels of H3K4me3 and H3K4me2 in FRDA cells as compared to unaffected cells. H3K4me3 in the coding region is associated with the efficiency of the post-initiation step in transcription (22,23) and acts as an anchor to recruit chromatin-modifying complexes (26,39). Thus, the significant decrease of H3K4me3 at the region upstream of the GAA repeats in FRDA cells may impair the recruitment of chromatin-modifying complexes that are necessary to produce full-length transcripts, which subsequently results in the impediment of FXN transcription elongation in FRDA patients.
Regulatory elements located in the first intron effect expression of several genes (40,41). Studies using reporter constructs have demonstrated that deletions of fragments of the FXN intron 1 significantly decrease luciferase activity (24). Results of our ChIP experiments show the most pronounced difference in the chromatin landscape between FRDA and control cell lines exists in the intronic region upstream of the hyperexpanded GAA repeats. This region is also preferentially hyper-methylated in FRDA patients (24) and the levels of DNA methylation correlate with the number of the GAA repeats (25). One of the methylated CpG residues at the region upstream of the GAA repeats is embedded in an E-box motif (CANNTG) that is recognized by bHLH transcription factors. Analyses of global distribution of the c-Myc, the bHLH transcription factor, demonstrated that this protein interacts with the E-box located in the region upstream of the GAA repeats (42). Moreover, the induction of c-Myc has been shown to upregulate FXN transcription (43). Myc proteins have been demonstrated to stimulate gene expression via release of the paused RNA pol II (28) or by interaction with histone modifying proteins such as histone acetyltransferases (44,45). In contrast to FRDA cells, the region upstream of the GAA repeats encompassing the E-box motif in unaffected cells is enriched in H3K4me3, H3K4me2 and H3K79me2 histone modifications, potentially facilitating the binding of myc proteins. Hypermethylated intronic regions (including the E-box CpG) in FRDA patient cells, perhaps affect the interactions between the E-box sequence and myc proteins (46,47). Taken together these data emphasize the importance of the region upstream of the repeats as the regulatory element of FXN expression. It is likely that long GAA repeats induced epigenetic changes in the first intron of the FXN gene and interfere with transcriptional enhancers leading to the reduced progression of transcriptional machinery and consequently to the silencing of FXN expression in FRDA.
Histone methylation status represents both an important regulatory element of gene expression and an indicator of the transcriptional status of a gene. Enzymes that add or remove methyl groups on specific sites of histones regulate the balance between methylation and demethylation in the cell. The activity of a large group of histone demethylases containing the Jumonji C-terminal domain is dependent on the presence of non-heme iron (48). Several reports demonstrate that low expression of FXN, an iron chaperon, results in the accumulation of iron in the mitochondria and the depletion of iron in other cellular compartments leading to a severe imbalance of iron in FRDA cells (2,3). This suggests that the concentration of iron is associated with the activity of an iron-dependent demethylase in FRDA patients. In the FRDA cells when compared to controls, we detected significant changes in various histone methylation marks including H3K27me3 that can potentially be affected by iron-dependent histone demethylases. In fact, overexpression of iron dependent UTX demethylase is responsible for H3K27me3 demethylation and has been demonstrated to increase expression of FXN mRNA in KYSE180 cancer cells (49). Additionally, FXN deficiency has been shown to downregulate expression of genes involved in DNA packaging and nucleosome assembly (50). Although, expansion of the GAA repeats is the primary factor driving chromatin changes, it is possible that metabolic changes resulting from FXN deficiency in FRDA cells contribute to the epigenetic landscape at the FXN gene. Global analyses of nucleosome position and histone modifications in FRDA are necessary to reveal the full spectrum of epigenetic consequences attributed to reduced FXN levels.
Dissecting the mechanism of transcriptional silencing induced by the hyperexpanded GAA repeats is crucial for the rational design of therapeutic strategies aimed to alleviate transcriptional blocks imposed by the repeat expansions. Currently, the most promising small molecules enhancing transcription of the FXN gene are HDAC inhibitors. HDAC inhibitors target the epigenetic consequences of the GAA expansions and alter the chromatin status in the vicinity of the repeat tract without affecting the chromatin landscape at the promoter region of the FXN gene (10). However, the initial event or signal instigating the cascade of epigenetic changes in the GAA repeat region remains an open question. Perhaps the hyperexpanded GAA sequences or non-canonical DNA structures transiently formed by the hyperexpanded GAA tracts recruit specific proteins that initiate modifications in the chromatin environment surrounding the repeats. Additionally, antisense transcription at the FXN locus (FAST-1) can be involved in triggering silencing of the FXN locus (11). Solving these questions will bring us significantly closer to developing an effective treatment for FRDA and other diseases associated with repeat expansion and gene silencing.
FUNDING
Schissler Foundation Fellowship in the Genetics of Human Disease at the University of Texas Health Science Center in Houston (to E.K.); Kyle Bryant Translational Research Award from Friedreich’s Ataxia Research Alliance and National Ataxia Foundation (to M.N.); UTMDACC Senior Trust Award (to S.Y.R.D.). Funding for open access charge: The Cynthia and George Mitchell Foundation Award (to S.Y.R.D.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Jill Butler, PhD for her helpful comments on this manuscript and Hilary Graham for editing the manuscript.
REFERENCES
- 1.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 2.Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 3.Adinolfi S, Iannuzzi C, Prischi F, Pastore C, Iametti S, Martin SR, Bonomi F, Pastore A. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 2009;16:390–396. doi: 10.1038/nsmb.1579. [DOI] [PubMed] [Google Scholar]
- 4.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 5.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 7.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 8.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soragni E, Herman D, Dent SY, Gottesfeld JM, Wells RD, Napierala M. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 11.De Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J. Biol. Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson RB, Roof DM. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat. Genet. 1997;16:352–357. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 14.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl Acad. Sci. USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 2008;6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Cortopassi G. Frataxin knockdown causes loss of cytoplasmic iron-sulfur cluster functions, redox alterations and induction of heme transcripts. Arch. Biochem. Biophys. 2007;457:111–122. doi: 10.1016/j.abb.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasilnikova MM, Kireeva ML, Petrovic V, Knijnikova N, Kashlev M, Mirkin SM. Effects of Friedreich's ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 2007;35:1075–1084. doi: 10.1093/nar/gkl1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehm T, Scheiber-Mojdehkar B, Kluge B, Goldenberg H, Laccone F, Sturm B. Variations of frataxin protein levels in normal individuals. Neurol. Sci. 2011;32:327–330. doi: 10.1007/s10072-010-0326-1. [DOI] [PubMed] [Google Scholar]
- 21.Punga T, Buhler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okitsu CY, Hsieh JC, Hsieh CL. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell Biol. 2010;30:2933–2946. doi: 10.1128/MCB.01478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castaldo I, Pinelli M, Monticelli A, Acquaviva F, Giacchetti M, Filla A, Sacchetti S, Keller S, Avvedimento VE, Chiariotti L, et al. DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J. Med. Genet. 2008;45:808–812. doi: 10.1136/jmg.2008.058594. [DOI] [PubMed] [Google Scholar]
- 26.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 2004;117:2491–2501. doi: 10.1242/jcs.01238. [DOI] [PubMed] [Google Scholar]
- 31.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 33.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 35.Balakrishnan L, Milavetz B. Histone hyperacetylation in the coding region of chromatin undergoing transcription in SV40 minichromosomes is a dynamic process regulated directly by the presence of RNA polymerase II. J. Mol. Biol. 2007;365:18–30. doi: 10.1016/j.jmb.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagata M, Ito T, Arimitsu N, Koyama H, Sekimizu K. Transcription arrest relief by S-II/TFIIS during gene expression in erythroblast differentiation. Genes Cells. 2009;14:371–380. doi: 10.1111/j.1365-2443.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 37.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 38.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Jaco A, Camp S, Taylor P. Influence of the 5′ intron in the control of acetylcholinesterase gene expression during myogenesis. Chem. Biol. Interact. 2005;157–158:372–373. doi: 10.1016/j.cbi.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 41.Lee JG, Dahi S, Mahimkar R, Tulloch NL, Alfonso-Jaume MA, Lovett DH, Sarkar R. Intronic regulation of matrix metalloproteinase-2 revealed by in vivo transcriptional analysis in ischemia. Proc. Natl Acad. Sci. USA. 2005;102:16345–16350. doi: 10.1073/pnas.0508085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flinn EM, Wallberg AE, Hermann S, Grant PA, Workman JL, Wright AP. Recruitment of Gcn5-containing complexes during c-Myc-dependent gene activation. Structure and function aspects. J. Biol. Chem. 2002;277:23399–23406. doi: 10.1074/jbc.M201704200. [DOI] [PubMed] [Google Scholar]
- 45.Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl Acad. Sci. USA. 2007;104:14917–14922. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 47.Perini G, Diolaiti D, Porro A, Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl Acad. Sci. USA. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn P, Bose J, Edler S, Lengeling A. Genomic structure and expression of Jmjd6 and evolutionary analysis in the context of related JmjC domain containing proteins. BMC Genomics. 2008;9:293–318. doi: 10.1186/1471-2164-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang ML, Becker EM, Whitnall M, Rahmanto YS, Ponka P, Richardson DR. Elucidation of the mechanism of mitochondrial iron loading in Friedreich’s ataxia by analysis of a mouse mutant. Proc. Natl Acad. Sci. USA. 2009;106:16381–16386. doi: 10.1073/pnas.0906784106. [DOI] [PMC free article] [PubMed] [Google Scholar]