Abstract
In a previous study, we presented the dimer structure of DNA gyrase B′ domain (GyrB C-terminal domain) from Mycobacterium tuberculosis and proposed a ‘sluice-like’ model for T-segment transport. However, the role of the dimer structure is still not well understood. Cross-linking and analytical ultracentrifugation experiments showed that the dimer structure exists both in the B′ protein and in the full-length GyrB in solution. The cross-linked dimer of GyrB bound GyrA very weakly, but bound dsDNA with a much higher affinity than that of the monomer state. Using cross-linking and far-western analyses, the dimer state of GyrB was found to be involved in the ternary GyrA–GyrB–DNA complex. The results of mutational studies reveal that the dimer structure represents a state before DNA cleavage. Additionally, these results suggest that the dimer might also be present between the cleavage and reunion steps during processive transport.
INTRODUCTION
Type II DNA topoisomerases are ‘marvelous molecular machines’ that catalyze the ATP-dependent transport of one DNA duplex (the ‘transport’ or ‘T’ segment) through a transient break in another segment (the ‘gate’ or ‘G’ segment) (1,2). With the exception of topo VI (topo IIB), type II topoisomerases, including prokaryotic DNA topoisomerase II (DNA gyrase and topo IV) and eukaryotic DNA topoisomerase II, are similar in sequence, and belong to the type IIA enzymes (2). Eukaryotic topoisomerase II is a homodimer whereas bacterial DNA gyrase exists as an A2B2 heterotetramer (Figure 1A). The B and A subunits of DNA gyrase are homologous to the N- and C-terminus, respectively, of eukaryotic topo II (Figure 1A). DNA gyrase is the primary target of many important antibacterial agents (3). It is of great benefit to drug design to investigate the mechanism of the enzyme, especially for those from infectious pathogens, such as Mycobacterium tuberculosis. DNA gyrase has been found as the sole type IIA topoisomerase in M. tuberculosis so far (4).
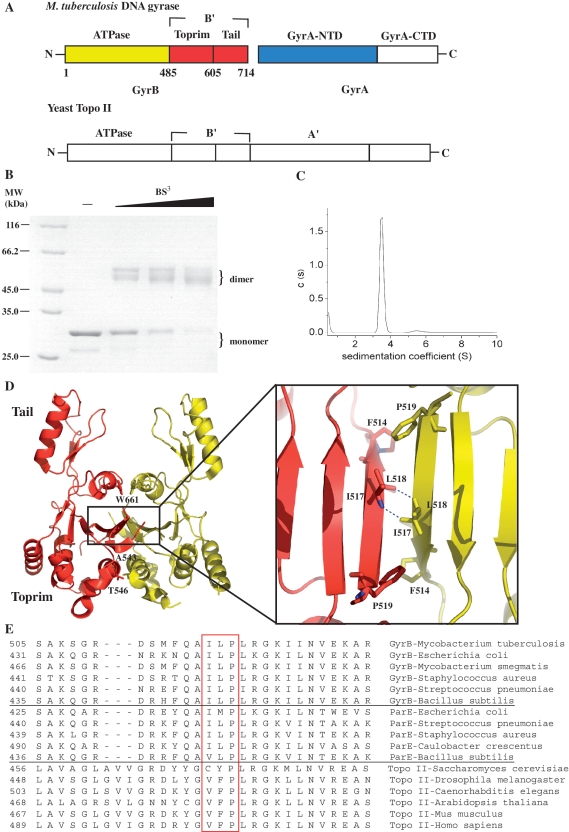
Figure 1.
(A) The primary structure diagram of M. tuberculosis gyrase and yeast topoisomerase II. The core enzyme of topoisomerase IIA is composed of B′ and A′ domains. The domains of M. tuberculosis gyrase and yeast topoisomerase II are homologous, except for the C-terminal domains. (B) A Coomassie blue-stained SDS–PAGE gel of the B′ protein was cross-linked by 0.11, 0.33 and 1 mM BS3. (C) Analytical ultracentrifugation analysis of the B′ protein at 0.5 mg/ml. The B′ protein used in (A and B) was purified by a Ni–NTA column and then was further purified by a HiLoad 16/60 Superdex 75. There are three peaks in the typical elution profile of gel filtration and the fractions of the second peak were collected and exchanged into the required buffer. (D) Crystal structure of B′ dimer (17). One monomer is colored in red and the other in yellow. Each monomer contains a Tail and a Toprim domain. The hydrophobic core is formed as indicated by the box and shown in detail in the inset. The residues (F514, I517, L518 and P519) contributing to this core are highlighted in ball-and-stick representation and H-bonds are shown in dashes. The residues A543, T546 and W661 are highlighted in ball-and-stick representation. The structure pictures were produced with PyMOL. (E) Multiple sequence alignment of the hydrophobic core within the dimer interface of B′ domain in the classes of prokaryotic gyrase, topo IV and eukaryotic topo II. From top to bottom, lines 1–6 contains sequences from prokaryotic gyrase, line 7–11 prokaryotic topo IV, line 12–17 eukaryotic topo II. Three residues (I517, L518 and P519) from M. tuberculosis and their homologous residues from the other species are highlighted in the red box.
Structural and functional studies, particularly with the enzymes from Escherichia coli and Saccharomyces cerevisiae, have established a two-gate mechanism for type IIA topoisomerase catalysis (5–8). According to this model, each enzyme is composed of two identical halves and two protein gates are formed on both sides of the enzyme (the entrance gate or ‘N’ gate and the DNA gate, where the G-segment DNA is cleaved). The T segment travels through these two gates, and in type IIA enzymes, finally exits through a third gate (the ‘exit’ or ‘C’ gate). A series of structures have provided direct evidence that the DNA gate opens by swiveling around the molecular dyad from a closed conformation to partially open and then to fully open conformation (9–11). The structures of the core enzyme of topo IIA–DNA complexes reveal the conformation of the cleavage complex and suggest large protein conformational changes and the sharp bending of the G segment (12,13).
In recent years, single-molecule analyses of type IIA topoisomerases have also provided invaluable insights into the mechanism of topoisomerases. Using a magnetic-tweezers assay, Nollmann et al. (14) demonstrated that gyrase can switch between three distinct modes of activity in response to small changes of tension in DNA. Under low mechanical stress, gyrase processively introduces negative supercoils, but this reaction becomes notably less processive as tension in the DNA increases slightly. A simple mechanochemical model explains that processivity depends on a kinetic competition between dissociation and rapid, tension-sensitive DNA wrapping (15). It was also shown that topo IV is highly processive on positively supercoiled DNA, but is distributive on negatively supercoiled DNA, giving rise to asymmetric supercoil relaxation (16). However, the molecular basis for the mechanism of processivity is not known.
Although the mechanism of type IIA topoisomerases is well known in outline, many fundamental questions remain obscure. For example, dramatic conformational changes are believed to be coupled to T-segment transport, which involves a series of sequential steps including the motion of the B′ domain, the opening of the DNA gate and strand passage through the separated gate (9), but how these conformation changes are coordinated to direct transport of the T segment and what the driving force behind these changes is are poorly understood. An important reason for this is that the structures of the reaction intermediates, which correspond to the various steps, have not yet been elucidated. To solve these and other problems, we determined the structure of M. tuberculosis GyrB C-terminal domain (hereafter termed B′) (Figure 1A); its roles in T-segment transport were suggested largely on the basis of structural analysis (17). We report here mechanistic studies of this domain. The functional state of this domain during the catalysis of DNA gyrase is determined and a model is constructed to discuss its exact role during strand passage.
MATERIALS AND METHODS
Materials
The GyrA, GyrB and B′ proteins were prepared as described previously unless indicated (17). Mutations were introduced by site-directed mutagenesis (18) and mutant proteins were purified in the same way as their wild-type equivalents. pET28–sbp–gyrA was constructed as follows: a Streptavidin-Binding Peptide (SBP) coding sequence plus a linker peptide coding sequence was subcloned into pET28a and the gyrA gene was inserted into the C-terminus of the linker sequence (19). The expressed fusion protein GyrA–SBP was purified using the same protocol as GyrA. The above proteins were further purified by gel filtration, where indicated. Relaxed pBR322, positively supercoiled pBR322 and kDNA were purchased from Inspiralis Limited, UK, negatively supercoiled pBR322 was from Fermentas, DTT, BS3 and BMH were from Pierce, 1,10-phenanthroline and AMPPNP were from Sigma.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed at 20°C using a Beckman Optima XL-I analytical ultracentrifuge. Purified GyrB and B′ protein at various concentrations in 50 mM Tris–HCl (pH 7.5), 55 mM KCl, 4 mM MgCl2 and 4 mM DTT were loaded into 12 mm path-length cells and centrifuged at 42 000 and 60 000 rpm, respectively. Data were recorded with absorbance detection at a wavelength of 280 nm and were analyzed with a c(s) distribution of the Lamm equation solutions calculated by the Sedfit program (20). The Lamm equation describes the evolution of the concentration distribution of a species with sedimentation coefficient and diffusion coefficient in a sector-shaped volume and in the centrifugal field. A distribution of sedimentation coefficient c(s) can be addressed with the Svedberg equation by assuming some prior knowledge (the partial specific volume and the friction ratio) of the macromolecules. The friction ratio can be used to calculate molecular weight. For each protein concentration, experimental profiles with 200 generated sets of data were modeled for s values between 1 S and 15 S (GyrB) or between 0.5 S and 10 S (B′) with a confidence level of 0.95 for the regularization procedure. This allowed us to evaluate the homogeneity of the sample and to determine a sedimentation coefficient (s) for the main species. We assume the partial specific volume to be 0.73 ml/g, the buffer density to be 1.00 g/ml, and the buffer viscosity to be 0.01002 poise, the best-fit friction ratio is 1.65 for GyrB and is 1.49 for the T546C–GyrBnc mutant.
Protein cross-linking
B′ protein and its mutant (5 μM) were exchanged extensively into 20 mM HEPES (pH 7.5), 20% glycerol and 50 mM NaCl and incubated with BS3 in 20 mM HEPES (pH 7.5), 55 mM KCl and 4 mM MgCl2 for 30 min at room temperature (22 ± 2°C). Reactions (10 μl) were terminated by the addition of 0.5 μl, 1 M Tris–HCl (pH 7.9) and then separated on 9% SDS-PAGE gels and stained with Coomassie blue. Disulfide bond formation in the presence of (1,10-phenanthroline)copper(II) and BMH cross-linking was performed in 50 mM Tris–HCl (pH 7.5), 55 mM KCl, 4 mM MgCl2 and was carried out as described previously (6). The cross-linked dimer of GyrB used for further research was prepared as following: T546C–GyrBnc (GyrBnc indicates the no-cysteine mutant of GyrB, see Results section) protein was purified with gel filtration and the disulfide bonds of its peak II were formed in the presence of (1,10-phenanthroline)copper(II). The reaction mixture was then exchanged into 50 mM Tris–HCl (pH 7.5), 20% glycerol and 50 mM NaCl.
Surface plasmon resonance assay
This assay was done with an surface plasmon resonance (SPR) instrument BIAcore 3000 (BIAcore AB, Uppsala, Sweden) as described previously (21). GyrA–SBP or a biotin-labeled dsDNA was coupled to a streptavidin-coated chip (SA sensor chip, BIAcore) according to the manufacturer's instructions. The biotin-labeled dsDNA fragment (363 bp) was amplified by PCR from pBR322 using two primers (5′- biotin-ATC GAT AAG CTT TAA TGC GGT AGT T-3′ and 5′-GTA GAG GAT CCA CAG GAC GGG TGT-3′). Different forms of GyrB at different concentrations were each passed across the chip in 50 mM Tris–HCl (pH 7.5), 55 mM KCl, 4 mM MgCl2 and 0.005% (v/v) Tween 20, at a flow-rate of 30 µl/min. Generally, five different analyte concentrations were used to determine the kinetic parameters for each interaction. Kinetic parameters were obtained by fitting of the sensorgrams to a 1:1 binding with mass transfer model (BIAevaluation 4.1 software).
Far-western analysis
Interactions between GyrA, GyrB and DNA were analyzed by far-western assay, as described, with minor modifications (19). The DNA was negatively supercoiled, relaxed or linear pBR322. Linear pBR322 was prepared by digestion of the supercoiled form with EcoRI. Buffer C was 25 mM Tris–HCl (pH 7.5), 150 mM NaCl, 55 mM KCl, 4 mM MgCl2, 5% (w/v) non-fat milk and buffer D was 25 mM Tris–HCl (pH 7.5), 150 mM NaCl, 55 mM KCl, 4 mM MgCl2. Where PBS buffer was used, 4 mM MgCl2 was added. To compare interactions between GyrA, GyrB and DNA, we quantified the far-western spots using the volume tools of Quantity One (Biorad, Version 4.6.2). Global background subtraction was used to determine positive signals at the lowest enzyme concentrations for each kind of interactions. The value of the signal versus the enzyme concentration can give a rough measure of interactions. The interaction between GyrB (II) and GyrA is normalized to be 1.
Enzyme assays
Supercoiling and decatenation were carried out as described previously (17), under the following conditions: 40 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 25 mM KCl, 200 mM potassium glutamate, 5 mM DTT, 5% glycerol, 2 mM ATP, 2 mM spermidine, 0.1 mg/ml yeast tRNA, 0.36 mg/ml BSA, 5 μg/ml relaxed pBR322 or kDNA used as the substrates; incubations were for 100 min at 37°C. Relaxation of positive supercoils reactions were carried out under the same conditions except that positively supercoiled pBR322 replaced relaxed pBR322 or kDNA. Relaxation (of negative supercoils) reactions were performed as described for supercoiling except that ATP and potassium glutamate were omitted and negatively supercoiled pBR322 replaced relaxed pBR322. Cleavage of relaxed pBR322 or positively supercoiled pBR322 reactions were performed as described for supercoiling except that ATP was omitted. Cleavage of negatively supercoiled pBR322 reactions were performed as described for relaxation (of negative supercoils). Quinolone-induced cleavage included 40 μg/ml norfloxacin for cleavage of relaxed pBR322 or positively supercoiled pBR322 and 10 μg/ml norfloxacin for cleavage of negatively supercoiled pBR322. In Ca2+-induced cleavage assays, 5 mM CaCl2 replaced 5 mM MgCl2. All the cleavage reactions were performed at 37°C for 30 min. Religation assays were performed as previously described with some modifications (22). Reactions were initiated with Ca2+-induced cleavage as described above. After 30 min incubations at 37°C, the reactions were transferred to 30°C. NaCl was then added to 300 mM to start time-courses. All the above reactions were terminated by the addition of SDS to 0.2% and proteinase K to 0.2 mg/ml at 37°C for 30 min and analyzed by electrophoresis.
Agarose gels were visualized and quantified using the AlphaEaseFC (AlphaImager 2200) software. Cleavage and decatenation activities were directly quantified by the analysis tools (spot denso) and expressed as a percentage of the wild-type enzyme activities after correcting for the different enzyme concentrations. To determine relative activities of relaxation (of negative or positive supercoils) and supercoiling, the concentration of a mutant enzyme needed to reach a half-maximum activity of the wild-type enzyme was determined first, then we calculated the ratio of the concentration of the wild-type enzyme for its half-maximum activity to the above concentration of the mutant enzyme. The ratio is the relative activity of the mutant enzyme versus the wild-type enzyme.
RESULTS
The existence of the dimer structure in full-length GyrB
We have shown previously that B′ is a dimer in the crystal structure and that the residues involved in interactions within the dimer interface are highly conserved (17). In order to determine the oligmeric state of the B′ protein in solution, chemical cross-linking and analytical ultracentrifugation assays were performed. Chemical cross-linking suggests that B′ is a dimer (Figure 1B). The cross-linked B′ protein shows more than one band, probably because this protein is liable to degradation (data not shown). The sedimentation coefficient is 3.5 S, consistent with a 55 kDa dimer (Figure 1C) (17). Since the B′ domain monomers are not detectable at sub-micromolar protein concentrations by analytical ultracentrifugation (Figure 1C), the dissociation constant of its dimer is likely to be in the sub-micromolar range. Consistent with this result, there is a hydrophobic core within the dimer interface and the buried area of the dimer contact is large (1100 Å2) (17), indicating that the interactions within the dimer interface are relatively strong. We have previously shown that the B′ dimer, together with GyrA, exhibits ATP-independent relaxation activity (17). Overall, these data suggest that the dimer of the B′ protein is likely to represent an important conformation.
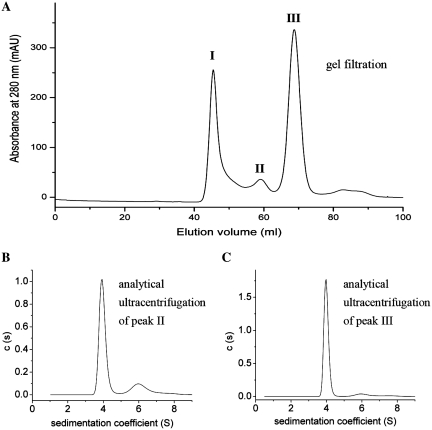
To determine whether this dimer exists in the full-length GyrB from M. tuberculosis, this protein was analyzed by gel filtration. The typical elution profile of the full-length GyrB reveals three distinct peaks: a void volume species (peak I, >200 kDa) and two other species (peaks II and III) under the reducing conditions (Figure 2A). SDS–PAGE analysis demonstrated that GyrB is present in all these peaks (data not shown). Peak II GyrB is always smaller than peak III and it becomes larger when DTT is omitted in the solution (data not shown). In the presence of GyrA, the peak I species exhibited the lowest topoisomerase activities (supercoiling, relaxation and decatenation), ~12–30% of peak III species (Table 1), consistent with a previous report (23), suggesting that this species is mis-folded or aggregated. The topoisomerase activities of peak II are very comparable to those of the peak III species, about one-half of the latter (Table 1) and they probably correspond to the dimer and monomer conformation of GyrB, respectively. Analytical ultracentrifugation experiments were performed to determine the oligomeric state of the peaks II and III species. The results of both species showed a main peak at 3.9 S ± 0.1 S and a secondary peak at 5.9 S ± 0.1 S, which correspond to an 80 kDa monomer and a 160 kDa dimer, respectively (Figure 2B and C). The dimer accounted ~18% of the total cell content for peak II at 0.6 mg/ml and ~6% for peak III at 0.6 mg/ml. The ratio of the latter approximates to that of GyrB during its purification by gel filtration (~8%, the peak I species is not included). These results indicate that M. tuberculosis GyrB alone exists as a monomer–dimer equilibrium in solution and this equilibrium process is slow, consistent with that of E. coli GyrB (24). Since the above experiments have been done under reducing conditions and the ratio of the dimer state is relatively significant, the slow equilibrium process may be physiologically relevant.
Figure 2.
Analysis of the oligomer of GyrB. (A) GyrB protein was purified by gel filtration. GyrB that was purified by a Ni–NTA column was applied to a HiLoad 16/60 Superdex 200 (GE Pharmacia) in 50 mM Tris–HCl (pH 7.5), 220 mM KCl, 4 mM MgCl2 and 4 mM DTT. Three peaks are shown (I, II and III). Fractions of peaks II and III were collected, respectively, and were exchanged extensively into 50 mM Tris–HCl (pH 7.5), 55 mM KCl, 4 mM MgCl2 and 4 mM DTT for analytical ultracentrifugation analysis. The sedimentation coefficient distribution for the peaks II (B) and III (C) of GyrB protein at concentration of 0.6 mg/ml are shown.
Table 1.
Relative topoisomerase activities
| Reactions | Peak I (%) | Peak II (%) | Peak III (%) | W661C | T546C | A543C |
|---|---|---|---|---|---|---|
| Supercoiling | 13 ± 4 | 42 ± 8 | 100 | <3 (% WT) | 6 ± 1 (%WT) | <3 (% WT) |
| Decatenation | 11 ± 2 | 55 ± 7 | 100 | NT | NT | NT |
| Relaxationa | 32 ± 8 | 70 ± 10 | 100 | 70 ± 10 (% WT) | 25 ± 4(% WT) | 20 ± 5(% WT) |
aRelaxation of negatively supercoiled pBR322.
All the activities are in the presence of wild-type GyrA. The topoisomerase activities of peaks I and II of GyrB are given as a percentage of the peak III; the topoisomerase activities of W661C, T546C and A543C mutants are given as a percentage of the wild-type enzyme. Data in table 1 are averaged from three separate experiments (mean ± standard deviations). NT, not tested.
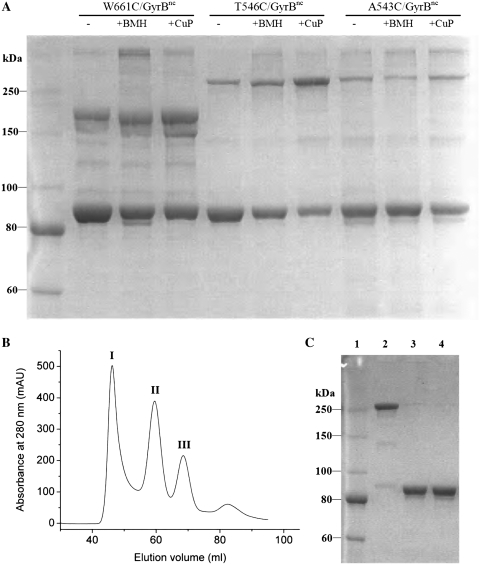
The existence of the dimer state of GyrB alone in solution was confirmed by cysteine cross-linking. In the presence of a mild oxidizing reagent (1,10-phenanthroline)copper(II), GyrB formed non-exclusive disulfide bonds (data not shown), which are likely to result from its three native cysteines. Therefore, a no-cysteine mutant of GyrB (C20A–C427A–C484A–GyrB, termed GyrBnc) was constructed and three cysteine mutations (W661C, T546C and A543C) were each introduced into the no-cysteine mutant (Figure 1D). In the presence of GyrA, the relaxation (of negative supercoils) activity of these mutants (W661C–GyrBnc, T546C–GyrBnc and A543C–GyrBnc) is between 25–70% of the wild-type enzyme, and supercoiling activity of the T546C–GyrBnc mutant is ~6% whereas supercoiling activity of the other two mutants is undetectable (Table 1). The above GyrB mutants were subjected to cross-linking analysis using disulfide bond formation and a cysteine-specific cross-linking reagent BMH (Figure 3A). It showed that spontaneous disulfide bond formation occurred in all three mutants under non-reducing conditions (Figure 3A). Both (1,10-phenanthroline)copper(II) and BMH caused intersubunit cross-linked dimers for all three GyrB mutants. More than one band is observed for each of the mutants. The main band of cross-linked dimer for W661C–GyrBnc showed faster mobility than those of the other two mutants, indicating different SDS-bound conformations. This can be attributed to different sites of cross-linking (Figure 1D). All these cross-linked dimers produced by (1,10-phenanthroline)copper(II) could be broken by incubation with 4 mM DTT during sample preparation that was applied to non-reducing SDS–PAGE (Supplementary Figure S2), indicating that they were disulfide bonds. In contrast, no specific cross-linked dimers were found in the no-cysteine mutant of GyrB under the same conditions (Supplementary Figure S2). Overall, these data reveal that the intersubunit cross-linking was formed at the cysteines that were introduced and confirm the dimer interaction within GyrB.
Figure 3.
(A) SDS–PAGE analysis of cross-linked GyrB. W661C–GyrBnc, T546C–GyrBnc and A543C–GyrBnc (4 µM) were incubated under non-reducing conditions alone or in the presence of BMH (25 µM) or (1,10-phenanthroline)copper(II) (CuP, 125 µM). Reactions were incubated for 1 h at 37°C for BMH cross-linking and quenched with DTT (4 mM). Reactions were incubated for 0.5 h at 37°C for (1,10-phenanthroline)copper(II) cross-linking and quenched with EDTA (1 mM). The above quenched mixtures were analyzed by SDS–PAGE under non-reducing conditions. T546C–GyrBnc protein was purified with gel filtration (B) and its peak II was cross-linked by (1,10-phenanthroline)copper(II) (C). Lane 1, protein marker; lane 2, the prepared cross-linked T546C–GyrBnc mutant protein by (1,10-phenanthroline)copper(II); lanes 3 and 4 were the cross-linked protein incubated with 5 and 10 mM DTT, respectively. Samples were analyzed by SDS–PAGE under nonreducing conditions.
The cross-linked dimer of GyrB blocks the binding to GyrA, but binds dsDNA with a much higher affinity than that of the monomer state
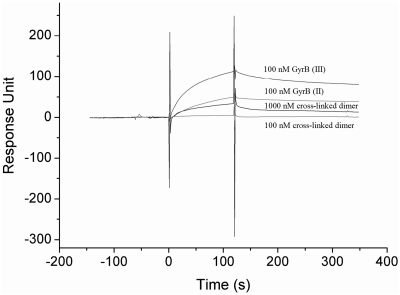
In order to probe the roles of the dimer and monomer of GyrB, the peaks II and III of GyrB [GyrB (II) and (III)] and the cross-linked dimer of GyrB (II) were used to compare their interactions with GyrA or DNA. The T546C–GyrBnc mutant was chosen to prepare the cross-linked dimer. Because the cross-linking efficiency of (1,10-phenanthroline)copper(II) is much higher than that of BMH for all three mutants, this mild oxidizing reagent was used to produce the cross-linked dimer of GyrB. The conversion into cross-linked dimer is high (>90%) (Figure 3C). In the presence of GyrA, the topoisomerase activities of the cross-linked dimer are relatively high in the presence of DTT compared to those of the wild-type enzyme (Supplementary Figure S3), indicating that the cross-linked dimer is active. This cross-linked dimer bound very weakly to immobilized GyrA compared to those of the GyrB (II) and GyrB (III), even at very high concentrations (Figure 4). The KD of GyrB (II) for binding to GyrA increased about 2-fold compared to that of GyrB (III) (Table 2). Unexpectedly, the peak II of the T546C–GyrBnc mutant also bound very weakly to GyrA (data not shown), very similar to its cross-linked dimer. It was observed that peak II became much higher than peak III in the above mutant whereas peak II was much smaller than peak III in wild-type GyrB under the same conditions of purification (Figure 2A and 3B), indicating that the dimer state becomes much more stable in the mutant. To our expectations, the results of analytical ultracentrifugation analysis revealed a main peak at 6.4 S ± 0.1 S and a secondary peak at 4.0 S ± 0.1 S (Supplementary Figure S4A), corresponding to a 134 (peak II) or 157 (peak III) kDa dimer and a 66 (peak II) or 71 (peak III) kDa monomer, respectively. The dimer accounted ~77% of the total cell content for peak II at 0.6 mg/ml and ~26% for peak III at 0.6 mg/ml (Supplementary Figure S4). These results suggest that the dimer state prevents GyrB from binding GyrA.
Figure 4.
SPR analysis of interactions between GyrA and different forms of GyrB. An amount of 12 µg/ml GyrA–SBP in 50 mM Tris–HCl (pH 7.5), 20% glycerol and 50 mM NaCl was injected at a flow rate of 10 µl/min to immobilize on a streptavidin-coated chip and about 1200 response units (RU) was generated. Different forms of GyrB at indicated concentrations were each passed across the chip at a flow-rate of 30 µl/min.
Table 2.
The kinetic constants between GyrB and GyrA
| Proteins | ka (M−1s−1) | kd (s−1) | KA (M−1) | KD (M) |
|---|---|---|---|---|
| GyrB (II) | 2.1 (±0.1) × 104 | 7.8 (±0.7) × 10−4 | 2.7 (±0.4) × 107 | 3.8 (±0.6) × 10−8 |
| GyrB (III) | 6.2 (±0.6) × 104 | 10.1 (±1.1) × 10−4 | 6.2 (±1.3) × 107 | 1.7 (±0.4) × 10−8 |
The kinetic constants between GyrB and dsDNA are presented in Table 3. It shows that the KD of the cross-linked dimer for binding to the immobilized DNA is very close to that of GyrB (II), but decreases about two orders of magnitude compared to that of GyrB (III). Peak II of the T546C–GyrBnc mutant [T546C–GyrBnc (II)] behaved like its cross-linked dimer and GyrB (II), showing a comparable KD, in accordance with the fact that the dimer state was a predominant species for peak II of this mutant. These data reveal that the dimer state of GyrB has a much higher affinity for dsDNA than its monomer state.
Table 3.
The kinetic constants between GyrB and dsDNA
| Proteins | ka (M−1s−1) | kd (s−1) | KA (M−1) | KD (M) |
|---|---|---|---|---|
| GyrB (II) | 9.8 (±2.3) × 103 | 4.0 (±2.5) × 10−5 | 2.4 (±0.9) × 108 | 4.6 (±1.8) × 10−9 |
| GyrB (III) | 2.5 (±0.7) × 103 | 5.5 (±0.9) × 10−4 | 4.8 (±2.2) × 106 | 2.4 (±1.1) × 10−7 |
| Cross-linked dimer | 8.3 (±2.1) × 103 | 3.9 (±1.9) × 10−5 | 2.3 (±0.6) × 108 | 4.5 (±1.1) × 10−9 |
| T546C–GyrBnc (II) | 1.7 (±0.4) × 104 | 2.5 (±0.1) × 10−4 | 7.0 (±2.0) × 107 | 1.5 (±0.4) × 10−8 |
The dimer structure of GyrB is involved in the complex of GyrB–GyrA–DNA
In order to probe whether the dimer structure of GyrB forms in the context of the functional heterotetramer of gyrase and DNA, cross-linking assays were used to analyze the interactions between GyrA, GyrB and DNA. The cross-linked dimer band of the T546C–GyrBnc mutant remained almost constant in the presence of AMPPNP, GyrA, DNA, GyrA–DNA or GyrA–DNA–AMPPNP (Supplementary Figure S5A). When the mutant was replaced with the mutants of A543C–GyrBnc and W661C–GyrBnc in the above assays, very similar results were obtained (Supplementary Figure S5B and C). It is reported that GyrB, GyrA and DNA can form a ternary complex (25). Thus the dimer state of GyrB may be present in the complex of GyrB–GyrA–DNA.
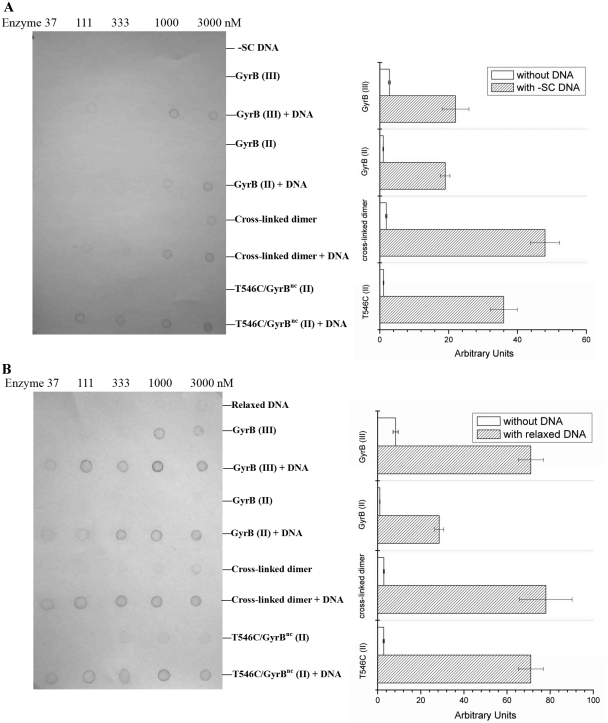
Far-western blot assays were also utilized to analyze interactions between different forms of GyrB and GyrA–DNA. Negatively supercoiled pBR322 was found to interact very weakly with GyrA–SBP under conditions tested (Figure 5A). A similar result was obtained when this DNA passed over a SA chip where GyrA–SBP was immobilized in the SPR analysis, even when the DNA concentration was up to 50 µg/ml (data not shown). When the concentration of negatively supercoiled pBR322 was kept just below the limit of detection in the far-western analysis of GyrA–DNA interaction, the interaction of GyrB–GyrA–DNA was ~9-fold stronger than that of GyrB–GyrA for both GyrB (II) and GyrB (III) (Figure 5A), indicating that a stable complex of GyrB–GyrA–DNA was formed. It increased to ~27-fold when the cross-linked dimer of the T546C–GyrBnc mutant was applied to the above assays (Figure 5A). Very similar results were shown for T546C–GyrBnc (II) (Figure 5A). Considering that GyrA–SBP bound GyrB dimer weakly and no obvious interaction was detected between GyrA–SBP and the negatively supercoiled DNA under the conditions tested, the above results suggest that GyrA–SBP can strongly bind the complex of DNA and the dimer state of GyrB. The interaction of GyrB–GyrA–DNA for the cross-linked dimer was nearly the same as the interaction for T546C–GyrBnc (II) and was stronger than for GyrB (II) and GyrB (III) (Figure 5A and B), indicating that the dimer state more readily forms the ternary complex than the monomer state. When negatively supercoiled pBR322 was replaced with relaxed pBR322 in all the above assays, the results were almost identical except that GyrA–SBP bound the relaxed DNA with a higher affinity than the negatively supercoiled DNA (~10-fold) (Figure 5A and B). Also, the results obtained with linear DNA were very similar (data not shown). Together, the above results suggest that the dimer structure of GyrB is involved in the ternary complex.
Figure 5.
Far-western analysis of interactions between GyrA, GyrB and DNA. In (A and B), different forms of DNA, GyrB (Peaks II and III of GyrB protein purified by gel filtration, peak II of T546C–GyrBnc protein purified by gel filtration and its cross-linked dimer) alone or in the presence of DNA were applied to a nitrocellulose membrane. These proteins were at different concentrations (37–3000 nM, from left to right) which are indicated. When the DNA was alone, it was at different concentrations: 1.2, 3.7, 11, 33, 100 µg/ml (from left to right) for negatively supercoiled pBR322 (–SC) (A) and 0.12, 0.37, 1.1, 3.3, 10 µg/ml (from left to right) for relaxed pBR322 (B). When the DNA was in the presence of GyrB protein, its concentration was kept constant: 100 µg/ml for negatively supercoiled and 10 µg/ml for relaxed pBR322. The membrane was then incubated with 10 µg/ml GyrA–SBP after blocking with non-fat milk. After successive washing, GyrA–SBP remained bound to the membrane was detected by incubating with streptavidin alkaline phosphatase conjugate and BCIP–NBT sequentially. Quantification of interactions from far-western blotting pictures is on the right side of the pictures. Each of interactions between different forms of GyrB and GyrA in the presence and absence of DNA were quantified (see ‘Materials and Methods’ section).
The dimer structure of GyrB is involved in a step before the cleavage
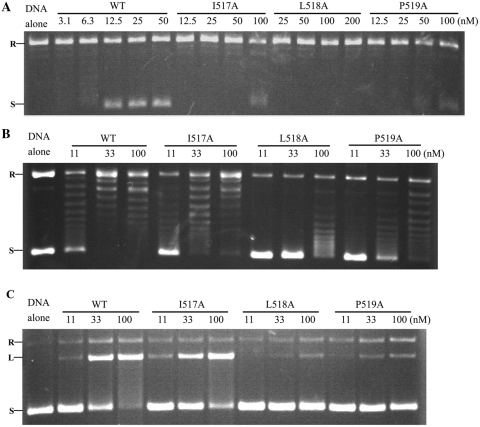
Since the activity of the cleavage and reunion steps can be determined separately, structures of the catalytic intermediates that correspond to these steps can be evaluated. In order to discover at which steps the dimer structure is involved, site-directed mutagenesis was applied to destabilize the dimer interface. We focused on the most conservative residues that contribute to the hydrophobic core (Figure 1D and E); three single mutants, I517A, L518A and P519A, were constructed. DNA gyrase bearing the I517A mutation shows about half the activity of the wild-type enzyme in decatenation, relaxation (of negative supercoils) and quinolone-induced cleavage, whereas this further decreases to ~8% in supercoiling and Ca2+-induced cleavage of relaxed pBR322 (Table 4, Figure 6 and Supplementary Figure S5). The L518A and P519A mutants demonstrate a very similar pattern except that all the above activities of the latter reduce to almost the same degree (Table 4, Figure 6 and Supplementary Figure S5). These mutants, especially L518A, greatly reduced all activities, supporting the idea that the dimer structure may represent an important conformation. Gel filtration analysis showed that most of the B′ dimer, which was the predominant species in wild-type B′, converts into polymers in the L518A-B′ mutant (Supplementary Figure S7A and B). The cross-linking analysis of this mutant also showed no cross-linked dimer band (Supplementary Figure S7C). Overall, these observations suggest that the mutation does destabilize the dimer interaction, thereby favoring the monomer which polymerizes probably because of the exposure of the hydrophobic core in the monomer form. Similar gel filtration elution profiles were obtained for each of the mutants of full-length GyrB (Supplementary Figure S8). The effect of those three mutations on the structure of GyrB was investigated by the CD analysis and limited proteolysis. The results of the CD analysis show that these three GyrB mutants are similar to wild-type GyrB (Supplementary Figure S9). Likewise, the proteolytic fingerprints of these three GyrB mutants alone and in the presence of AMPPNP, GyrA, DNA, AMPPNP–GyrA, GyrA–DNA, GyrA–DNA–AMPPNP or GyrA–DNA–norfloxacin were almost identical to the wild-type (data not shown). These results indicate that the secondary structures of the mutants are likely to be the same as the wild-type. Therefore, the reduced activities of the mutants are unlikely due to misfolding of the mutant proteins.
Table 4.
Relative topoisomerase activities of mutants
| Reactions | I517A | L518A | P519A |
|---|---|---|---|
| Supercoiling (% WT) | 8 ± 1 | <3 | 23 ± 10 |
| Relaxation (% WT)a | <3 | <3 | 29 ± 6 |
| Decatenation (% WT) | 50 ± 10 | 12 ± 2 | 28 ± 4 |
| Relaxation (% WT)b | 50 ± 8 | 10 ± 2 | 29 ± 6 |
| Quinolone (% WT)c | 64 ± 9 | 11 ± 1 | 33 ± 5 |
| Quinolone (% WT)d | 49 ± 7 | 8 ± 1 | 35 ± 3 |
| Quinolone (% WT)e | 37 ± 11 | 12 ± 4 | 54 ± 13 |
| Ca2+ (% WT)d | 9 ± 1 | <2 | 36 ± 10 |
| Ca2+ (% WT)e | <3 | <2 | 44 ± 4 |
aRelaxation of positively supercoiled pBR322.
bRelaxation of negatively supercoiled pBR322.
cCleavage of negatively supercoiled pBR322.
dCleavage of relaxed pBR322.
eCleavage of positively supercoiled pBR322.
All the activities are in the presence of wild-type GyrA. Generally data in Table 4 are averaged from three separate experiments (mean ± standard deviations).
Figure 6.
Representative assays for WT gyrase and its various mutants: (A) supercoiling and (B) relaxation of negative supercoils; (C) representative norfloxacin-induced cleavage of negatively supercoiled pBR322 by WT gyrase and its various mutants under relaxation (of negative supercoils) reaction conditions. Negative controls of DNA alone, WT and its various mutants are shown. Concentrations of WT and its various mutants (GyrB in the presence of equal molar GyrA) are indicated. R, L and S denote relaxed, linear and supercoiled pBR322, respectively.
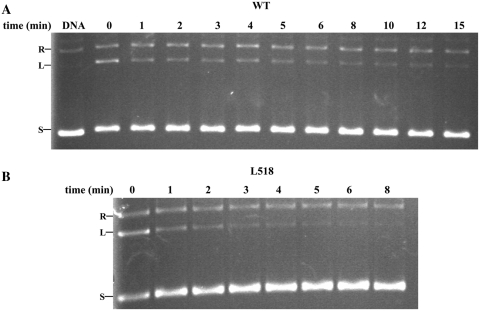
The effect of quinlones on gyrase is thought to trap the enzyme on DNA and stabilize the cleavage complex (12,13,26). Quinolone-induced cleavage is therefore a good reaction that can evaluate the cleavage reaction of topoisomerase activities. The fact that quinolone-induced cleavage, relaxation (of negative supercoils) and decatenation activities decreased to almost the same extent for each of the three mutants (Table 4) indicates that a common reaction, cleavage, is likely to be deficient in the above reactions. Consistent with this idea, the mutant L518A had a religation rate that was comparable to that of wild-type (Figure 7), indicating that the mutation has not directly affected the religation step of the reaction and thus it is more likely to affect the cleavage–religation reaction of the enzyme by decreasing the cleavage activity. There are two possibilities that can lower the cleavage reaction: a defect of the cleavage step per se or a step before cleavage. The latter possibility is favored because the cleavage-competent conformation of B′ is not a dimer (12,13). The configuration of the DXD motif in the dimer structure is obviously quite different from its conformation in the cleavage complex (17). The defect therefore can be attributed to a step before the cleavage. Given that the mutations destabilize the dimerization of the L518A and P519A mutants, the dimer structure of GyrB is probably involved in the step before the cleavage step of relaxation (of negative supercoils) and decatenation reactions.
Figure 7.
Religation assay for WT gyrase (A) and its mutant L518A (B). An amount of 100 ng of negatively supercoiled pBR322 was added in all reactions. An amount of 11 and 100 nM enzyme were added for WT and its mutant L518A (GyrB in the presence of equal molar GyrA), respectively. DNA alone is a negative control. Reaction times (in minutes) are indicated. R, relaxed pBR322; L, linear pBR322; S, supercoiled pBR322.
Notably, the supercoiling activity of the I517A and L518A mutants reduced much more than their relaxation (of negative supercoils), decatenation and quinolone-induced cleavage activities compared to the wild-type enzyme (Table 4, Figure 6 and Supplementary Figure S5), implying that the B′ dimer may have a different role in the supercoiling reaction compared to the other reactions. Interestingly, supercoiling activity decreased to the same extent as Ca2+-induced cleavage activity in the I517A and L518A mutants (Table 4, Figure 6A and Supplementary Figure S5C), suggesting that the defects in both reactions are likely to be the same. The fact that the decrease in quinolone-induced cleavage of relaxed DNA is almost the same as quinolone-induced cleavage of negatively supercoiled DNA for each of the mutants (Table 4, Figure 6C and Supplementary Figure S5B) implies that one defect in the supercoiling reactions of the I517A and L518A mutants, the defect of the step before cleavage, is common to the other reactions; i.e. the dimer structure of GyrB is involved in the step before the cleavage of supercoiling. Another defect may exist that leads to the further decrease in supercoiling and Ca2+-induced cleavage activities of the mutants. Given that Ca2+-induced cleavage reflects a shift within the cleavage–religation equilibrium, this defect in the supercoiling reaction of the mutants, which shifts the equilibrium towards religation, may occur between these two steps. Although the relaxation of positive supercoils is a topologically equivalent reaction to (negative) supercoiling, the former implies a different mode of activity at high force from the latter (14). The above two mutants therefore were also subjected to the tests of positive supercoil relaxation and their quinolone- and Ca2+-induced cleavage. As expected, the losses of these activities compared to the wild-type enzyme are very similar to those of their counterpart reactions of supercoiling, except that relaxation of positive supercoils activity in the I517A mutant and its Ca2+-induced cleavage activity were not detectable (Table 4). In contrast to the other topoisomerase activities, both relaxation of positive supercoils and supercoiling reactions of DNA gyrase are processive. Therefore, the dimer structure is possible to play an important role in the processivity of DNA gyrase (see Discussion section).
Combining the above biochemical data with the alignment results for the residues forming the hydrophobic core of the dimer structure (Figure 1E), two of the most important residues, I517 and L518, both in terms of structure and activities for M. tuberculosis DNA gyrase, are kept constant among DNA gyrases and are highly conserved among topo IV enzymes. However, in eukaryotic topo II enzymes these residues are less conserved, generally being substituted with Val and Phe; in yeast topo II these residues are substituted with Cys and Tyr, which are even not hydrophobic residues. Another residue, P519, is invariant among type IIA topoisomerases (Figure 1E). In contrast to the I517A and L518A mutants, the supercoiling and positive supercoil relaxation activities of the P519A mutant decreased to almost the same extent as their quinolone- and Ca2+-induced cleavage activities (Table 4). It is evident that P519 is situated at the edge of the hydrophobic core (Figure 1D) and thus contributes less than I517 and L518 to the stability of the dimer structure, which is confirmed by the gel filtration analysis of the three mutants (Supplementary Figure S8). Overall, these results suggest that the dimer state of the B′ domain may well be ubiquitous in prokaryotic type II topoisomerases, but possibly does not exist, or is less stable, in eukaryotic type II topoisomerases.
DISCUSSION
The dimer state of GyrB is an active form
Mycobacterium tuberculosis GyrB appears to exist as a slow monomer–dimer equilibrium in solution, which is the case for the E. coli counterpart (24). The dimer interaction of M. tuberculosis GyrB was confirmed within B′ by cross-linking analysis. Moreover, it has been shown that the interactions within the dimer interface of B′ are highly conserved and are vital to the enzyme activities of DNA gyrase. Therefore, the slow monomer–dimer equilibrium for GyrB may well be common to DNA gyrases from different species.
Although we have shown that the dimer state prevents GyrB from binding GyrA, it does not block GyrB to bind the complex of GyrA–DNA. Conversely, the dimer state more readily forms the ternary complex than the monomer state. The mutations at the dimer interface of GyrB, destabilizing the dimer interaction, which did not obviously affect the native secondary structure of GyrB, reduce all the topoisomerases activities. Overall, these data suggests that the dimer state of GyrB is an active form.
A model for the initial complex of DNA gyrase
We have shown that the formation of the dimer structure of GyrB occurs before the cleavage step and the dimer structure is involved in the complex of GyrB–GyrA–DNA. Moreover, the dimer structure is an active form. The dimer structure therefore is likely to be involved in a complex before cleavage, i.e. an initial complex. Several lines of evidence support this idea. The fact that the ternary complex forms in the presence of either negatively supercoiled or relaxed DNA suggests that the complex forms not only in the relaxation (of negative supercoils) reaction, but also in the supercoiling reaction, in agreement with the idea that the formation of the dimer structure occurs before the cleavage step of both relaxation (of negative supercoils) and supercoiling reactions. The formation of the ternary complex is independent of ATP in the presence of relaxed DNA and can also occur in the presence of linear DNA. Thus, the complex should correspond to a catalytic intermediate before the reaction. The dimer state more readily forms the complex of GyrB–GyrA–DNA than the monomer state, indicating that the dimer state is required for the formation of the ternary complex.
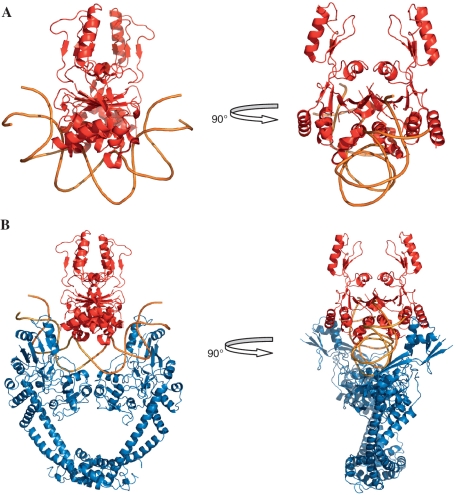
The E. coli GyrA–NTD structure is thought to have a DNA gate-closed conformation at the beginning of the strand-passage (9,11). Therefore, a hypothetical model for the initial complex was generated by manually docking the dimer structure of the B′ domains to this structure (11). The steric hindrance between the dimer of the B′ domain and the GyrA–NTD–DNA complex was minimized (Figure 8B). The region of the B′ dimer that docks to the DNA is a groove formed by the toprim domains of the dimer, the size of which is a good fit for dsDNA (Figure 8A). Interestingly, this docking result shows that the long axis of the negatively charged groove is perpendicular to the G segment, and a simple rotation of the B′ domain around the molecular dyad of the dimer can produce the orientation of B′ domain in the structure of the cleavage complex (12,13,17). We have shown that GyrA can bind relaxed DNA with a higher affinity than negatively supercoiled DNA, making the initial complex form more easily in the presence of relaxed DNA than negatively supercoiled DNA. This result implies that the initial complex is more prone to function before the replication fork where DNA is positively supercoiled, in accordance with the functionality of DNA gyrase (27). The presence of the initial complex in yeast topo II is impossible because the G segment cannot enter this initial complex of yeast topo II due to the homodimer of this enzyme in vivo. This may also explain why the dimer structure of the B′ domain is unlikely to occur in eukaryotic type II topoisomerases.
Figure 8.
Schematic illustration of the dimer structure of the B′ domain docking to distorted DNA (A) and the initial complex of DNA gyrase (B). The complex structure of GyrA–NTD–DNA (GyrA–NTD in blue and DNA in orange) is based on the structure of the yeast topo II–DNA complex and E. coli GyrA–NTD (11,12). The dimer structure of the B′ domain (in red) (17) was docked manually to distorted DNA and the complex of GyrA–NTD–DNA. For clarity the ATPase domain and GyrA–CTD are not included in the initial complex. The structure pictures were produced with PyMOL.
A working model for the mechanism of processivity in DNA gyrase
Processivity is an important property of molecular motors, many of which achieve their processivity by efficient translocation (28,29). However, translocation in DNA topoisomerases is not obvious and they must utilize a different mechanism (30). Processivity in DNA gyrase means that the enzyme can carry out many consecutive strand-passage reactions without releasing the DNA substrate. There are two possibilities for the mechanism of processivity: (i) cleavage, strand passage and reunion occur once in each enzymatic cycle, and a processive reaction contains many of these cycles; (ii) the enzyme cleaves and religates the G-segment only once and performs many successive strand passages in a processive reaction. In contrast to the first possibility, the second possibility can be exempt from many cleavage and reunion steps, thereby enhancing the efficiency of processivity, and can also avoid potential dsDNA breakage due to the failure of reunion for the first possibility. As regards the second possibility, the enzyme is required to recover its conformation at the very beginning of the reaction, forming a cycle of enzymatic processes within the reaction. We have shown that one defect in the processive reactions is common to the other reactions, i.e. the dimer state of GyrB is involved in the formation of the initial complex of processive reactions. Another defect may occur between cleavage and religation. Given that the dimer state of GyrB is involved in the formation of the GyrB–GyrA–DNA complex and the dimer structure may play an important role in the processivity of DNA gyrase, we propose that the dimer structure can also occur in the step between the cleavage and reunion during processive strand passages. This idea may provide a structural basis for the second possibility of the processive reactions (see below).
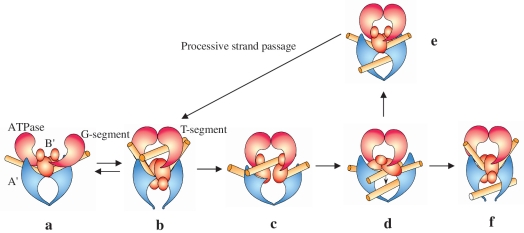
Based on previous studies and the new results reported here, we propose a working model for the role of the B′ domain within the global catalytic mechanism of DNA gyrase (Figure 9). The two subunits of DNA gyrase first assemble on the substrate, forming the initial complex (Figures 8B and 9a). The basic grooves of the ‘head’ dimer of GyrA–NTD bind to a side of one segment of DNA (G-segment) (11). The B′ dimer sits at the saddle-like region of the GyrA–NTD–DNA complex with the bottom groove of the B′ dimer binding to the opposing side of the G-segment. The G-segment is slightly distorted at this moment. Another segment (T-segment) is wrapped by the C-terminal domain of GyrA and is delivered to the ATP-operated clamp (31) (not shown in Figure 9). This clamp (the ATPase domains) dimerizes upon the binding of ATP and captures the T segment within the closed N gate (Figure 9b). The Tail domains swivel around the molecular dyad of the B′ dimer (17), thereby pulling apart the dimer structure. The B′ domains then rotate to form the configuration observed in structures of the core enzyme of topo IIA–DNA complex (12,13) and the G segment is cleaved by forming a covalent enzyme–DNA complex (Figure 9b). Then the B′ domains may also rotate back to the conformation of the initial state, but they do not dimerize as the T segment comes between them (Figure 9c). When the T segment is held within the central hole (Figure 9d), two processes probably occur. For non-processive transport, the B′ domain first rotates to a conformation that resembles the cleavage configuration, and then the cleaved G-segment is religated and the exit gate opens to release the T-segment (Figure 9f). For processive transport, the B′ domain recovers the dimer arrangement as it has in the initial state (Figure 9e). The B′ dimer again rotates to open by swiveling around the molecular dyad and the T segment can be released (Figure 9b; the release of the T segment is not shown). ATP hydrolysis and product release reset the enzyme in the initial complex for another round of strand passage, where the cleaved G segment is not resealed (Figure 9a). In this way, the enzyme can perform consecutive strand passages without religating the cleaved G-segment (the circle of a–b–c–d–e–b–a in Figure 9) and only religates in the final cycle (a–b–c–d–f in Figure 9). This putative mechanism provides an explanation for the much greater reduction in supercoiling and relaxation of positive supercoils activities than in other activities for the mutants described above. Thus it possibly provides a structural basis for the processive transport in DNA gyrase, and we speculate that this mechanism may be also applied to processivity in topo IV.
Figure 9.
A working model for strand passage in DNA gyrase. The ATPase, B′ and A′ domains are in pink, red and blue, respectively; the G and T segment are both shown in yellow. For clarity GyrA–CTD is not included. In (a) the dimer structures of GyrB (ATPase and B′ domains) and the A′ domain bind the G segment to form the initial complex. The ATPase domains dimerize upon the binding of ATP, thereby capturing the T segment; the B′ dimer is opened, accompanying the distortion of the G segment, and finally converts to the cleavage–competent complex (b), where the G segment is cleaved by the active centre formed by the B′ and A′ domains. A series of conformational changes occur and the cleaved G segment is separated (c), including a conformational change shown in (d). Then an electrostatic potential gradient within the enzyme might be generated, thereby drawing the T segment through the break and towards the central hole (d). In (e) the enzyme recovers the conformation of the initial complex as shown in (a) and thus b–c–d–e–b forms a cycle of consecutive strand passage. The G segment is resealed and the T segment released from the exit gate (f).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Chinese Academy of Sciences (KSCX2-YW-R-164 to L.B.); the National Natural Science Foundation of China (Grant No: 30970590 to L.B.); National Basic Research Program of China from the ministry of science and technology of China (2009CB825402 to L.B., 2009CB522605 to J.D.); the Major Special Program on Infectious Diseases from Ministry of Health of China (2008ZX10003-005 to L.B.); Biotechnology and Biological Sciences Research Council (BBSRC), UK (to A.M.). Funding for open access charge: The Major Special Program on Infectious Diseases from Ministry of Health of China.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank James Berger, Andy Bates and Zhenfeng Liu for their comments and discussion, Shiqiang Lin for reading the manuscript, Xiaoxia Yu and for technical support and Huisheng Wang for drawing the pictures of the working model.
REFERENCES
- 1.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5:102–109. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 4.Aubry A, Fisher LM, Jarlier V, Cambau E. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2006;348:158–165. doi: 10.1016/j.bbrc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams NL, Maxwell A. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999;38:13502–13511. doi: 10.1021/bi9912488. [DOI] [PubMed] [Google Scholar]
- 7.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 8.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 9.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 10.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 11.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 12.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 13.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 14.Nollmann M, Stone MD, Bryant Z, Gore J, Crisona NJ, Hong SC, Mitelheiser S, Maxwell A, Bustamante C, Cozzarelli NR. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 15.Gore J, Bryant Z, Stone MD, Nollmann M, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu G, Wu J, Liu W, Zhu D, Hu Y, Deng J, Zhang XE, Bi L, Wang DC. Crystal structure of DNA gyrase B' domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009;37:5908–5916. doi: 10.1093/nar/gkp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shenoy AR, Visweswariah SS. Site-directed mutagenesis using a single mutagenic oligonucleotide and DpnI digestion of template DNA. Anal. Biochem. 2003;319:335–336. doi: 10.1016/s0003-2697(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 19.Li F, Liu Q, Chen YY, Yu ZN, Zhang ZP, Zhou YF, Deng JY, Bi LJ, Zhang XE. Escherichia coli mismatch repair protein MutL interacts with the clamp loader subunits of DNA polymerase III. Mutat. Res. 2008;637:101–110. doi: 10.1016/j.mrfmmm.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang YY, Deng JY, Gu J, Zhang ZP, Maxwell A, Bi LJ, Chen YY, Zhou YF, Yu ZN, Zhang XE. The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA) Nucleic Acids Res. 2006;34:5650–5659. doi: 10.1093/nar/gkl695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hockings SC, Maxwell A. Identification of four GyrA residues involved in the DNA breakage-reunion reaction of DNA gyrase. J. Mol. Biol. 2002;318:351–359. doi: 10.1016/S0022-2836(02)00048-7. [DOI] [PubMed] [Google Scholar]
- 23.Williams NL, Howells AJ, Maxwell A. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 2001;306:969–984. doi: 10.1006/jmbi.2001.4468. [DOI] [PubMed] [Google Scholar]
- 24.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Modular structure of the full-length DNA gyrase B subunit revealed by small-angle X-ray scattering. Structure. 2007;15:329–339. doi: 10.1016/j.str.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Kampranis SC, Maxwell A. Conformational changes in DNA gyrase revealed by limited proteolysis. J. Biol. Chem. 1998;273:22606–22614. doi: 10.1074/jbc.273.35.22606. [DOI] [PubMed] [Google Scholar]
- 26.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 28.Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 29.Seidel R, Dekker C. Single-molecule studies of nucleic acid motors. Curr. Opin. Struct. Biol. 2007;17:80–86. doi: 10.1016/j.sbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl Acad. Sci. USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]