Abstract
RNA structure plays a fundamental role in internal initiation of translation. Picornavirus internal ribosome entry site (IRES) are long, efficient cis-acting elements that recruit the ribosome to internal mRNA sites. However, little is known about long-range constraints determining the IRES RNA structure. Here, we sought to investigate the functional and structural relevance of the invariant apical stem of a picornavirus IRES. Mutation of this apical stem revealed better performance of G:C compared with C:G base pairs, demonstrating that the secondary structure solely is not sufficient for IRES function. In turn, mutations designed to disrupt the stem abolished IRES activity. Lack of tolerance to accept genetic variability in the apical stem was supported by the presence of coupled covariations within the adjacent stem–loops. SHAPE structural analysis, gel mobility-shift and microarrays-based RNA accessibility revealed that the apical stem contributes to maintain IRES RNA structure through the generation of distant interactions between two adjacent stem–loops. Our results demonstrate that a highly interactive structure constrained by distant interactions involving invariant G:C base pairs plays a key role in maintaining the RNA conformation necessary for IRES-mediated translation.
INTRODUCTION
Translation initiation is a key step in the process of protein synthesis (1). In some RNA viruses, exemplified by picornaviruses and hepatitis C virus (HCV), and in a subset of cellular mRNAs which are translated during stress, internal ribosome entry site (IRES) elements drive translation initiation using a cap-independent mechanism (2,3). IRES elements differ in nucleotide sequence, RNA secondary structure and trans-acting factors requirement (2,4,5). This complexity compromises the general understanding of the mechanism of internal initiation, leading to the view that IRES elements may promote translation initiation using a large variety of mechanisms (6–8).
RNA structure plays a fundamental role in IRES-dependent translation initiation (4,9–11), as well as in other processes guided by RNA regulatory elements (12–14). This is illustrated by the fact that compensatory substitutions in base-paired regions tend to conserve the secondary structure during RNA evolution (2,15–17).
Foot-and-mouth disease virus (FMDV) is a picornavirus characterized by a high genetic variability (18). This feature provides crucial information regarding the tolerance to accept nucleotide substitutions all along the viral genome, and particularly, within the highly structured untranslated regions (UTRs) located at each end of the genome (19). The IRES element located at the 5′-UTR of FMDV is organized in structural domains, termed 2–5 in 5′- to 3′-end, which appear to have a division of functions (20). Domains 2, 4 and 5 determine the interaction with RNA binding proteins and various translation initiation factors (eIFs), with the exception of eIF4E (21,22). Domains 4 and 5 are responsible for the recruitment of eIF4G, eIF4B, eIF3 and other IRES binding factors (23,24). Remarkably, domains 4 and 5 do not possess IRES activity by themselves (25), indicating that interaction with these factors is necessary but not sufficient for IRES function.
In contrast to the large number of factors interacting with the distal domains, the central one, termed domain 3, interacts with a reduced number of host factors, such as PCBP2, EBP1, hnRNPK and DAZ-1 (26). This result could be interpreted as the consequence of having a large proportion of its nucleobases hidden within a compact RNA 3D structure. In fact, secondary structure determination of the FMDV IRES by RNA probing (9,25) revealed that the apical region of domain 3 contains four GC rich stems that join at a cruciform structure. At the base of this domain, a long stem interrupted by internal bulges serves as a bridge to upstream and downstream IRES domains. Mutational analysis has shown that conserved motifs in the RNA structure of domain 3 play a crucial role in IRES-dependent translation (20,27–29). However, the 3D structure of this domain is still unknown.
Picornavirus IRES elements are remarkably long regions compared to other viral IRES, including those of HCV and dicistroviruses (2,30,31). Determining the structural organization of long IRES elements is the first step to begin to understand the efficient mechanism of internal initiation driven by these elements. Their long size and flexibility is well suited to structural analysis by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (32). Although SHAPE probing does not reach the atomic level resolution of NMR or X-ray structural analysis, it has the advantage over the latter approaches of allowing the analysis of long RNA molecules (33) and hence, it sheds light on RNA organization of the entire RNA regulatory element. Recently, we have performed a combined study involving SHAPE probing and microarray-based oligonucleotide accessibility of the FMDV IRES element (34) confirming its modular organization in structural domains.
In this study, we sought to investigate the involvement in IRES function of RNA structural motifs within domain 3 possibly constrained by distant interactions. To this end, we took advantage of the large variability of FMDV RNA sequence to identify invariant regions as well as coupled covariations all along the IRES element, paying special attention to nucleotides within domain 3. Mutational analysis was carried out to verify the biological relevance of an invariant GC-rich apical stem for IRES activity. Subsequently, structural analysis revealed the involvement of this invariant region in maintaining the RNA structure through the generation of distant interactions between two adjacent and conserved stem–loops of domain 3. The reciprocity of the structural changes observed in mutants affected in these stem–loops further supported the existence of a highly interactive RNA structure within the apical region of domain 3 that performs an essential function during internal initiation.
MATERIALS AND METHODS
Constructs
The constructs expressing the IRES RNA (nucleotides 1–462) of FMDV C-S8 or its domain 3 alone (nucleotides 84–297) were previously described (35). Mutations in G195, G196, G203, C204 and C205 were generated by mutagenic PCR using oligonucleotides S1 (CTTTTGGTTCCGTGGGTCCTTGTTAC), S2 (GTGGGCGTAACTTTTGGCCCCGTG), S3 (GGCGTCCCTTTTGGCCCCGTG), S4 (GTCCCTTTTGGGGCCGTG), S5 (GGCGTGGTTTTTGGCCCCGTG) as described (29).
Sequence covariation analysis of the IRES element
Sequence alignment
FMDV sequences were retrieved from the GenBank using Blast (http//blast.ncbi.nlm.nih.gov/Blast.cgi) using the FMDV C-S8 IRES sequence as query. Duplicates and incomplete sequences were removed and each sequence was given a unique identifier. FASTA-formatted sequences were first aligned using CLUSTALW (http//www-ebi.ac.uk/Tools/clustaw2/index.html). RNA secondary structure features were used to align the sequences.
Alignment position classification and error estimation
Columns of the alignment were classified as fixed (F), variable (V) or gapped (G), depending on whether they contain the same nucleotide, different nucleotides or a gap, respectively. From all paired bases in stems that were protected by SHAPE in both positions, we considered those composed of one fixed and one variable column (VF or FV). For these pairs, we counted the substitution frequencies of the variable position along the alignment. An error rate was estimated by calculating the fraction of non-conservative changes, i.e. substitutions that would disrupt the canonical base pairing (A:U, G:C or G:U). This resulted in an average of three non-canonical changes in base-paired SHAPE-protected positions. Thus, we can assume that variable positions with three or less non-conservative changes are a result of sequencing errors and can be considered as fixed. Accordingly, 95 variable columns were re-classified as fixed, as their variation was within the estimated error rate.
Mutual information
In order to determine the covariation between pairs of positions, we computed a measure of association, called mutual information (MI):
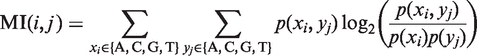 |
where 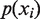 is the probability of finding the nucleotide
is the probability of finding the nucleotide 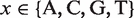 in position
in position  ,
, 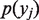 is the probability of finding the nucleotide
is the probability of finding the nucleotide 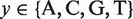 in position
in position 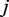 and
and 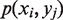 is the joint probability of simultaneously finding a particular combination of nucleotides
is the joint probability of simultaneously finding a particular combination of nucleotides 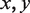 in positions
in positions 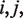 respectively. MI was calculated for all possible pairs of variable columns (VV) in the multiple alignment.
respectively. MI was calculated for all possible pairs of variable columns (VV) in the multiple alignment.
Random expectation of covariation
In order to determine which pairs had an MI value significantly different from random expectation, we generated 300 randomized sets by vertical shuffling all the columns of the alignment. This maintains the proportion of nucleotides for each column, but shuffles the co-occurrences in each row. The distribution of the MI values in these randomized alignments was used to calculate the corresponding z-scores. Variable pairs (VV) were therefore classified as covariant, if z-score > 2.96, or independent, otherwise. Moreover, each variable pair (VV) was further classified as conservative or non-conservative, according to whether the fraction of changes that preserved the canonical base pairing was >50% or <50%, respectively.
RNA structure modeling
PDB RNA structure models were performed using Mc-Fold (www.major.iris.ca/MC-Pipeline) to obtain the secondary structure in bracket format, followed of 3D MC-SYM generator to produce the PDB RNA structure. RNA structure was visualized using Swiss-PdbViewer. The first PDB structure obtained with the MC-SYM generator using the most stable energy RNA structure according to Mc-Fold was used to depict the model of each IRES sequence.
IRES activity assays
Relative IRES activity was quantified as the expression of luciferase normalized to that of chloramphenicol acetyltransferase (CAT) from bicistronic mRNAs as described (36) in transfected BHK-21 monolayers. Experiments were performed on triplicate wells and each experiment was repeated at least three times.
In vitro transcription
Plasmids were linearized to generate domain 3 or the full-length IRES using SmaI or XhoI, respectively. Transcription was performed for 1 h at 37°C using 1000–3000 U of highly purified T7 RNA polymerase in the presence of 10–15 µg of linearized DNA template, 40 mM Tris–HCl, 50 mM DTT, 0.5 mM rNTPs, as described (34).
SHAPE analysis
RNA (0.5 pmols) was treated with N-methylisatoic anhydride (NMIA) as described (34,37). For primer extension, equal amounts of NMIA-treated and untreated RNAs (10 µl) were incubated with 0.5 µl of the appropriate antisense 5′-end 32P-labeled primer (5′-CTACGAAGCAACAGTG, 5′-CCCGGGTGTGGGTACC, 5′-GGAATGGGATCCTCGAGCTCAGGGTC). Primer extension was conducted in a final volume of 15 µl containing reverse transcriptase (RT) buffer (50 mM Tris–HCl, pH 8.3, 3 mM MgCl2, 75 mM KCl, 8 mM DTT) and 1 mM of each dNTP. The mix was heated at 52°C for 1 min, prior to addition of 100 U of Superscript III RT (Invitrogen) and incubation at 52°C for 30 min. cDNA products were fractionated in 6% acrylamide, 7 M urea gels, in parallel to a sequence obtained with the same primer. For SHAPE data processing, the intensities of RT-stops were quantified as described (34). Data from three independent assays were used to calculate the mean (±SD) SHAPE reactivity.
Gel-shift assays
For RNA–RNA interactions, the uniformely [α-32P]-CTP labeled GNRA hairpin RNA (nucleotides 160–196) (38) was incubated with increasing concentrations of unlabeled domain 3 RNAs (50–1000 nM) in 50 mM sodium cacodylate, pH 7.5, 300 mM KCl, 10 mM MgCl2 (35,39). RNA–RNA complexes were allowed to form for 90 min at 37°C and immediately analyzed by electrophoresis in native acrylamide gels supplemented with 2.5 mM MgCl2 as described (38).
Microarray hybridization and data analysis
DNA oligonucleotides complementary to the IRES region of FMDV were described in (34). Mutant RNAs encompassing domain 3 were fluorescently labeled with Alexa 647 using the Ulysis 647 kit (Invitrogen). Microarrays were prehybridized and hybridized as described (34). Data were retrieved using the Genepix pro 6.0 software. The differential capacity of transcripts for antisense oligonucleotide hybridization was measured in three independent experiments as described (34).
RESULTS
Identification of an invariant apical stem within the IRES element
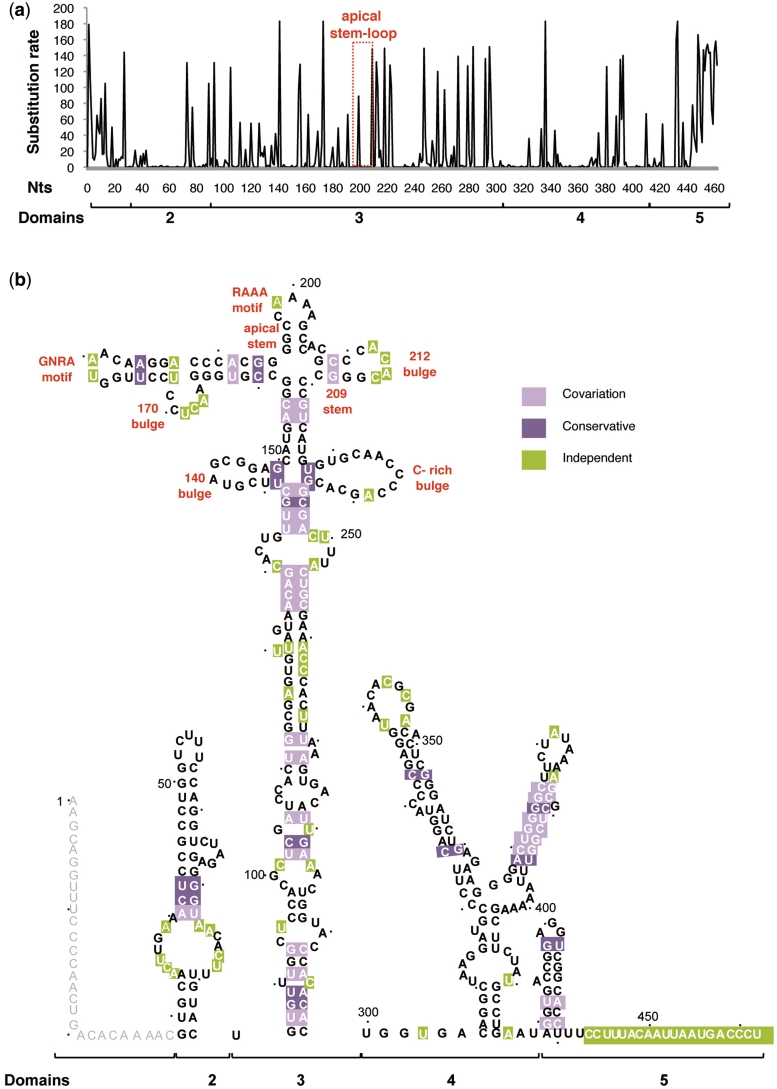
The alignment of nucleotide sequences belonging to 183 FMDV RNA isolates deposited in databases readily indicated an extensive degree of sequence heterogeneity across the IRES element (Figure 1a). In addition to regions that accumulated a large number of substitutions, we observed others in which coupled nucleotide covariation led to compensatory changes (Table 1) that maintained the RNA secondary structure and, interestingly, specific nucleotide tracts that were less tolerant to substitutions (Figure 1b).
Figure 1.
Sequence variability of the IRES element in FMDV RNA. (a) The total number of changes found in 183 aligned IRES sequences is plotted against the nucleotide position. Location of the residues conforming the apical stem–loop, including the variable R residue of the RAAA motif, is indicated by a rectangle. (b) Sequence heterogeneity maintains the secondary structure of the FMDV IRES. Invariant nucleotides in the IRES secondary structure are marked in bold. Covariant and conservative nucleotide changes are depicted by light and dark violet squares, respectively. Independent substitutions are marked by green squares. Nucleotide positions are denoted by dots 10 nt apart. The most relevant motifs referred to in the text are indicated. Distribution of the IRES in domains (2–5) is indicated below each panel.
Table 1.
Nucleotide substitutions leading to covariations and conservative changes
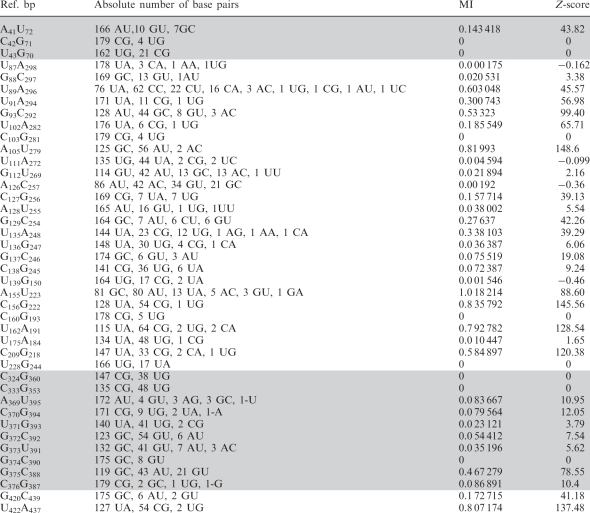 |
Ref. bp indicates the sequence of the base pair in the reference IRES sequence. MI indicates the probability of finding simultaneously a particular combination of nucleotide at a given position. Z-score > 2.96 indicates the pairs with MI significantly different from random changes.
The region that accommodated the higher number of changes was mapped to the single-stranded region at the 3′-end of domain 5 (Figure 1a), although the polypyrimidine tract is conserved (Supplementary Table S1). Similarly, other nucleotide variations located in loops within the IRES secondary structure (Figure 1b) corresponded to conserved motifs, as illustrated by the GNRA and the RAAA motifs in the apical region of domain 3 as well as the pyrimidine-rich motif in the apical loop of domain 2.
Analysis of covariation between pairs of positions with MI values significantly different from random expectation (z-score > 2.96) (Table 1) readily demonstrated the presence of paired regions in the IRES element (Figure 1b) that matched stems according to RNA probing (25,34). Covariation between variable positions within domain 3 occurs more frequently than between these and variable positions of the other domains (Supplementary Table S2), reinforcing the idea of a modular organization in the IRES element. In this regard, covariation pairs for which any of the positions had 3 or less changes were discarded, since these could be fortuitous or due to sequencing errors.
We anticipate that the invariant regions may have been subjected to selection pressure to keep their primary sequence, because of their involvement in RNA–protein interactions or in maintaining the correct 3D RNA structure. Indeed, we observed that invariant regions tend to accumulate around nucleotides previously identified as the target site of RNA binding proteins involved in IRES activity. Thus, the upper stem–loop of domain 2 (nucleotides 45–68) and the stem at the base of domain 4 (nucleotides 306–320 and 396–416) (Figure 1b) provide the binding site for PTB and eIF4G, respectively (40,41).
The apical region of domain 3 contains three invariant regions (Figure 1b), encompassing nucleotides 140–150 (the 140 bulge), 195–205 (the apical stem–loop, that includes the apical stem and the RAAA motif), and 229–243 (the C-rich bulge). While the latter is a candidate sequence to interact with poly(rC) binding proteins and Ebp1 (26,42,43), proteins recognizing the other two invariant regions have not been identified. Furthermore, RNA probing carried out with substitution mutants of the conserved GNRA motif evidenced apical stem–loop reorganization leading to the disruption of the stem, despite the mutation being located in a distant sequence (34,38). These data, together with results of the covariation analysis, led us to focus our attention to the invariant sequence of the apical stem of domain 3 as a region contributing to an RNA structure crucial for IRES function.
The conserved apical stem uncovers a determinant of IRES activity
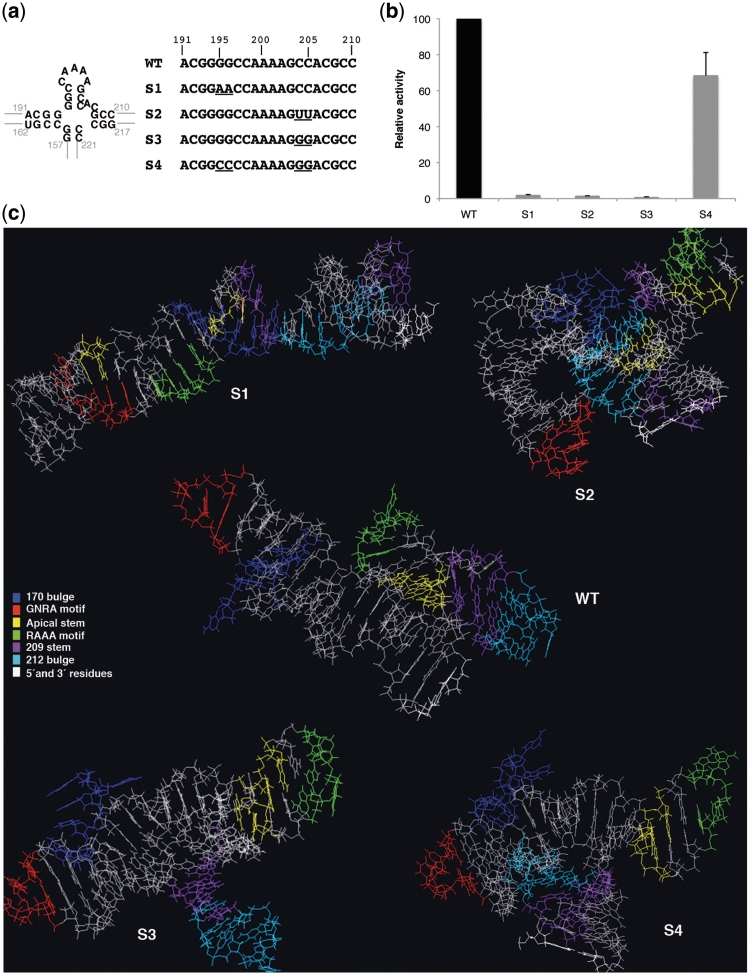
To determine the biological relevance of the invariant apical stem of domain 3 for IRES activity, we generated a set of mutants aimed at modifying both, the primary sequence and the secondary structure of this region (Figure 2a). The S1 mutant carrying a double substitution G195G196 to AA, designed to disrupt the canonical G:C base pairs of the wt IRES, led to a severe loss of IRES activity (1.8% with respect to the wt IRES) (Figure 2b). Similarly, a double substitution on the opposite arm of the stem, C204C205 to UU (mutant S2) also abrogated IRES activity (1.4%), suggesting that G:U base pairs were not formed or, conversely, they were not sufficient to stabilize this short stem.
Figure 2.
Mutational analysis of the invariant apical stem. (a) Nucleotide substitutions present in S1–S4 mutants are represented below the wild-type C-S8 FMDV IRES sequence (nucleotides 191–210). A diagram of the wild-type secondary structure of this region is shown (left). (b) Relative IRES activity, determined as the ratio of luciferase to CAT in BHK-21 cells transfected with plasmids of the form CAT-IRES-luciferase made relative to the activity obtained with the wt IRES. Values correspond to the mean of three independent assays. Errors bars, SD. (c) PDB RNA structure prediction by Mc-fold pipeline of the apical region of domain 3 (nucleotides 155–223) bearing wt and S1–S4 mutant sequences. Location of the loops and stems referred to in the text are depicted by the color code listed in the legend.
To confirm that disruption of the G:C base pairs of this stem lead to IRES inactivity, we generated a mutant carrying the double substitution C204C205 to GG (S3). This mutant turned out to be severely defective (0.7%) (Figure 2b). The strong reduction of IRES activity exhibited by these three sets of mutations indicated that the primary sequence on each side of the stem and, most likely, its secondary RNA structure are important for IRES activity. To further confirm this point, we generated a fourth mutant, S4, with a compensatory mutation relative to S3. As shown in Figure 2b, the second-site mutant S4, which replaced two consecutive G:C base pairs by C:G, partially restored IRES activity (68.4% of the wt IRES). This result suggests that the maintenance of the RNA secondary structure of this apical stem is required for IRES function, although the full activity is also dependent on its correct primary sequence. As a control, a construct that harbors a single nucleotide substitution of G203A (S5) was as active as the wt IRES (data not shown), demonstrating that increasing the A-rich loop size is compatible with IRES activity.
RNA structure modeling of the apical region of domain 3 fully supports that the nucleotide substitutions induced important changes in the RNA organization of this region (Figure 2c). Specifically, mutations in S1 and S2 IRES disrupted the apical stem as well as the 209 stem. Mutations in S3 IRES led to a rearrangement of the apical stem in which C197C198 are base paired to the mutated G204G205. Additional changes in the relative orientation of the GNRA motif and the 170 bulge occurred in all mutants, with the peculiarity that the GNRA motif in S1 RNA is close to nucleotides 195–196, and residues belonging to 170 bulge are in the vicinity of nucleotides C204-G208.
We conclude that the invariant stem of the apical stem–loop plays a crucial role for IRES activity, and that it has a strong preference for G:C base pairs. Their substitution by C:G retained activity although this IRES mutant did not reach the same activity levels than the wt. These results are in strong agreement with the lack of genetic variability found in the apical stem–loop within the IRES sequence alignment.
RNA structural analysis reveals the specific need for G:C base-pairs for IRES function
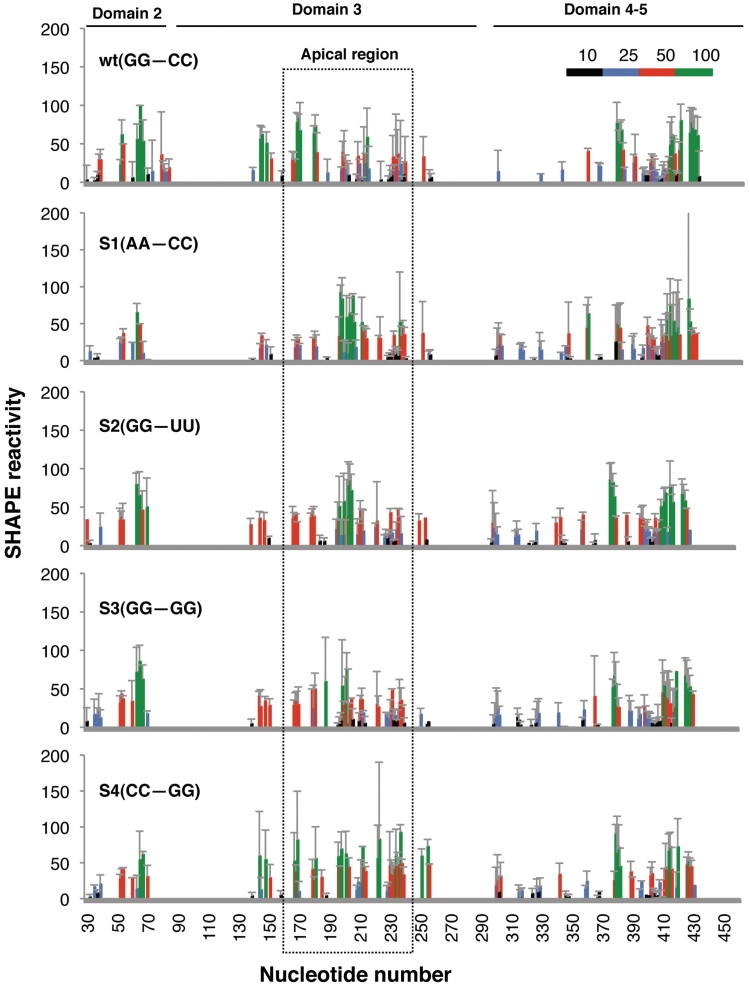
Next, to establish a direct relation between RNA structure and activity of IRES mutants carrying mutations in the apical stem, we performed structural analysis by SHAPE, which allows the study of long, flexible RNA molecules in solution (32,33). RT extension products obtained with primers spaced along the RNA sequence allowed overlapping readings of the entire IRES region (34). For this, cDNA products derived from the untreated and NMIA-treated RNAs were resolved in denaturing acrylamide–urea gels in parallel to a sequence ladder obtained with the same primer. The intensity of each RT-stop band was normalized to that of the full-length product in the corresponding gel lane, and the background values of the untreated RNA were subtracted from the respective RT-stop intensity yielded by NMIA-treated RNA. In all cases, the mean of three independent assays was calculated from the reactivity of each nucleotide, relative to the most reactive nucleotide (set to 100%). The resulting color-coded profiles (Figure 3) revealed gross modifications of SHAPE reactivity in domain 3 of the mutants S1–S4 with respect to that of the wt RNA. This was particularly evident in residues 150–250 encompassing the apical region, which showed increased SHAPE reactivity. However, SHAPE reactivity was only slightly modified in domains 2, 4 or 5. Furthermore, changes in the SHAPE reactivity of domain 3 accumulated in similar positions when the transcripts encompassing either the entire IRES or the domain 3 alone (data not shown) were analyzed, suggesting that the RNA reorganization mainly affected the central domain.
Figure 3.
SHAPE reactivity of apical stem IRES mutants. Values of SHAPE reactivity at each individual nucleotide position correspond to the mean reactivity (±SD) of three independent assays. RNAs, treated with NMIA or untreated, were subjected to primer extension analysis conducted with 5′-end labeled primers. The intensity of each band was normalized to the full-length cDNA product detected in the corresponding gel lane after subtraction of the corresponding background RT-stop signal in the untreated RNA. Nucleotide positions are indicated on the x-axis. The SHAPE reactivity is depicted using color-coded bars. Position of the apical region is indicated by a broken line rectangle.
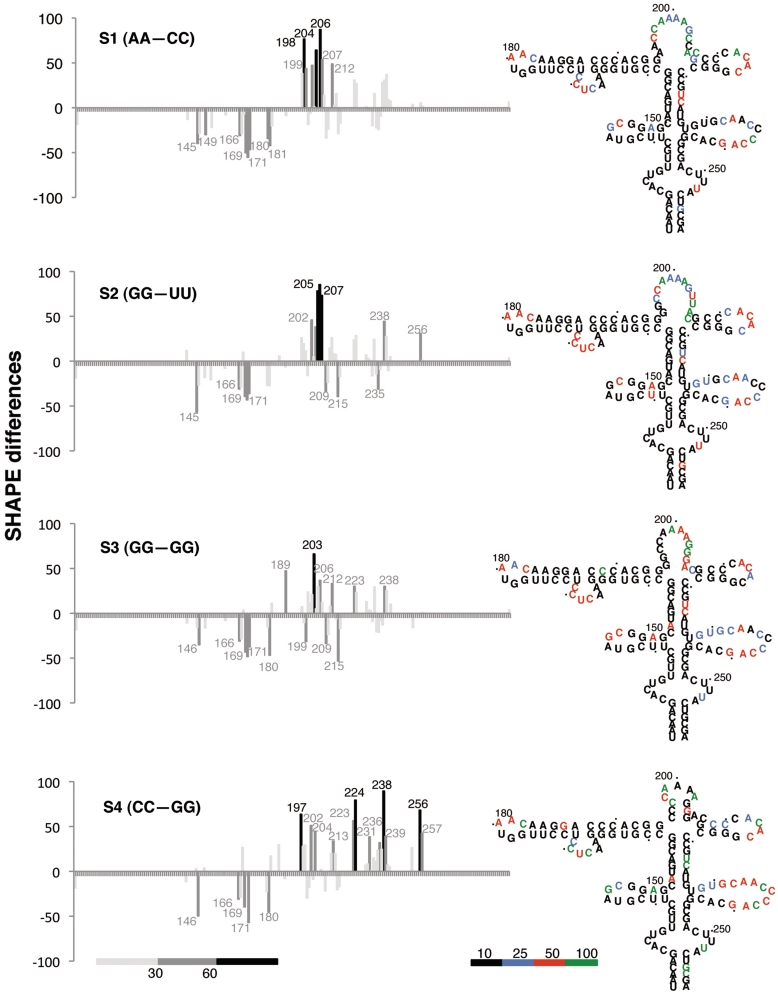
The differences in SHAPE reactivity of the wt RNA (Supplementary Figure S1) with respect to each IRES mutant are depicted in Figure 4. Specifically concerning the S1 mutant RNA, there was a remarkable increase in SHAPE reactivity in residues 198–207, supporting the notion that nucleotides corresponding to the apical stem were not engaged in base pairs (Figure 4). Concomitant with this increase, a strong decrease of reactivity was observed in nucleotides 169–171 and 181, revealing a protection of the 170 bulge and the GNRA motif, respectively. Therefore, these results suggest a functional interaction between residues located in two stem–loops of the apical region, with a distinctive feature: an increase in SHAPE reactivity of the apical stem–loop was accompanied by a decrease in reactivity of nucleotides belonging to the GNRA hairpin, including the 170 bulge. These results are also in agreement with the RNA structure model of S1 IRES (Figure 2c) that not only depicts the mutated nucleotides 195–196 apart from nucleotides 204 to 205, but also predicts a reorganization of the GNRA motif and the 170 bulge.
Figure 4.
Impact of the apical stem sequence composition on SHAPE reactivity differences. SHAPE difference plots of each mutant domain 3 relative to the wt RNA. Nucleotides with absolute changes in SHAPE reactivity >60% are depicted in black, while those between 30 and 60% are marked in dark grey. The secondary structure model of each IRES mutant, with nucleotides colored as in Figure 3 to reflect their mean SHAPE reactivity, is shown on the right. The reference mean SHAPE reactivity of the wt RNA is shown in Supplementary Figure S1.
According to SHAPE differences of the S2 mutant RNA, nucleotides 202–207 were more reactive to NMIA (Figure 4), indicating that the U residues inserted by site-directed mutagenesis were unpaired, which is in agreement with the RNA structure model of S2 IRES mutant (Figure 2c). In addition, and similar to S1 RNA, a decrease in the reactivity of the 170 bulge was observed (Figure 4). Thus, we conclude that while the substitutions introduced in S1 and S2 RNAs affected opposite sides of the stem, they induced similar changes in RNA structure organization that correlated to analogous negative effects on IRES activity.
The results derived from SHAPE reactivity of the S3 RNA pointed again to the apical region (Figure 4). It is worth noting that the decrease in SHAPE reactivity of nucleotides belonging to the GNRA hairpin was similar to the effect observed in S1 and S2 RNA. Hence, disruption of the G:C base pairs within the apical stem led to a distant effect that reached the adjacent stem–loops, reinforcing the occurrence of long-distant interactions between these stem–loops of the apical region. Further supporting this conclusion, SHAPE analysis of the S4 mutant RNA revealed that the RNA structure found in the wt IRES (Supplementary Figure S1) was partially restored, although SHAPE differences were observed within the region 146–257 (Figure 4). Therefore, destabilization of the invariant apical stem (mutants S1, S2 and S3) impaired IRES activity. Conversely, compensatory mutations that preserved the secondary RNA structure (mutant S4) only partially restored IRES activity, since the wt IRES carrying the GC stem performed better than that of the S4 mutant IRES.
We conclude that the invariant bases that compose the apical stem of the IRES play a crucial role in determining the RNA structural organization affecting the adjacent GNRA hairpin that, in turn, has also a strong influence in the ability of this IRES element to drive internal initiation of translation.
Long-distance interactions within domain 3 and local RNA accessibility depends on the sequence composition of the apical stem
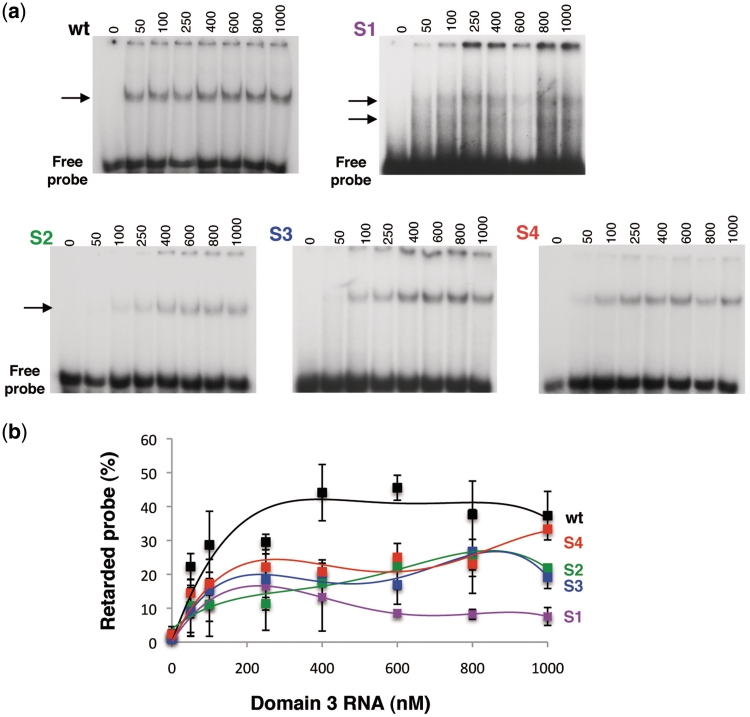
We have previously identified long-range RNA–RNA interactions within domain 3, which are sequence specific, as well as Mg2+ and RNA concentration dependent (38). The changes in RNA structure observed in the GNRA hairpin and the apical stem of S1–S4 mutants strongly suggested that the mutations introduced in the apical stem could generate different conformations of domain 3, profoundly modifying long-range RNA–RNA interactions. To test the hypothesis that distant RNA–RNA interactions could be affected in the S1–S4 mutants, we determined by gel mobility-shift assays the capacity of a labeled transcript encompassing the GNRA hairpin (corresponding to nucleotides 160–196) to interact with unlabelled transcripts of domain 3 bearing either the wt or the mutated S1–S4 sequences (Figure 5a). The mutant RNAs responded in different manners (Figure 5b), with Kd ranging between 112.3 nM (wt), 271.7 (S1), 234.3 (S2), 279.0 (S3) and 173.1 (S4). Strikingly, S1 RNA showed a reduced capacity to establish long-range RNA interaction with the GNRA hairpin, although, at the same time, it has acquired the property to form two retarded complexes clearly distinguishable in long exposures (Figure 5a). This result may imply the presence of at least two conformations of this mutant RNA, which very weakly interact with the GNRA hairpin. S2 and S3 mutant RNAs also showed a reduced capacity to bind the labeled GNRA hairpin probe all along the concentration range tested. In turn, the S4 RNA that bears the compensatory mutation evidenced an intermediate capacity to interact with the GNRA hairpin probe (Figure 5b). Consistent with IRES activity and RNA structural analysis, the binding efficiency of the S4 domain 3 was greater than that of the other mutants, achieving values similar to the wt RNA only at the highest concentration (1000 nM) used in the assay.
Figure 5.
Mobility-shift binding assays of domain 3 RNAs with the GNRA hairpin. (a) Representative examples of gel-shift assays for each construct, wt and S1 to S4 mutant RNAs. Gel-shift assays were carried out using 32P-labeled GNRA hairpin (corresponding to nucleotides 160–196 of domain 3) (50 nM) as probe and the wild-type or mutant unlabeled domain 3 RNAs (0–1000 nM). The position of the retarded complex is depicted by an arrow. Note that a longer exposure is shown for S1 RNA to allow the detection of two weak retarded complexes. (b) The percentage of retarded probe calculated from duplicate assays was plotted against the unlabeled RNA concentration. Error bars, SD.
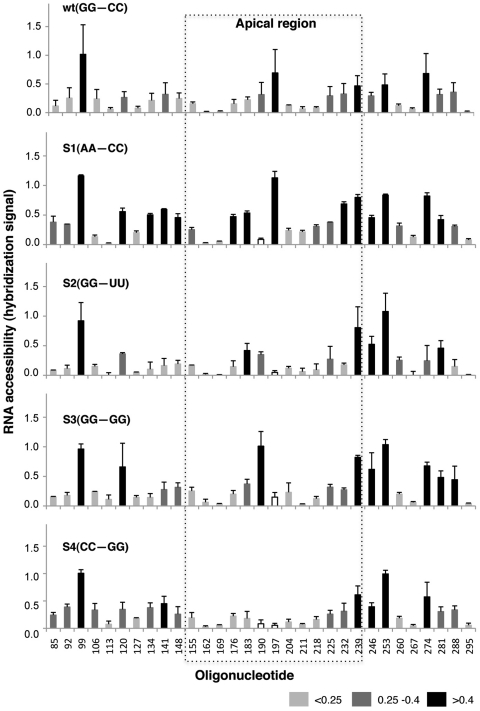
The results observed with S1 mutant suggested that this IRES region could adopt different conformation depending on the sequence of the apical stem. Thus, to gain additional information about the local RNA structure of the IRES mutants, we measured the accessibility of domain 3 RNAs to a panel of customized overlapping antisense DNA oligonucleotides covering the entire IRES sequence printed on microarrays (34). RNA hybridization was conducted using fluorescent-labeled transcripts carrying the indicated mutations in the apical stem under the same ionic conditions and temperature used for SHAPE probing. The hybridization signal displayed by each mutant RNA revealed changes in RNA accessibility (Figure 6). Interestingly, S1 RNA exhibited an overall higher accessibility in the microarray-based assay than the wt RNA, demonstrating a less tight RNA structure that particularly affected residues complementary to oligonucleotides 134–148, 176–183 and 232–253. In contrast, changes in RNA accessibility of the S2, S3 and S4 mutants were concentrated in the region complementary to oligonucleotide 253. A decrease in accessibility to oligonucleotides 190 and 197, complementary to nucleotides 190–211, can be explained by the presence of two substitutions in the RNA sequence of these IRES mutants. These results strongly suggest that domain 3 consists of a highly interactive structure in which its apical stem and the GNRA stem–loop are in close structural and functional connection.
Figure 6.
Hybridization of IRES transcripts to antisense oligonucleotides printed on microarrays. Hybridization signal of the fluorescent-labeled domain 3 transcripts plotted against each oligonucleotide (mean ± SD) averaged from three independent assays. The array contains 14-nt long oligonucleotides (termed 1–449, by the position on the IRES sequence complementary to the 3′-end of the primer), overlapping each 7 nt within domain 3 (34). Changes in accessibility of mutant IRES to oligonucleotides 190 (S1, S4) and 197 (S2, S3, S4), depicted with white bars, are likely due to mismatches in the sequence of these RNAs with the sequence printed in the microarray. Position of the apical region is indicated by a broken line rectangle. Differences in accessibility are depicted in black (>0.4), grey (range 0.4–0.25) and light grey (<0.25).
In summary, our results reinforce the notion that the nucleotide composition of the apical stem performs a crucial role in maintaining the correct RNA conformation of domain 3. Using complementary approaches such as functional analysis, SHAPE reactivity, gel-shift and microarray-based RNA accessibility, we have established the relevance of the RNA structural organization for the function of the invariant stem of domain 3 in IRES-mediated translation.
DISCUSSION
The modular organization of picornavirus IRES is consistent with a distribution of functions among the different domains. The central domain of the IRES, termed domain 3, is a self-folding region since it adopts a very similar RNA structure organization when it is expressed alone or in the context of the entire IRES (34). Moreover, the local RNA structure of this domain plays a critical role during IRES-dependent translation (9,38). SHAPE reactivity data provided evidence for the existence of stem–loops whose structural conformation depends on distant interactions within this domain, involving residues of the GNRA motif (34). These results point to the existence of a structural element in the apical region of domain 3 that performs an essential role during internal initiation. In support of a general implication of this structural feature to picornavirus IRES performance, not only FMDV but also encephalomyocarditis virus (EMCV) IRES-dependent translation is impaired by nucleotide substitutions in the conserved GNRA motif (27,28). Thus, the RNA structure adopted by the apical region of domain 3 could constitute a signature of type II IRES elements, typically found in picornavirus genomes.
The genetic variability of the FMDV IRES analyzed in 183 isolates indicated certain tolerance to accept changes in the sequence of domain 3 (Figure 1b). Despite this overall variability, the GNRA and the RAAA motifs are fully conserved (Supplementary Table S1). We report here that the G:C base pairs that hold these motifs are also invariant. This suggested to us that they perform a key role in maintaining the functional IRES structure. Furthermore, the sequence of the internal 170 bulge within the GNRA hairpin tolerates substitutions, but not insertions or deletions (Supplementary Table S1).
We have shown that the G:C base pairs that conform the apical stem–loop have a primary relevance for IRES activity. Importantly, disruption of this stem occurred concomitantly with a structural rearrangement of the apical region, affecting two adjacent stem–loops (Figures 2c and 4). The results of SHAPE reactivity of wild-type and mutant IRES RNAs strongly support a direct effect of residues G195–G196 and C204–C205 in governing the RNA conformation of the active IRES element. Specifically, SHAPE reactivity, gel mobility-shift and RNA accessibility displayed by the S1 mutant (where the G195G196 were substituted by AA) supported a change in RNA conformation of the apical region affecting distant regions in the secondary structure of domain 3. The results derived from the study of S2 (CC substituted by UU) and S3 (CC changed to GG) RNAs demonstrated that the disruption of the apical stem induced the reorganization of the entire apical region of domain 3, including the GNRA hairpin. Unexpectedly, although the substitutions in S4 RNA partially restored IRES activity, the organization of the apical region was modified, as indicated by differences in SHAPE reactivity of the GNRA stem–loop and the apical stem–loop. Therefore, this result further confirmed the biological relevance of the GG:CC composition of the apical stem, in agreement with the lack of tolerance to accept sequence changes.
A common feature observed in the IRES mutants studied here is that an increase in reactivity of nucleotides belonging to the apical stem–loop (195–205) is coupled to a decrease in reactivity of nucleotides 169–171 and 181, that belong to the 170 bulge and the GNRA motif, respectively. This is specifically consistent with the reorganization predicted in the S1 RNA structure (Figure 2c). The implications of our mutational analysis suggest a cross-talk between distant nucleotides of domain 3, located at the apical stem and the GNRA hairpin, that share the property of being conserved at the structural level between picornavirus IRES classified as type II (2,43). Specifically, the observation that S1 mutant RNA severely affected the capacity to form distant interactions (Figure 5a), and that S4 mutant only partially restored IRES activity (Figure 2b) strongly suggests that the base pair composition of this stem determines the architecture of this IRES region. This line of evidence prompted us to consider the possibility of whether this reorganization is due to the disruption of distant interactions.
Changes in the efficiency and pattern of long-distant interactions within domain 3 have been demonstrated by gel-shift analysis of the GNRA hairpin with specific mutant IRES transcripts (Figure 5a and b) (38) and by RNA structure probing of mutants in the conserved GNRA motif (9,34). Previous studies have shown that the GNRA motif of FMDV, EMCV and poliovirus IRES adopts a tetraloop conformation at the tip of a stem–loop (9,44,45). GNRA tetraloops are frequently involved in tertiary interactions (46,47). In particular, it has been shown that GYRA tetraloops interact with GG:CC receptors when R corresponds to A (as in the GUAA sequence of the wt IRES sequence used here), whereas it can accept CU:AG receptors when the R is a G (48). The GNRA motif of the FMDV IRES shows a strong preference for GUAA, but it tolerates sequence variation to GUGA, GCAA, GAGA and GCGA (Supplementary Table S1) (34). Thus, covariation analysis of the IRES region could give us a hint about the location of the putative receptor. However, we could not detect signals of a GNRA receptor based solely on the genetic variability analysis. Although it can be argued that this could be due to insufficient number of analyzed samples, we cannot rule out the possibility of alternative binding sites or the existence of different RNA conformations as it has been observed in various RNAs (12,49).
The lack of variability of the apical stem, supported by the covariation observed in the adjacent stems, pointed toward its relevant contribution to IRES activity. Our results of mutational analysis of this invariant stem revealed a better performance of G:C than C:G base pairs, demonstrating that it is not only the secondary structure of this region, but its 3D RNA conformation is what determines efficient IRES activity. Furthermore, changes in IRES activity were accompanied by a structural reorganization, as revealed by SHAPE reactivity and gel mobility-shift interactions. A region effectively modified, in addition to the mutated bases, includes the 170 bulge, the GNRA motif, the stem 209, loop 140 and 238. Interestingly, genetic variability of the 170 bulge showed that whereas its sequence accepts certain nucleotide substitution, insertions or deletions were not observed (Supplementary Table S1). It is worth noting that the overall RNA organization of the 170 bulge within the GNRA hairpin may resemble that of the HCV IRES domain II (50).
Importantly, we have found a modification of RNA structure affecting multiple nucleotides of the apical region in all mutants analyzed here. Additionally, we have shown that conformational changes in RNA structure play a pivotal role in IRES function. Taken together, our results demonstrate that the active conformation of domain 3 consists of an interactive structure constrained by distant interactions, in which two invariant G:C base pairs within the apical stem–loop play a key role in maintaining the correct RNA structure, likely interacting with the GNRA hairpin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants from Ministerio de Ciencia e Innovación (BFU2008-02159, CSD2009-00080 to E.M.S.); Institutional grant from Fundación Ramón Areces. Work at Universitat Pompeu Fabra was supported by grant BIO2008-01091. Work at Centro de Astrobiología was supported by grants BIO2007-67523, BIO2010-20696, EUI2008-00158; CSIC (grant 200920I040); European Union and Comunidad de Madrid. CIBERehd is funded by the Instituto de Salud Carlos III. Funding for open access charge: CSD-2009-0080, BFU2008-02159.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to I. Luque for help in the generation of PDB RNA structure model, and to C. Gutierrez for critical reading of the manuscript.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallejos M, Deforges J, Plank TD, Letelier A, Ramdohr P, Abraham CG, Valiente-Echeverria F, Kieft JS, Sargueil B, Lopez-Lastra M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr189. April 10; epub ahead of print; doi:10.1093/nar/gkr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 7.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 8.Locker N, Chamond N, Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo X, Wang J, Yu P, Eyler D, Xu H, Starich MR, Tiede DM, Simon AE, Kasprzak W, Schwieters CD, et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc. Natl Acad. Sci. USA. 2010;107:1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat. Struct. Mol. Biol. 2009;16:255–264. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorodkin J, Hofacker IL, Torarinsson E, Yao Z, Havgaard JH, Ruzzo WL. De novo prediction of structured RNAs from genomic sequences. Trends Biotechnol. 2010;28:9–19. doi: 10.1016/j.tibtech.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkhout B, Das AT. Virus evolution as a tool to study HIV-1 biology. Methods Mol. Biol. 2009;485:436–451. doi: 10.1007/978-1-59745-170-3_30. [DOI] [PubMed] [Google Scholar]
- 17.Smith DB, Mellor J, Jarvis LM, Davidson F, Kolberg J, Urdea M, Yap PL, Simmonds P. Variation of the hepatitis C virus 5' non-coding region: implications for secondary structure, virus detection and typing. The International HCV Collaborative Study Group. J. Gen. Virol. 1995;76 (Pt 7):1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 18.Domingo E, Escarmis C, Martinez MA, Martinez-Salas E, Mateu MG. Foot-and-mouth disease virus populations are quasispecies. Curr. Top Microbiol. Immunol. 1992;176:33–47. doi: 10.1007/978-3-642-77011-1_3. [DOI] [PubMed] [Google Scholar]
- 19.Serrano P, Pulido MR, Saiz M, Martinez-Salas E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- 20.Serrano P, Ramajo J, Martinez-Salas E. Rescue of internal initiation of translation by RNA complementation provides evidence for a distribution of functions between individual IRES domains. Virology. 2009;388:221–229. doi: 10.1016/j.virol.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 22.Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Quinto S, Lafuente E, Martinez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pacheco A, Lopez de Quinto S, Ramajo J, Fernandez N, Martinez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009;37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Miragall O, Lopez de Quinto S, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 2009;139:172–182. doi: 10.1016/j.virusres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Pacheco A, Reigadas S, Martinez-Salas E. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics. 2008;8:4782–4790. doi: 10.1002/pmic.200800338. [DOI] [PubMed] [Google Scholar]
- 27.Lopez de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson ME, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Salas E, Regalado MP, Domingo E. Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. J. Virol. 1996;70:992–998. doi: 10.1128/jvi.70.2.992-998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 33.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez N, Garcia-Sacristan A, Ramajo J, Briones C, Martinez-Salas E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology. 2011;409:251–261. doi: 10.1016/j.virol.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Ramos R, Martinez-Salas E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5:1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Salas E, Saiz JC, Davila M, Belsham GJ, Domingo E. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol. 1993;67:3748–3755. doi: 10.1128/jvi.67.7.3748-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1991;65:6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phelan M, Banks RJ, Conn G, Ramesh V. NMR studies of the structure and Mg2+ binding properties of a conserved RNA motif of EMCV picornavirus IRES element. Nucleic Acids Res. 2004;32:4715–4724. doi: 10.1093/nar/gkh805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du Z, Ulyanov NB, Yu J, Andino R, James TL. NMR structures of loop B RNAs from the stem-loop IV domain of the enterovirus internal ribosome entry site: a single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry. 2004;43:5757–5771. doi: 10.1021/bi0363228. [DOI] [PubMed] [Google Scholar]
- 46.Correll CC, Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geary C, Chworos A, Jaeger L. Promoting RNA helical stacking via A-minor junctions. Nucleic Acids Res. 2011;39:1066–1080. doi: 10.1093/nar/gkq748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gultyaev AP, Franch T, Gerdes K. Coupled nucleotide covariations reveal dynamic RNA interaction patterns. RNA. 2000;6:1483–1491. doi: 10.1017/s1355838200990708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.