Abstract
Cytotoxicity of 5-fluorouracil (FU) and 5-fluoro-2′-deoxyuridine (FdUrd) due to DNA fragmentation during DNA repair has been proposed as an alternative to effects from thymidylate synthase (TS) inhibition or RNA incorporation. The goal of the present study was to investigate the relative contribution of the proposed mechanisms for cytotoxicity of 5-fluoropyrimidines. We demonstrate that in human cancer cells, base excision repair (BER) initiated by the uracil–DNA glycosylase UNG is the major route for FU–DNA repair in vitro and in vivo. SMUG1, TDG and MBD4 contributed modestly in vitro and not detectably in vivo. Contribution from mismatch repair was limited to FU:G contexts at best. Surprisingly, knockdown of individual uracil–DNA glycosylases or MSH2 did not affect sensitivity to FU or FdUrd. Inhibitors of common steps of BER or DNA damage signalling affected sensitivity to FdUrd and HmdUrd, but not to FU. In support of predominantly RNA-mediated cytotoxicity, FU-treated cells accumulated ~3000- to 15 000-fold more FU in RNA than in DNA. Moreover, FU-cytotoxicity was partially reversed by ribonucleosides, but not deoxyribonucleosides and FU displayed modest TS-inhibition compared to FdUrd. In conclusion, UNG-initiated BER is the major route for FU–DNA repair, but cytotoxicity of FU is predominantly RNA-mediated, while DNA-mediated effects are limited to FdUrd.
INTRODUCTION
5-fluorouracil (FU) and 5-fluoro-2′-deoxyuridine (FdUrd) are widely used in the treatment of solid cancers, particularly gastrointestinal malignancies. Most commonly used, FU was introduced clinically five decades ago and presently some two million patients are treated each year. However, its major cytotoxic mechanism remains unclear and approximately one-half of the patients fail to respond positively to FU therapy. FU is converted to several active metabolites thought to mediate cytotoxicity directly and indirectly by interfering with RNA and DNA functions (1). Incorporation of 5-fluorouridine triphosphate (FUTP) into RNA causes disruption of rRNAs (2,3), tRNAs (4), snRNA processing (5), RNA exosome function (6) and inhibits the conversion of uridine to pseudouridine in RNA (7). DNA metabolism is perturbed by 5-fluoro-2′-deoxyuridine monophosphate (FdUMP), which inhibits thymidylate synthase (TS) and thereby de novo synthesis of dTMP. This may result in imbalanced nucleotide pools and increased incorporation of dUTP and FdUTP into DNA (8), where FU may pair with either A or G. Genomic uracil and FU are subject to repair by base excision repair (BER) or mismatch repair (MMR). BER of FU in DNA may be initiated by five human uracil–DNA glycosylases. These are uracil-N-glycosylase 1 and 2 (mitochondrial UNG1 and nuclear UNG2), single-strand selective monofunctional uracil–DNA glycosylase 1 (SMUG1), thymine–DNA glycosylase (TDG) and methyl-binding domain 4 protein (MBD4) (9–12). In addition, MMR can process FU:G in a nicked plasmid in vitro and it has also been implicated in repair FU:A base pairs (13). However, the quantitative contribution of MMR and BER, as well as the possible role of individual DNA glycosylases in fluoropyrimidine cytotoxicity remain obscure.
Deficiency in DNA repair is associated with tolerance to fluoropyrimidines in several cell systems, indeed suggesting a role of DNA repair in cytotoxicity. Mechanistically, this may be explained by accumulation of BER intermediates, such as abasic sites (AP-sites) and cleaved DNA strands that are more cytotoxic than the original base lesion (14). Furthermore, long repair tracts produced during MMR may be cytotoxic and mutagenic in cells having imbalanced nucleotide pools (1,15). MMR may also act as DNA damage sensor, inducing G2 arrest following FdUrd treatment (16). Consistently, a FU-tolerant phenotype has been reported for both human and murine cells deficient in MMR (16,17). The evidence linking BER to fluoropyrimidine cytotoxicity is more ambiguous. Mouse embryonic fibroblast (MEF) knockouts of genes encoding TDG or MBD4 display FU tolerance (18,19), and POLβ knockout MEFs showed increased tolerance to FdUrd and other TS-inhibitors (20). Overexpression of a dominant negative APE1 mutant in hamster CHO cells confers 25-fold tolerance to FdUrd and 5-fold to FU (21). In contrast, siRNA knockdown of SMUG1 in MEFs increased sensitivity to FU while Ung−/– MEFs and Ung−/– chicken B cells (DT40) were essentially identical to wild type (22–24). As for human cancer cell lines, the expression levels of UNG was not correlated with sensitivity to TS-inhibitors (25). Furthermore, expression of the UNG-specific inhibitor Ugi did not affect FdUrd or FU sensitivity (26). Also, down-regulation of POLβ had no effect on FU cytotoxicity (27). Whether non-human MEF, CHO and DT40 cells are good models to study the mechanism of fluoropyrimidines in human cancer cells is an open question.
In this article we analyse the relative contribution of BER, including individual DNA glycosylases and MMR to FU–DNA repair in human cancer cell lines. In addition, we investigate the overall significance of the BER pathway in 5-fluoropyrimidine cytotoxicity using BER- and DNA damage signalling inhibitors. The cytotoxic mechanism of FU, FdUrd and 5-fluorouridine (FUrd) were further elucidated by quantifying FU levels in DNA and RNA after exposure, measuring reversal effects by normal deoxyribo- and ribo-nucleosides/-nucleotides, and by analysing inhibition of TS. We found that BER initiated by UNG was the major contributor to FU–DNA repair in vitro and in vivo. The contribution from MMR was surprisingly modest in vivo and limited to FU:G contexts in vitro. However, BER processes did not significantly affect overall FU cytotoxicity, consistent with the majority of earlier reports (23–27). Rather, the cytotoxic mechanisms of FU may be dominated by perturbation of RNA functions, with DNA-mediated effects apparently limited to FdUrd.
MATERIALS AND METHODS
Cell lines, chemicals and enzymes
Human cell lines HeLa S3 (cervical adenocarcinoma), SW480 (colon adenocarcinoma), CX-1 (colon adenocarcinoma), HCT-8 (ileocecal adenocarcinoma), HBL-100 (epithelial non-tumorigenic breast) and AGS (gastric adenocarcinoma) were purchased from ATCC. Cells were cultured in DMEM (4500 mg/l glucose) with 10% FCS, 0.03% l-glutamine, 0.1 mg/ml gentamicin and 2.3 µg/ml fungizone at 37°C and 5% CO2. MEFs were cultured as described (23). FU, FdUrd, FUrd, HmdUrd, methoxyamine (MX), 4-amino-1,8-naphthalimide (4-AN), caffeine, vanillin, nucleosides, nucleotides and oligodeoxynucleotides were from Sigma-Aldrich. ATM Kinase inhibitor (sc202963) was from Santa Cruz Biotech. siRNA targeting UNG (Assay ID: 36376), SMUG1 (AM16708A, ID: 21193, 140141, 21109), TDG (Assay ID: 12923), MSH2 (siRNA ID: s8966) and MBD4 (siRNA ID: s17077) were from Ambion. Radionucleotides were from Perkin-Elmer. Restriction endonucleases were from New England Biolabs. Recombinant human His-tagged APE1, UNG2 and SMUG1 were purified as described (9,28). Human TDG cDNA from the construct pPRS202b (10) was subcloned into the BamHI and SalI sites of the pET28A vector (Novagene), generating pET28a-hTDG. His-tagged recombinant TDG protein was produced in Escherichia coli BL21 CodonPlus (DE3)-RIPL (Stratagene), purified using Dynabeads Talon (BD Biosciences), and further purified by MonoQ (GE Healthcare) chromatography.
Combined MMR and BER assay
Cultured cells were harvested by trypsination at 50–70% confluence. Nuclear extracts were prepared as described (29). To generate a substrate for both BER and MMR, a unique Nt.BbvCI site was introduced into the substrate plasmid (pGEM-3Zf+) at position 388 using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol, allowing generation of a nick serving as a strand selection signal for MMR. Substrates containing FU opposite A or G for BER and MMR were prepared essentially as described (29). Substrate (300 ng cccDNA treated with 5 U Nt.BbvCI when indicated) was incubated with 40 µg nuclear extract (TDG depleted and pre-incubated with Ugi or neutralizing SMUG1 antibodies when indicated) in BER buffer (29) supplemented with 250 µM NAD and 10 µM of each dNTP at 37°C for indicated time periods. Reactions were stopped by addition of 25 mM EDTA, 0.5% SDS, 150 µg/µl proteinase K (final concentrations) and incubation at 55°C for 30 min. DNA was purified by phenol–chloroform extraction and ethanol precipitation using 10 µg glycogen as carrier. DNA was then treated with purified recombinant human UNG (0.1 µg/µl) (30) (U- and FU-substrates) or TDG (0.5 µg/µl) (T:G substrates), as well as 50 mM MX and 0.2 µg/µl RNaseA (NEB buffer 2 and 0.1 µg/µl BSA) for 1 h at 37°C, followed by treatment with restriction endonucleases XmnI and HincII (5 U each) for 1 h. Restriction fragments were analysed on 2% agarose gels, stained with ethidium bromide and band intensities were quantified using ImageJ software (http://rsb.info.nlh.gov/ij/).
In vitro BER-, DNA-glycosylase- and AP-site cleavage activity assays
U, FU- and HmU-DNA excision activities were measured using a 5′-end labelled FAM or 33P-labelled 22-mer oligodeoxynucleotide containing a centrally positioned modified base (5′-GATCCTCTAGAGT-X-GACCTGCA-3′, where X = FU, HmU or U). Excision activity by nuclear extract (5 µg) or total cell extract (10 µg) was measured using the indicated oligomer substrate, as described (9). Glycosylases (in oligomer and BER activity assays) were inhibited by pre-incubating the cell extracts on ice with 0.1 µg Ugi, 0.1 µg neutralizing SMUG1 antibody (PSM1) (9) and 1 µl anti-TDG antiserum (diluted 1:3) (31) when indicated. Excision activities by purified proteins were measured using recombinant human His-tagged UNG2, SMUG1, or TDG, 0.1 pmol oligonucleotide substrate in UDG buffer (9) containing 50 mM NaCl and 0.1 pmol recombinant hAPE1 (28) after incubation at 37°C for 30 min. BER incorporation assays were carried out in the same buffer as BER/MMR assays, supplemented with 3 µCi dCTP or dTTP (3000 Ci/mmol, Perkin-Elmer) essentially as described (29). AP-site incision (Figure 5A) assays after MX-treatment of AP-site-containing oligonucleotide as substrate were carried out essentially as described (28). UDG-activity assays using [3H]uracil-containing calf thymus DNA substrates were as described (9) using 3 µg whole-cell extract (Supplementary Figure S1B). Kinetic properties of UNG2 and SMUG1 on FU:A and FU:G oligomer substrates were examined using excess substrate as described (28) (Supplementary Figure S1A).
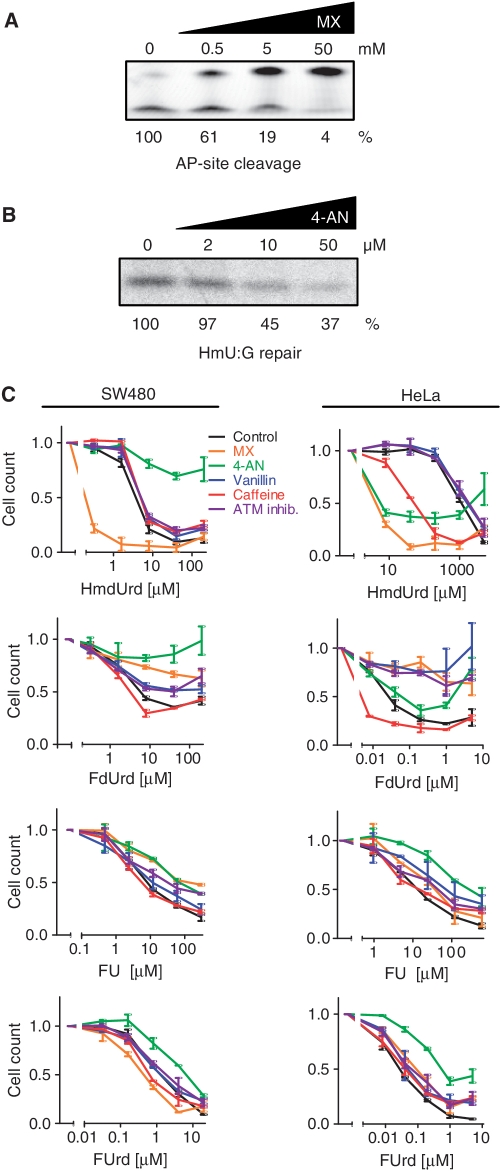
Figure 5.
Effects of inhibitors of BER, ATM, ATR and DNA-PK on survival after exposure of cells to HmdUrd, FdUrd, FU and FUrd. (A) Inhibition of APE1 cleavage of methoxyamine-modified AP-sites. A double-stranded oligonucleotide containing an AP-site was pre-treated for 20 min with various concentrations of MX, and then incubated with recombinant APE1. The upper bands observed after denaturing PAGE represent uncleaved 19-mer substrate, and the lower bands represent cleaved products. Bottom row numbers represent quantification of AP-site cleavage (lower gel band) as a percentage of control for various concentrations of MX (top row). (B) BER assay of repair of cccDNA substrate containing a single 5-hmU:G base pair in the presence of increasing concentrations of the PARP-1 inhibitor 4-AN. Nuclear extracts from the SW480 cells were pre-incubated with various concentration of 4-AN. Bands represent incorporation of [α33P]dCTP at the position of HmU. Bottom row numbers represent quantification of band signal as a percentage of control for various concentrations of 4-AN (top row). (C) SW480 and HeLa cell survival measured by the MTT assay after four days of continuous exposure to varying concentrations of HmdUrd, FdUrd, FU or FUrd in the presence or absence (black) of either 50 mM MX (orange), 20 µM 4-AN (green), 2 mM vanillin (blue), 2 mM caffeine (red), or 10 µM ATM kinase inhibitor (violet). The curves are normalized to untreated cells in the presence of the indicated molecular inhibitors. The data represent the mean ± SD of at least two parallel experiments.
Transfection with siRNA and verification of silencing
Cells were plated in six-well dishes (150 000 cells/well) in 1.6 ml antibiotic-free medium and cultured overnight. Cells were then transfected using Dharmafect (Dharmacon) transfection agent (4 µl/well) and siRNAs dissolved in OptiMEM (Invitrogen). siRNAs targeted SMUG1 (a mix of three siRNAs, final concentration 30 nM each), UNG (60 nM final), TDG (60 nM final), MSH2 (100 nM final), or MBD4 (60 nM final). After 24 h cells were either trypsinated and replated (for survival assays) or incubated for another 24 h in complete medium prior to drug exposure (for survival assays, FACS and LC/MS/MS) or harvesting (western blots, UDG activity assays). Whole-cell extracts for verification of UDG silencing (activity assays) were prepared 48 h after transfection by dissolving harvested cells in 50 µl lysis buffer [10 mM Tris–HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 1× Complete protease inhibitor (Roche) and 0.5% NP-40] and sonication for 3 × 45 s at 4°C. UNG activity (3 µg extract protein) was measured by the release of [3H]uracil from nick-translated calf thymus DNA (U:A substrate). SMUG1 activity (10 µg extract) was measured by oligomer cleavage assays on a U:G 22-mer in the presence of Ugi (0.1 µg) and 1 µl anti-TDG antiserum (diluted 1:3). TDG activity was measured in 10 µg extract on the same oligomer substrate, but in the presence of Ugi (0.1 µg) and neutralizing SMUG1 antibodies (PSM1) (9).
For western blot verifications of protein knock downs, 50–100 µg protein in whole-cell extract was treated with 0.5 µl each of Omnicleave Endonuclease (200 U/µl Epicentre Technologies), DNase (10 U/µl; Roche), micrococcal nuclease (100–300 U/mg; Sigma-Aldrich) and 1 µl RNaseA (20 mg/ml; Sigma-Aldrich) for 10 min at room temperature. Extracts were then subjected to gel electrophoresis and western blot detection of UNG [UNG antibody PU059 (30)], SMUG1 [PSM1 (9)], TDG [hTDG-antiserum (31)], MSH2 [mouse monoclonal MSH2 antibody, 3A2B8C (ab52266) (AbCam), 1:500], MBD4 [rabbit polyclonal MBD4 antibody, ab12187, AbCam) and β-actin (mouse monoclonal ab8226 (AbCam), 1:2000], essentially as described (9).
Survival assays after drug exposure
For continuous drug exposure assays, 2000–4000 cells/well in 96-well culture dishes were exposed 24 h after plating (48 h post-transfection for siRNA tranfected cells) and cultured for four days with continuous drug exposure in the presence or absence of molecular inhibitors when indicated (MX, 4-AN, vanillin, caffeine, ATM kinase inhibitor, ATM/ATR kinase inhibitor). For transient exposure, 150–300 cells/well were exposed for three days, after which medium was removed, cells washed with PBS and allowed to grow unexposed or in the presence of the indicated molecular inhibitors (MX, 4-AN) for seven days. For colony formation assays, 200–4000 cells were plated in six-well plates, exposed for three days, washed with PBS and allowed to grow unexposed in complete medium for seven days. Colonies larger than ~20 cells were counted. For the MTT-assay, growth medium was replaced with 100 µl fresh medium containing 0.5 mg/ml MTT (Sigma), and incubated at 37°C for 4 h. An amount of 50 µl of medium was subsequently removed, 100 µl 2-propanol with HCl (0.1 M) was added and MTT–formazan dissolved using a mechanical shaker. Absorption at 588 nm was recorded using a Titertek Multiscan Plus Reader.
FACS analysis of cell-cycle
At 48 h post-transfection, cells were exposed to the indicated drugs for 48 h prior to harvesting by trypsination. Cells were fixed in 70% methanol, washed twice with PBS and then treated with 50 µl RNaseA (100 µg/ml in PBS) at 37°C for 30 min prior to DNA staining with 200 µl propidium iodide (50 µg/ml in PBS) at 37°C for 30 min. Cell-cycle analyses were performed using a FACS Canto flow cytometer (BD-Life Science).
Quantification of FU in DNA and RNA by LC/MS/MS
Nucleic acids were isolated from fluoropyrimidine-treated cells by the DNeasy Blood and cell culture DNA isolation kit (Qiagen) or by the mirVana RNA-isolation kit (Ambion). DNA or RNA samples were enzymatically hydrolyzed to nucleosides by nuclease P1, snake venom phosphodiesterase and alkaline phosphatase as described (32). Then 3 volumes of methanol were added and tubes centrifuged (16 000 g, 30 min). The supernatants were dried and dissolved in 50 µl 5% methanol in water (v/v) for LC/MS/MS analysis of FdUrd and FUrd. Potential contamination by residual free FdUrd and FUrd was excluded by running parallel control samples treated with alkaline phosphatase only, in which no FdUrd and FUrd were detectable. A portion of each sample was diluted for the quantification of the unmodified nucleosides (dAdo, dCyd, dGuo, dThd, Ado, Cyd, Guo and Urd). Chromatographic separation was performed on a Shimadzu Prominence HPLC system with a Zorbax SB-C18 2.1 × 150 mm i.d. (3.5 µm) column equipped with an Eclipse XDB-C8 2.1 × 12.5 mm i.d. (5 µm) guard column (Agilent Technologies). For FdUrd and FUrd, the mobile phase consisted of water and methanol, starting with a 3.5 min gradient of 5–70% methanol, followed by 1 min with 70% methanol and 6.5 min re-equilibration with 5% methanol. Unmodified nucleosides were chromatographed isocratically with water:methanol:formic acid in ratio 85:15:0.1. Mass spectrometry detection was performed using an Applied Biosystems/MDS Sciex 5000 triple quadrupole (Applied Biosystems) operating in negative electrospray ionization mode for FdUrd and FUrd, or positive electrospray ionization mode for unmodified nucleosides. LC/MS/MS chromatograms showing FdUrd in DNA and FUrd in RNA hydrolysates are shown in the Supplementary Figure S4.
TS assay
TS activity was measured as previously described (33) with minor modifications. Cells were seeded in 24-well plates (70 000 cells/well) and treated with the indicated 5-fluoropyrimidine for 1 h. An amount of 1 µCi of [5-3H]deoxyuridine (20 Ci/mmol, Moravek Biochemicals Inc.) was added (500 µl final volume) and incubated for 90 min. The reaction was stopped by transferring 400 µl growth medium to 400 µl (150 mg/ml) activated charcoal suspended in 5% trichloroacetic acid. The samples were vortexed and centrifuged (16 000 g, 4°C), and 400 µl aliquots of the supernatant were counted using a liquid scintillation counter. Values were corrected for background counts.
RESULTS
Repair of FU in DNA is mainly carried out by BER in human cancer cell lines
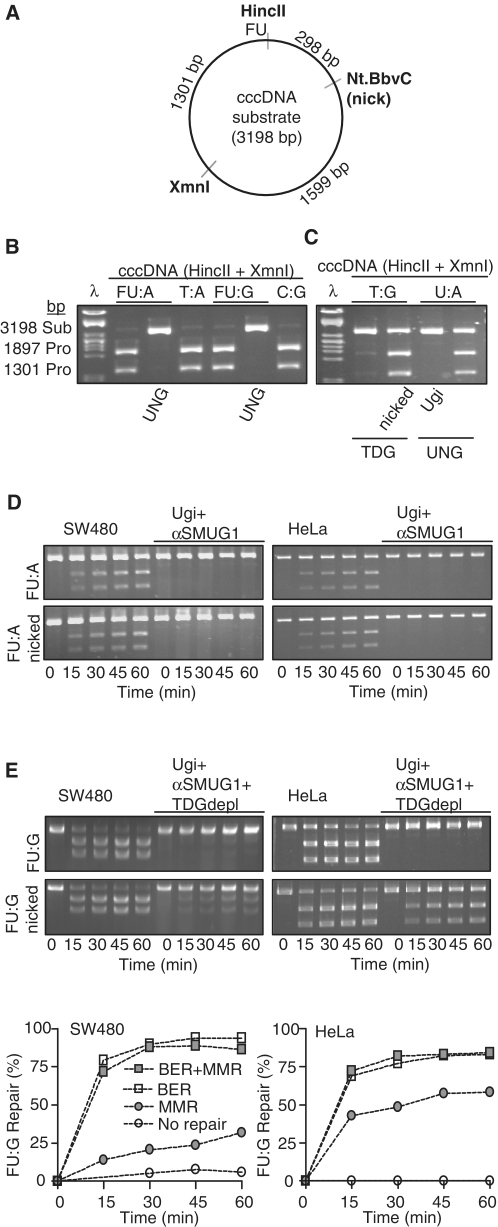
In vitro studies suggest that both BER and MMR contribute to repair of FU in DNA. However, their relative contribution has not been established (13). We employed a novel plasmid assay to monitor DNA repair of FU in nuclear extracts from human cancer cell lines. We introduced FU opposite adenine or guanine within the HincII (GTYRAC, Y = C/T, R = A/G) site of pGEM-3Zf(+) generating FU:A and FU:G cccDNA substrates, respectively. To distinguish between BER and MMR we also introduced a Nt.BbvCI nicking site in the pGEM-3Zf(+) vector, since MMR employs a nick to distinguish damaged and template strands (Figure 1A). FU:A base pairs resembles T:A product base pairs to such an extent that restriction enzymes are not capable of distinguishing between them (13). However, treatment with uracil–DNA glycosylase readily converts FU:A and FU:G into AP-sites (AP:A and AP:G) which remain unprocessed after HincII digestion (Figure 1B). Thus, by excising unrepaired FU from the plasmid with recombinant UNG after the repair reactions and prior to HincII digestion, we could distinguish unrepaired substrates from repaired substrates. To validate the assay we used nuclear extract from SW480 and verified repair of a T:G mismatch substrate in a nick-dependent manner, consistent with an active MMR system. The same extract under identical assay conditions also carried out repair of a non-nicked-U:A substrate, but this process was completely inhibited by the UNG inhibitor Ugi, consistent with BER initiated by UNG (Figure 1C). Thus, these substrates can be used to measure both BER and MMR. Importantly, one or both pathways can be specifically inactivated; BER by directly inhibiting the initiating glycosylase(s) and MMR by not introducing the nick that MMR is critically dependent on.
Figure 1.
Repair of FU–DNA by BER and MMR in nuclear extracts. (A) Cartoon showing the cccDNA substrate designed to measure FU:A and FU:G repair by both BER and MMR. FU is positioned in the HincII recognition sequence. The nicking endonuclease Nt.BbvCI cleaves one strand 298 bp 3′ to the lesion, thus providing a strand-discrimination signal for MMR. (B) Controls for validating the repair assay. Agarose gel showing HincII+XmnI treated cccDNA substrates containing either FU:A, T:A, FU:G or C:G in the HincII recognition site. Distinction between substrates (FU:A, FU:G) and products (T:A, C:G) are performed by FU excision by UNG generating AP-sites that are uncleavable by HincII. (C) Positive controls for MMR and BER and their inhibition. cccDNA substrate incubated with SW480 nuclear extract (40 µg), followed by treatment with recombinant TDG (T:G) or UNG (U:A) and MX before purification and HincII + XmnI digestion. (D) FU:A repair by SW480 and HeLa nuclear extracts (40 µg) incubated with cccDNA (FU:A, FU:A-nicked) substrates. Ugi and anti-SMUG1 antibodies were added to the reactions when indicated. (E) FU:G repair by SW480 and HeLa nuclear extracts and TDG depleted nuclear extracts incubated with cccDNA (FU:G, FU:G nicked) substrates. Ugi and anti-SMUG1 antibodies were added to the reactions when indicated. Graphs represent quantifications of above FU:G repair assays: ‘BER + MMR’ (FU:G nicked substrate), ‘BER’ (un-nicked FU:G substrate), ‘MMR’ (FU:G nicked substrate, TDG-depleted nuclear extract with Ugi and neutralizing SMUG1 antibody), and ‘No repair’ (un-nicked FU:G substrate and TDG-depleted nuclear extract with Ugi and neutralizing SMUG1 antibody).
Repair of FU:A in both nicked and intact cccDNA substrates was completely inhibited by the presence of the UNG inhibitor Ugi and anti-SMUG1 antibodies in nuclear extracts from HeLa and SW480 (Figure 1D). This indicates that BER is the main, possibly sole, pathway for repair of FU:A. Surprisingly, we could not detect any contribution from TDG on FU:A repair in either extract. The repair of FU:G was also mainly performed by BER, as most of the FU:G substrate was repaired after 15 min whether nicked or not (Figure 1E). Inhibition of BER by adding Ugi and anti-SMUG1 antibody to a TDG-depleted extract (13) completely abolished detectable repair of FU:G from the non-nicked substrate, while a marked reduction was observed when using nicked substrate. These results indicate a dominant role for BER in repair of FU in DNA in both base pairing contexts, with a smaller, but significant, contribution of MMR to FU:G repair.
Uracil–DNA glycosylase UNG is the major contributor to FU–DNA repair in human cancer cells, while SMUG1 and TDG contribute more in mouse cells
Purified recombinant UNG2, SMUG1, TDG and MBD4 have all been reported to excise FU from DNA in vitro (9–11,13). However, their relative importance in FU–DNA repair in different cell types has so far not been investigated. We analysed the contribution of uracil–DNA glycosylases to the excision of FU from DNA in nuclear extracts from several human cancer cell lines (SW480, HeLa, CX-1, HCT-8, HBL-100, AGS), as well as in wild type and Ung−/– MEFs. Nuclear extracts were incubated with duplex oligonucleotides with a central FU paired with adenine (FU:A), guanine (FU:G), or as single-stranded DNA (FU). SMUG1 and TDG activities in the extracts were inhibited using neutralising antibodies as described (9,31), while UNG2 activity was inhibited with Ugi. Notably, MBD4 did not appear to be significantly involved, as inhibition of UNG2, SMUG1 and TDG was sufficient to inhibit essentially all measurable FU excision activity in the extracts (Figure 2A and Supplementary Figure S1A, lanes 5). UNG2 represented the major activity against all FU substrates in most extracts from human cancer cell lines (SW480, HeLa, CX1, HCT-8, HBL-100), while SMUG1 and TDG activities were measurable only with the FU:G substrate (Figure 2A and Supplementary Figure S1A). This was corroborated by kinetic analysis of human UNG2 and SMUG1, which revealed an almost 2-fold higher activity (kcat/KM) of SMUG1 compared to UNG2 against FU:G oligo substrates, while UNG had an almost 10-fold higher FU:A activity (kcat/KM) compared to SMUG1 (Supplementary Table S1). To compare substrate preference and specific activity, experiments with purified recombinant human UNG2, SMUG1 and TDG were carried out. The results confirmed that FU is substrate for all three UDGs, with UNG2 as the most efficient enzyme on FU:A and especially on FU in a single-stranded context, while SMUG1 was the most efficient enzyme on FU:G (Figure 2B and Supplementary Figure S1D). As expected, TDG excised FU efficiently from a FU:G context (Figure 2B), in accordance with the analysis of FU-excision in nuclear extracts. A dominant role for UNG2 in BER of FU:A was also substantiated by assays measuring complete BER in SW480 and HeLa extracts, since all detectable FU:A repair activity was abolished when Ugi was added to the nuclear extracts (Figure 2C). UNG2, SMUG1 and TDG were all able to initiate FU:G repair, although with varying efficiency.
Figure 2.
FU excision from DNA by human uracil–DNA glycosylases. (A) FU excision by uracil–DNA glycosylases in nuclear extracts from human cancer cell lines (SW480, HeLa, CX1) and MEFs. Nuclear extracts (5 µg) were pre-incubated with Ugi, neutralizing SMUG1 (αSMUG1), or neutralizing TDG (αTDG) antibodies as indicated and assayed with double-stranded oligonucleotide substrates with FU in FU:A or FU:G context, or in a single-stranded context (FU). U, S and T indicate the individual activities of UNG2, SMUG1 and TDG, respectively. (B) Varying amounts (0–1000 fmol) of purified recombinant hUNG2, hSMUG1 and hTDG assayed with oligonucleotide substrates containing FU in different contexts (FU:A, FU:G, FU). (C) BER incorporation assay using a cccDNA substrate containing FU opposite A (FU:A) or G (FU:G). Nuclear extracts (10 µg) from SW480 and HeLa were pre-incubated with Ugi, neutralizing SMUG1-(αSMUG1) and neutralizing TDG (αTDG) antibodies as indicated. BER was detected by measuring incorporation of radio-labelled nucleotides.
FU accumulates in DNA after UNG knockdown, while knockdown of MSH2, TDG and SMUG1 have minor effects
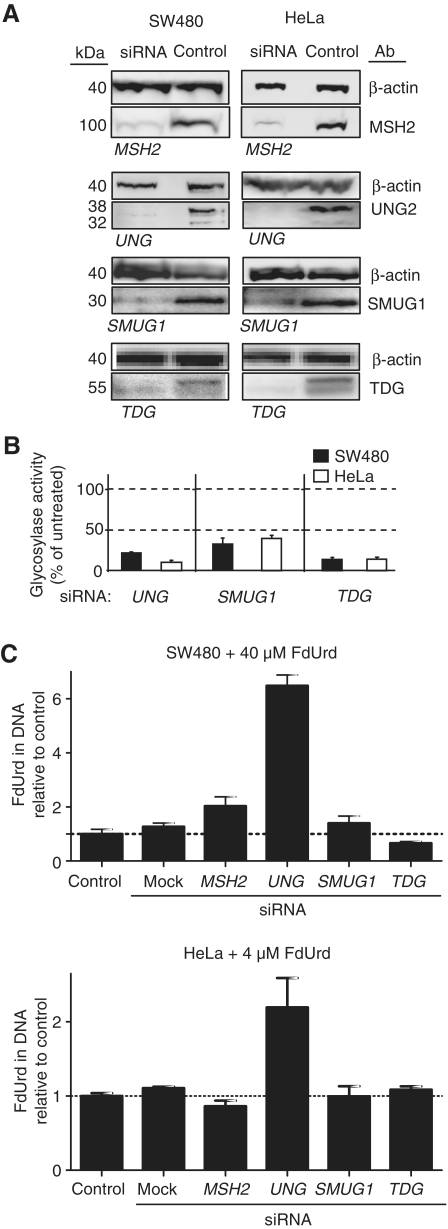
Our in vitro data from nuclear extracts suggested that FU in DNA was predominantly repaired by BER initiated by UNG2, SMUG1 or TDG. We therefore used siRNA knockdown to examine the in vivo effects of these UDGs in FU–DNA repair and in mediating overall 5-fluoropyrimidine cytotoxicity. UNG, SMUG1 and TDG excision activities were reduced 60–90% at 48 h after transfection and western blots verified knockdown at the protein level (Figure 3A and B). The protein levels recovered to 50–80% of control 4–6 days after transfection. At 48 h after transfection, SW480 and HeLa were exposed to 40 µM and 4 µM FdUrd, respectively, and incubated further for 24 h. DNA was then isolated for quantification of genomic FU (FdUrd) by LC/MS/MS. We observed a 2- to 6-fold increase in FU levels in DNA in HeLa and SW480 UNG-silenced cells (Figure 3C). In contrast, no significant differences in genomic FU were detected for SMUG1- and TDG-silenced cells compared to control. MSH2 knockdown increased FU levels in DNA ~2-fold in SW480 cells, while insignificantly decreasing genomic FU in HeLa cells. This was in agreement with our in vitro results, indicating a major role of UNG in FU–DNA repair, which apparently could not be compensated for by other UDGs or MMR. As expected, FU (FUrd) levels in co-purified RNA were unaffected by siRNA knockdown (data not shown).
Figure 3.
Verification of siRNA knockdown of UNG, SMUG1, TDG and MSH2 and their effect on FdUrd DNA incorporation. (A) Quantification of siRNA knockdown by western blots from whole-cell extracts of SW480 and HeLa MSH2, UNG, SMUG1, TDG siRNA silenced cells harvested 48 h post-transfection. β-actin was used as loading control. (B) Quantification of glycosylase knockdown by specific enzyme activity assays from whole-cell extracts 48 h after transfection. UNG excision activity was measured by the release of [3H]uracil from labelled calf thymus DNA (U:A substrate). SMUG1 and TDG activity were measured using a U:G oligomer substrate in the presence of either Ugi and neutralizing TDG antibodies, or Ugi and neutralizing SMUG1 antibodies, respectively. (C) Quantification by LC/MS/MS of incorporated FdUrd per nucleotide DNA after 24 h FdUrd exposure of MSH2, UNG, SMUG1 and TDG silenced SW480 (40 µM) and HeLa (4 µM) cells 48 h after transfection. Cells were harvested, and DNA isolated, hydrolysed and analysed for FdUrd content. FdUrd levels are normalized to the measured total number of normal deoxynucleosides in each sample. The data points represent fold change compared to control as is the mean ± SD of two to four parallel experiments.
Knockdown of UNG, SMUG1, TDG, MBD4 or MSH2 negligibly affects overall fluoropyrimidine cytotoxicity and cell-cycle perturbations
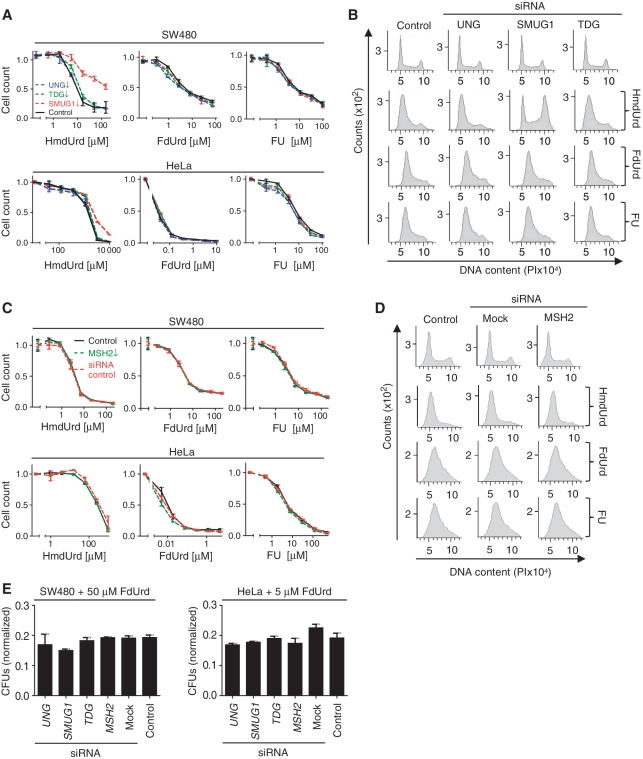
If fluoropyrimidine cytotoxicity is mediated by incorporation and/or repair of FU in DNA, sensitivity of cells to fluoropyrimidines should be altered by modulating repair activity, in particular the quantitatively dominant UNG2. However, knockdown of UNG, SMUG1, TDG, MBD4 or MSH2 by siRNA did not significantly alter the sensitivity of the cancer cell lines after 96 h exposure to FU or FdUrd (Figure 4A, C and Supplementary Figure S5). Moreover, the sensitivity of HeLa cells and SW480 cells to FU were essentially similar. Conversely, SW480 cells were far more resistant to FdUrd than HeLa cells (Figure 4A, middle panel) suggesting that FU and FdUrd cytotoxicity was mediated by distinct mechanisms. To distinguish between the different cytotoxic mechanisms of FU and its metabolites, we introduced a positive control for the DNA-directed effects of fluoropyrimidines, which would be analogous to those reported for 5-hydroxymethyl-2′-deoxyuridine (HmdUrd). HmdUrd metabolites are readily incorporated into DNA, but not RNA (34), and do not inhibit TS (35). HmU in DNA is in turn excised by SMUG1, leading to DNA strand breaks and apoptosis (9,36–39). HmdUrd therefore constitutes an ideal positive control for the concept of DNA repair-directed cytotoxicity. We monitored HmU-excision in nuclear extracts from SW480, HeLa and CX-1 and verified that SMUG1 constituted the only detectable excision activity, as excision was abolished by inhibition with SMUG1 antibodies (Supplementary Figure S1C). In accordance with this, both SW480 and HeLa SMUG1 knockdowns were significantly more tolerant to HmdUrd than control cells (Figure 4A). These results demonstrate that BER can in principle modulate cytotoxicity of pyrimidine anti-metabolites incorporated into DNA. The high tolerance of HeLa cells to HmdUrd was possibly due to phosphorolysis resulting in cleavage of the N-glycosylic bond in HmdUrd, thus inhibiting its incorporation into DNA (40,41). In agreement with this, addition of the HmU base increased sensitivity to HmdUrd several fold (data not shown). The differential response of knockdown cells to fluoropyrimidines and HmdUrd was also apparent in cell-cycle distributions, in which HmdUrd-exposed cells were shifted from G1/S arrest towards G2/M arrest subsequent to SMUG1 knockdown, while the cell-cycle profiles after FU and FdUrd treatment were apparently unaffected by BER and MMR knockdowns (Figure 4B and D).
Figure 4.
HmdUrd, FdUrd and FU survival and flow cytometry of SMUG1, TDG, UNG and MSH2 siRNA knockdown cells. (A/C) HeLa and SW480 cells were transfected with siRNA targeting (A) UNG, SMUG1, TDG, or (C) MSH2 and treated for four days with various concentrations of HmdUrd, FdUrd or FU. Controls were mock-transfected. Survival was measured by the MTT assay. The curves are normalized to untreated cells. Data points represent the mean ± SD from at least two parallel experiments. (B) and (D) FACS analysis (cell-cycle profiles) of SW480 (B) UNG, SMUG1, TDG, or (D) MSH2 siRNA knockdowns and control cells after treatment with 100 µM HmdUrd, 25 µM FU, or 25 µM FdUrd for 48 h. (E) CFU assays in which cells, 48 h post-siRNA transfection, were exposed for three days in six-well plates to 50 µM (SW480) or 5 µM (HeLa) FdUrd, and subsequently washed in PBS and allowed to grow for seven days in normal medium, before staining and manual counting of colonies (greater than ~20 cells). The data represent the mean ± SD of two parallel experiments normalized to the plating efficiency for the respective cells.
A shorter two day incubation time resulted in a more pronounced HmdUrd tolerance in SMUG1 knockdowns (Supplementary Figure S2B), compared to four day incubation (Figure 4A). This indicated that long incubation was not suited for transient siRNA silencing of glycosylases, as protein levels recover during the course of the assay. Therefore, we cannot exclude the possibility that long term effects on survival of cells permanently deficient in DNA repair proteins may be different from those observed here. Nevertheless, the lack of effect of glycosylase- and MSH2-knockdown on FdUrd tolerance was confirmed by CFU assays where colonies were counted manually in six-well plates (Figure 4E and Supplementary Figure S5).
BER and DNA damage response inhibitors affect sensitivity to HmdUrd and FdUrd but negligibly affect sensitivity to FU and FUrd
The inability of individual DNA glycosylase silencing to affect cytotoxicity of fluoropyrimidines could in principle be explained by functional redundancy of the individual uracil–DNA glycosylases. We attempted to achieve simultaneous knockdown of UNG, TDG and SMUG1. However, this resulted in unsatisfactory knockdown of at least one glycosylase. To circumvent this problem we employed molecular inhibitors that target the core BER proteins or DNA damage signalling. Methoxyamine (MX) reacts with AP-sites and inhibits their processing by APE1 (42) while 4-amino-1,8-naphtalimide (4-AN) is a potent inhibitor of PARP-1 polyribosylation (43) and BER (44,45). The inhibitory effect of MX and 4-AN was confirmed in vitro by APE1 activity measurements and BER incorporation assays, respectively (Figure 5A and B). We also inhibited DNA damage response proteins ATM/ATR by caffeine, DNA-PK by vanillin, and ATM by the specific inhibitor sc202963 (46). Inhibition of DNA damage response proteins significantly affected HmdUrd and FdUrd survival, although differently in HeLa and SW480 cells (Figure 5C). Relatively small effects were seen for FU and FUrd in both HeLa and SW480 cells (Figure 5C), suggesting that FU and FUrd have cytotoxic mechanisms different from FdUrd. Consistently, while sensitivities to HmdUrd and FdUrd were significantly affected by BER inhibitors (MX and 4-AN), much smaller effects were observed for FU and FUrd (Figure 5C). 4-AN seemed to protect the cells from HmdUrd and FdUrd cytotoxicity, while MX increased survival after FdUrd exposure, but severely aggravated HmdUrd cytotoxicity. To investigate this further, we also carried out transient three day exposure and seven days recovery in the presence or absence of lower doses of MX or 4-AN for the full 10 days period. Consistent with the shorter continuous incubation assays, only minor effects of MX and 4-AN were seen after FU and FUrd treatment. Interestingly, the initial protective effect of BER inhibitors from HmdUrd and FdUrd cytotoxicity turned into an aggravating effect after prolonged incubation times (Supplementary Figure S3). This was also observed for MX on HmdUrd and FdUrd cytotoxicity in SW480, but not in HeLa (Supplementary Figure S3). Possibly, the initial protective effects of inhibitors of BER and DNA damage signalling may have turned into a cytotoxic effect after cells again were allowed to multiply, due to replication of DNA with accumulated damage.
Taken together, these results demonstrate that BER intermediates are involved in HmdUrd cytotoxicity, but contribute negligibly to the cytotoxicity of FU. As opposed to FU and FUrd, FdUrd cytotoxicity seemed to be mediated through DNA damage, although the mechanism is apparently complex (Figure 5C and Supplementary Figure S3).
Cytotoxicity of FUrd and FU may be mediated predominantly through incorporation into RNA, while FdUrd toxicity is mediated by DNA effects
Since inhibition of BER and DNA damage signalling did not significantly affect FU cytotoxicity, we wanted to explore the mechanism further. We found that FU accumulated in RNA at 3000- to 15 000-fold higher levels than in DNA after 24 h FU drug exposure. While FdUrd-exposure resulted in considerable incorporation of FU into DNA (although lower than into RNA), incorporation of FU into DNA was very low (Figure 6A). Moreover, we found that fluoropyrimidines inhibited TS-activity in the following order for both SW480 and HeLa cells: FdUrd>FUrd>FU (Figure 6B). TS-inhibition by FdUrd required ~102-fold and 104-fold lower concentration to achieve 50% inhibition (IC50), as compared to FUrd and FU, respectively. While there were small differences between the cell lines with respect to TS-inhibition, the effects on cell survival were quantitatively very different, suggesting that at least two mechanisms must contribute to cytotoxicity of fluoropyrimidines. In order to distinguish the mechanisms of FdUrd-, FUrd- and FU-cytotoxicity, thought to arise through a lack of nucleotides and/or incorporation into nucleic acids, we attempted to reverse cytotoxicity by supplying normal nucleosides (dThd, dUrd, Urd) (Figure 6D and Supplementary Figure S2A) and nucleotides (dTMP, dUMP, UMP) (Figure 6E). We do appreciate that these experiments can give only a broad overview, since both cellular uptake and metabolic interconversions may be cell line- and compound-dependent. Nevertheless, while the cytotoxic effect of FdUrd was readily reversed by dThd and dTMP, and partially by dUrd and dUMP, it was relatively unaffected by Urd and UMP (Figure 6D, E and Supplementary Figure S2A). This indicated a significant contribution of TS-inhibition to FdUrd cytoxicity. The cytotoxic effect of high concentrations of dThd is most likely due to its well known cell-cycle blocking effect frequently exploited for cell-cycle synchronization. In contrast, FUrd cytotoxicity was efficiently reversed by Urd and UMP, but not by dUrd/dUMP or dThd/dTMP. Consistently, we found that the reversal of FUrd cytotoxicity by Urd was accompanied by a very strong reduction of FU in RNA (Figure 6C). This suggests that FUrd cytotoxicity was largely mediated through RNA incorporation. Interestingly, FU toxicity was also unaffected or slightly aggravated by dUrd and dThd, and, like FUrd, reversible by Urd and UMP, though to a lesser extent. FU cytotoxicity was slightly reversed by dTMP, although only at concentrations ~100- to 1000-fold higher than those required for reversal by UMP. The lack of reversibility of FU effects by dThd agrees well with the ~104-fold weaker TS-inhibition by FU than by FdUrd (Figure 6B), suggesting an RNA-mediated cytotoxicity similar to that of FUrd.
Figure 6.
Quantification of FU accumulation in RNA and DNA, relative TS-inhibition by FdUrd, FUrd and FU and reversal of their cytotoxicities and RNA incorporation by nucleosides/nucleotides. (A) Quantification of incorporated FUrd per nucleotide RNA and FdUrd per nucleotide DNA after 24 h FU or FdUrd exposure of SW480 and HeLa cells. Cells were harvested and RNA or DNA isolated, hydrolysed and analysed for FUrd or FdUrd content by LC/MS/MS. FUrd and FdUrd levels are normalized relative to the total number of normal nucleosides or deoxynucleosides measured. (B) TS activity measured by adding 1 µCi [5-3H]deoxyuridine and counting [3H]H2O released into the growth medium after 90 min in HeLa or SW480 cells pretreated with indicated concentrations of FdUrd, FUrd or FU. Measurements are plotted relative to the activity of untreated samples. (C) Urd reversal of FUrd incorporation into RNA. HeLa or SW480 cells were treated with 2.5 µM FUrd for 24 h in the presence or absence of Urd at indicated concentrations. RNA was isolated, hydrolysed and FUrd levels quantified by LC/MS/MS. The data represent the mean ± SD of at least two parallel measurements. (D) Effect on survival after exposure to FdUrd, FU and FUrd by concurrent treatment with various concentrations of nucleosides (dThd, dUrd or Urd). Increasing concentrations of nucleosides were added to SW480 or HeLa cells simultaneously treated with a fixed dose of either FdUrd (SW480: 25 µM, HeLa: 0.25 µM), FU (SW480: 15 µM, HeLa: 50 µM), or FUrd (2.5 µM). Survival was measured by the MTT assay after four days exposure. (E) Effect on FdUrd, FU and FUrd survival by concurrent treatment with nucleotides (dTMP, dUMP, UMP). Increasing concentrations of nucleotides were added to SW480 or HeLa cells simultaneously treated with either FdUrd (SW480: 50 µM, HeLa: 5 µM), FU (50 µM), or FUrd (5 µM). Survival was measured by the MTT assay after four days exposure. The survival data represent the mean ± SD of at least two parallel measurements.
DISCUSSION
FU is converted to different metabolites that may directly or indirectly affect both DNA and RNA structure and transactions (Figure 7A). We found that UNG-initiated BER was the main contributor to DNA repair of FU in vitro, while MMR was only active on FU:G base pairs. UNG was the sole glycosylase in nuclear extracts initiating repair in a FU:A context, and a major contributor in a FU:G context (Figure 2A and C). In support of our in vitro results, UNG was the only siRNA target that consistently caused an increase in genomic FU levels in both cell lines (Figure 3C). Still, neither knockdown of UNG, SMUG1, TDG, MBD4 nor MSH2 affected overall fluoropyrimidine sensitivity. In contrast, SMUG1 knockdown specifically reduced cytotoxicity of HmdUrd, demonstrating that the concept of cytotoxicity enhancement by DNA fragmentation is feasible in a ‘clean’ model case in which the phosphorylated metabolite is incorporated into DNA, followed by base excision and generation of strand breaks. HmdUrd has no known RNA effects and is not converted to a TS-inhibitor (34,35). Inhibition of the common downstream BER steps or DNA damage signalling modulated the sensitivity to FdUrd, indicating that BER contributes to cytotoxicity to some extent. However, this did not depend on a single glycosylase, suggesting that they are functionally redundant in this respect. Notably, BER and DNA damage signalling inhibitors did not modulate sensitivity to FU or FUrd, except for a small, but significant effect of the PARP-1 inhibitor 4-AN. This is, however, not necessarily BER related as PARP-1 is involved in multiple cellular processes apart from BER (47). Indeed, earlier generation PARP-1 inhibitors have been shown to enhance FU cytotoxicity at high concentrations, due to increased FU incorporation into RNA (48). Thus, our results are consistent with previous reports indicating that FdUrd cytotoxicity may be partially mediated by BER (20,21). In contrast, BER seems not to affect FU and FUrd cytotoxicity significantly.
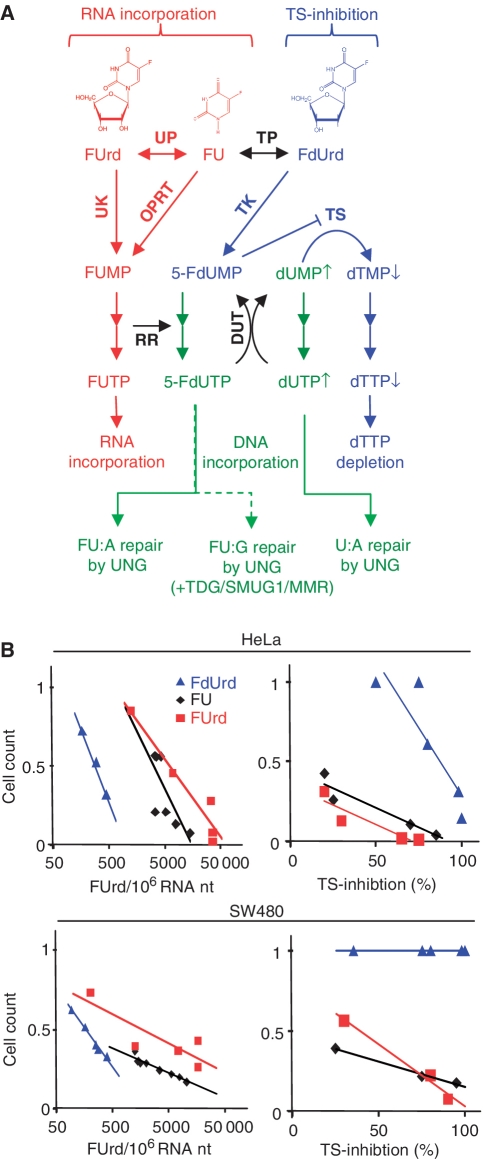
Figure 7.
Overview of metabolism of 5-fluoropyrimidines and correlations between survival and RNA incorporation or TS-inhibition. (A) Schematic overview of 5-fluoropyrimidine metabolism the three main routes to FU cytotoxicity: RNA incorporation of FUTP (red), TS-inhibition by FdUMP (blue) and DNA incorporation of FdUTP and dUTP (green). FdUTP incorporated into DNA end up as FU:A (mainly) or FU:G base pairs, which are predominantly repaired by the BER pathway initiated by UNG, with minor contributions from SMUG1, TDG and MMR for repair of FU:G. FU and FUrd cytotoxicity is predominantly mediated through RNA incorporation (red) and FdUrd through dTTP depletion (blue), while misincorporated 5-FdUTP and dUTP (green) contribute negligibly to overall 5-fluoropyrimidine cytotoxicity. DUT, deoxyuridine triphosphatase; DPD, dihydropyrimidine dehydrogenase; OPRT, orotic acid phosphoribosyl transferase; RR, ribonucleotide reductase; TK, thymidine kinase; TP, thymidine phosphorylase; UK, uridine kinase; UP, uridine phosphorylase. (B) Analysis of correlation of RNA incorporation versus survival and TS-inhibition versus survival after FdUrd, FUrd and FU treatment. Values for FUrd RNA levels (Figure 6A and C) or TS-inhibition {100% – [TS-activity (Figure 6B)]} are plotted against cell survival (Figure 4) after treatment with the nearest corresponding 5-fluoropyrimidine concentration. Lines represent linear trend lines in each plot.
Our results are in apparent conflict with a previous study reporting that SMUG1-knockdown MEFs were more sensitive to killing by FU and cells overexpressing SMUG1 less sensitive, when compared with wild type (22). We find that SMUG1 represents the main glycosylase activity excising FU from DNA in MEF nuclear extracts, while UNG had a negligible role in mouse cells (Figure 2A and Supplementary Figure S1). Thus, species differences in FU–DNA repair may contribute to these apparently conflicting results. This difference is of interest because results from FU treatment of MEF cells are often assumed to be valid for human cells. However, recently published data from our laboratory indicate that such extrapolations can be misleading, as there are generally significant differences in uracil excision activities between human and mouse cell lines (49). Species differences may also explain the discrepancy between our results and an earlier finding that Tdg−/– MEFs have increased tolerance to FU, as well as the their observation of a more modest effect of TDG knockdown in HeLa cells (18). The reported FdUrd-resistance in UNG-silenced HeLa (50) is more difficult to reconcile with our results. However, in agreement with our results, several studies report that UNG deficiency (22–24), UNG inhibition (26) or UNG expression (25) has no effect on cytotoxicity of FU, FdUrd or specific TS inhibitors. We conclude that BER contributes only modestly to cytotoxicity of FU in human cancer cells.
We found a clear contribution of MMR in repair in a FU:G context in vitro, although the rate was far lower than that of BER. We cannot exclude the possibility that the MMR contribution is underestimated in our in vitro assay, for example as a consequence of alternative repair of the nicked MMR DNA substrate by a DNA ligase. However, the modest contribution of MMR to FU removal in intact SW480 cells and the absence of significant effect in HeLa cells after knockdown of MSH2 support our in vitro results. Furthermore, our results and the inability of MMR proteins to recognise FU:A in gel shift assays (13,16), indicate that MMR most likely recognizes only FU:G mismatches. However, certain MMR mutants deficient in repair but proficient in DNA damage signalling are still able to mediate cytotoxicity of genotoxic drugs (51). Consequently MMR might mediate cytotoxicity of FU by mechanisms other than DNA repair. Notably, some 10–15% of colon cancers are MMR-deficient due to inactivating mutations or epigenetic silencing (52). Cancer cell lines deficient in the MMR components MSH2 (17) or MLH1 (19,53) are reported to display increased resistance to fluoropyrimidines. FU resistance and its correlation to microsatellite instability (MSI) or MMR-deficiency has been ambiguous (54,55). However, randomized controlled trials seem to agree that MSI correlates with less benefit from adjuvant FU (56,57). Thus, while our experiments indicate a modest role for MMR repair of FU in vitro and in cultured cells, they do not exclude a significant involvement of MMR in the FU resistant patients. One such mechanism could be that the MMR mutator phenotype enhances generation of FU resistant cells. An alternative possibility may be that the fraction of FU:G compared with FU:A is so small that its repair by MMR does not contribute measurably to overall levels of FU in DNA, but long repair patches generated by MMR of these quantitatively minor lesions might still contribute to cytotoxicity. The strongest arguments against these possibilities would be that we could not demonstrate increased survival in MSH2 knockdown cells (Figure 4E).
Generally, our results demonstrate that the 5-fluoropyrimidine nucleosides FdUrd, FUrd and the base FU exert their effects by quantitatively different mechanisms, suggesting that FU was predominantly metabolised to ribonucleotides and consequently incorporated into RNA (Figure 7A). Thus, we found that inhibition of TS-activity by FdUrd was achieved at ~100-fold lower concentrations than for FUrd, and inhibition by FU required another ~100-fold higher concentration compared with FUrd. These results and the RNA incorporation measurements demonstrate that there is significant cross metabolism between FU, FdUrd and FUrd (Figure 6A). In support of RNA incorporation as a major mechanism of cytotoxicity, we observed a correlation between cytotoxicity and RNA incorporation for all fluoropyrimidines (Figure 7B). It should be noted that although RNA incorporation is massive compared to DNA incorporation, we have not proven a causative relationship to cytotoxicity, merely a clear association. Further work on the possible mechanism of cytotoxicity from RNA incorporation would seem warranted. However, the level of RNA incorporation associated with a substantial decrease in survival was several fold lower for FdUrd than for FUrd and FU, indicating that factors other than RNA incorporation contribute to FdUrd cytotoxicity. It should be mentioned that FU cytotoxicity was slightly reversed by dTMP, although at relatively high concentrations. Earlier studies on FU cytotoxicity in other cell lines reported both reversal and aggravation by thymidine, indicating cell-specific differences in the mechanism of cytotoxicity (58). The failure of thymidine to reverse FU and FUrd cytotoxicity was, however, mirrored in correlation plots of TS-inhibition versus survival (Figure 7B), where FUrd and FU cytotoxity was significant even at low TS-inhibition levels. Again, this suggests mechanisms of action other than TS-inhibition for FUrd and FU. In accordance with this, in a comprehensive drug activity gene expression study, FU clustered with RNA synthesis inhibitors, suggesting that a major mechanism of action is RNA-directed (59). Finally, microarray profiling of FU resistant cell lines tend not to find BER genes to be differentially regulated, as one might expect if BER were an important mediator of cytotoxicity (60–63).
In conclusion, we find that cytotoxicity from excision repair, whether BER or MMR, contributes only in a minor way to the mechanism of action of FU. Furthermore, the cytotoxic contribution from BER of FU–DNA would be limited to FdUrd, which is less commonly used in the clinic. TS-inhibition also seems to contribute substantially to the mechanism of FdUrd, but the dominant mechanism of FU cytotoxicity seems to be more closely associated with RNA incorporation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Programme for Research in Functional Genomics in Norway (grant 159019) in the Research Council of Norway (grant 185308); the Norwegian Cancer Association (grant 418925); the Cancer Fund at St Olav’s Hospital Trondheim and the Svanhild and Arne Must Fund for Medical Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to express their gratitude to Professor Primo Schär (Basel, Switzerland), who provided TDG expression constructs and anti-sera against TDG, Olena Dyka (Trondheim, Norway) for the purification of recombinant human TDG, Nina Beate Liabakk (Trondheim, Norway) for FACS analysis and Hilde Nilsen (Oslo, Norway) for informative discussions.
REFERENCES
- 1.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 2.Ghoshal K, Jacob ST. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 1994;54:632–636. [PubMed] [Google Scholar]
- 3.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 4.Gustavsson M, Ronne H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA. 2008;14:666–674. doi: 10.1261/rna.966208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patton JR. Ribonucleoprotein particle assembly and modification of U2 small nuclear RNA containing 5-fluorouridine. Biochemistry. 1993;32:8939–8944. doi: 10.1021/bi00085a027. [DOI] [PubMed] [Google Scholar]
- 6.Silverstein RA, Gonzalez de Valdivia E, Visa N. The incorporation of 5-fluorouracil into RNA affects the ribonucleolytic activity of the exosome subunit Rrp6. Mol. Cancer Res. 2011;9:332–40. doi: 10.1158/1541-7786.MCR-10-0084. [DOI] [PubMed] [Google Scholar]
- 7.Samuelsson T. Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Res. 1991;19:6139–6144. doi: 10.1093/nar/19.22.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Laar JA, Rustum YM, Ackland SP, van Groeningen CJ, Peters GJ. Comparison of 5-fluoro-2′-deoxyuridine with 5-fluorouracil and their role in the treatment of colorectal cancer. Eur. J. Cancer. 1998;34:296–306. doi: 10.1016/s0959-8049(97)00366-3. [DOI] [PubMed] [Google Scholar]
- 9.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 10.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 11.Turner DP, Cortellino S, Schupp JE, Caretti E, Loh T, Kinsella TJ, Bellacosa A. The DNA N-glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine. Cancer Res. 2006;66:7686–7693. doi: 10.1158/0008-5472.CAN-05-4488. [DOI] [PubMed] [Google Scholar]
- 12.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA–general mutagen, but normal intermediate in acquired immunity. DNA Repair. 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–1868. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Seiple L, Jaruga P, Dizdaroglu M, Stivers JT. Linking uracil base excision repair and 5-fluorouracil toxicity in yeast. Nucleic Acids Res. 2006;34:140–151. doi: 10.1093/nar/gkj430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt MD, Wilson DM., 3rd Participation of DNA repair in the response to 5-fluorouracil. Cell Mol. Life Sci. 2009;66:788–799. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu A, Yoshioka K, Salerno V, Hsieh P. The mismatch repair-mediated cell cycle checkpoint response to fluorodeoxyuridine. J. Cell Biochem. 2008;105:245–254. doi: 10.1002/jcb.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J. Biol. Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 18.Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J, Schar P. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sansom OJ, Zabkiewicz J, Bishop SM, Guy J, Bird A, Clarke AR. MBD4 deficiency reduces the apoptotic response to DNA-damaging agents in the murine small intestine. Oncogene. 2003;22:7130–7136. doi: 10.1038/sj.onc.1206850. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Berger SH, Wyatt MD. Involvement of base excision repair in response to therapy targeted at thymidylate synthase. Mol. Cancer Ther. 2004;3:747–753. [PubMed] [Google Scholar]
- 21.McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM., 3rd Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol. Cancer Res. 2009;7:897–906. doi: 10.1158/1541-7786.MCR-08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–945. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 23.Andersen S, Heine T, Sneve R, Konig I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 24.Grogan BC, Parker JB, Guminski AF, Stivers JT. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-Fluorouracil in DNA. Biochemistry. 2011;50:618–627. doi: 10.1021/bi102046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsh SJ, Hobbs S, Aherne GW. Expression of uracil DNA glycosylase (UDG) does not affect cellular sensitivity to thymidylate synthase (TS) inhibition. Eur. J. Cancer. 2003;39:378–387. doi: 10.1016/s0959-8049(02)00610-x. [DOI] [PubMed] [Google Scholar]
- 26.Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair. 2008;7:162–169. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albertella MR, Lau A, O’Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair. 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE. The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair. 2008;7:1869–1881. doi: 10.1016/j.dnarep.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 31.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 32.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 33.Yalowich JC, Kalman TI. Rapid determination of thymidylate synthase activity and its inhibition in intact L1210 leukemia cells in vitro. Biochem. Pharmacol. 1985;34:2319–2324. doi: 10.1016/0006-2952(85)90788-9. [DOI] [PubMed] [Google Scholar]
- 34.Vilpo JA, Vilpo LM. Metabolism, incorporation into DNA, and interactions with 1-beta-D-arabinofuranosylcytosine of 5-hydroxymethyl-2′-deoxyuridine in human promyelocytic leukemia cells (HL-60) Cancer Res. 1988;48:3117–3122. [PubMed] [Google Scholar]
- 35.Wataya Y, Santi DV, Hansch C. Inhibition of Lactobacillus casei thymidylate synthetase by 5-substituted 2′-deoxyuridylates. Preliminary quantitative structure-activity relationship. J. Med. Chem. 1977;20:1469–1473. doi: 10.1021/jm00221a021. [DOI] [PubMed] [Google Scholar]
- 36.Mi LJ, Chaung W, Horowitz R, Teebor GW, Boorstein RJ. Excessive base excision repair of 5-hydroxymethyluracil from DNA induces apoptosis in Chinese hamster V79 cells containing mutant p53. Carcinogenesis. 2001;22:179–186. doi: 10.1093/carcin/22.1.179. [DOI] [PubMed] [Google Scholar]
- 37.Boorstein RJ, Chiu LN, Teebor GW. A mammalian cell line deficient in activity of the DNA repair enzyme 5-hydroxymethyluracil-DNA glycosylase is resistant to the toxic effects of the thymidine analog 5-hydroxymethyl-2′-deoxyuridine. Mol. Cell. Biol. 1992;12:5536–5540. doi: 10.1128/mcb.12.12.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 39.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair. 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 40.Kaufman ER. Biochemical analysis of toxic effects of 5-hydroxymethyl-2′-deoxyuridine in mammalian cells. Somat. Cell Mol. Genet. 1986;12:501–512. doi: 10.1007/BF01539921. [DOI] [PubMed] [Google Scholar]
- 41.Boorstein RJ, Teebor GW. Effects of 5-hydroxymethyluracil and 3-aminobenzamide on the repair and toxicity of 5-hydroxymethyl-2′-deoxyuridine in mammalian cells. Cancer Res. 1989;49:1509–1514. [PubMed] [Google Scholar]
- 42.Liuzzi M, Talpaert-Borle M. A new approach to the study of the base-excision repair pathway using methoxyamine. J. Biol. Chem. 1985;260:5252–5258. [PubMed] [Google Scholar]
- 43.Schlicker A, Peschke P, Burkle A, Hahn EW, Kim JH. 4-Amino-1,8-naphthalimide: a novel inhibitor of poly(ADP-ribose) polymerase and radiation sensitizer. Int. J. Radiat. Biol. 1999;75:91–100. doi: 10.1080/095530099140843. [DOI] [PubMed] [Google Scholar]
- 44.Boorstein RJ, Haldar J, Poirier G, Putnam D. DNA base excision repair of 5-hydroxymethyl-2′-deoxyuridine stimulates poly(ADP-ribose) synthesis in Chinese hamster cells. Carcinogenesis. 1995;16:1173–1179. doi: 10.1093/carcin/16.5.1173. [DOI] [PubMed] [Google Scholar]
- 45.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair. 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 47.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willmore E, Durkacz BW. Cytotoxic mechanisms of 5-fluoropyrimidines. Relationships with poly(ADP-ribose) polymerase activity, DNA strand breakage and incorporation into nucleic acids. Biochem. Pharmacol. 1993;46:205–211. doi: 10.1016/0006-2952(93)90405-l. [DOI] [PubMed] [Google Scholar]
- 49.Doseth B, Visnes T, Wallenius A, Ericsson I, Sarno A, Pettersen HS, Flatberg A, Catterall T, Slupphaug G, Krokan HE, et al. Uracil-DNA glycosylase in base excision repair and adaptive immunity: species differences between man and mouse. J. Biol. Chem. 2011;286:16669–16680. doi: 10.1074/jbc.M111.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer JA, Muller-Weeks S, Caradonna SJ. Fluorodeoxyuridine modulates cellular expression of the DNA base excision repair enzyme uracil-DNA glycosylase. Cancer Res. 2006;66:8829–8837. doi: 10.1158/0008-5472.CAN-06-0540. [DOI] [PubMed] [Google Scholar]
- 51.Lin DP, Wang YX, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin B, Werling U, Parris T, Kurihara N, et al. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 52.Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomaki P, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat. Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 53.Meyers M, Wagner MW, Hwang HS, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA mismatch repair protein in fluoropyrimidine-mediated cell death and cell cycle responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- 54.Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, Allegra CJ. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J. Clin. Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 55.Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 56.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010;28:3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valeriote F, Santelli G. 5-Fluorouracil (FUra) Pharmacol. Ther. 1984;24:107–132. doi: 10.1016/0163-7258(84)90030-5. [DOI] [PubMed] [Google Scholar]
- 59.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, et al. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 60.de Angelis PM, Fjell B, Kravik KL, Haug T, Tunheim SH, Reichelt W, Beigi M, Clausen OP, Galteland E, Stokke T. Molecular characterizations of derivatives of HCT116 colorectal cancer cells that are resistant to the chemotherapeutic agent 5-fluorouracil. Int. J. Oncol. 2004;24:1279–1288. [PubMed] [Google Scholar]
- 61.Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin. Cancer Res. 2004;10:272–284. doi: 10.1158/1078-0432.ccr-1025-3. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt WM, Kalipciyan M, Dornstauder E, Rizovski B, Steger GG, Sedivy R, Mueller MW, Mader RM. Dissecting progressive stages of 5-fluorouracil resistance in vitro using RNA expression profiling. Int. J. Cancer. 2004;112:200–212. doi: 10.1002/ijc.20401. [DOI] [PubMed] [Google Scholar]
- 63.Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63:8791–8812. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.