Abstract
Single-stranded (ss) transposition, a recently identified mechanism adopted by members of the widespread IS200/IS605 family of insertion sequences (IS), is catalysed by the transposase, TnpA. The transposase of IS608, recognizes subterminal imperfect palindromes (IP) at both IS ends and cleaves at sites located at some distance. The cleavage sites, C, are not recognized directly by the protein but by short sequences 5′ to the foot of each IP, guide (G) sequences, using a network of canonical (‘Watson–Crick’) base interactions. In addition a set of non-canonical base interactions similar to those found in RNA structures are also involved. We have reconstituted a biologically relevant complex, the transpososome, including both left and right ends and TnpA, which catalyses excision of a ss DNA circle intermediate. We provide a detailed picture of the way in which the IS608 transpososome is assembled and demonstrate that both C and G sequences are essential for forming a robust transpososome detectable by EMSA. We also address several questions central to the organization and function of the ss transpososome and demonstrate the essential role of non-canonical base interactions in the IS608 ends for its stability by using point mutations which destroy individual non-canonical base interactions.
INTRODUCTION
Insertion sequences (IS) play a preponderant role in shaping prokaryotic genomes. They are ubiquitous and have been identified in high numbers in many bacteria and archaea (1). Indeed, transposases, the enzymes which catalyse their movement, are by far the most numerous and ubiquitous genes in nature (2).
We recently described an unusual type of bacterial insertion sequence, the IS200/IS605 family, whose members undergo single-strand (ss) transposition. They use obligatory ssDNA intermediates and insert into an ssDNA target (3–5) such as the lagging strand template of replication forks (6). The paradigm of this family, IS608 from Helicobacter pylori (7) transposes efficiently in the heterologous host, Escherichia coli. IS200/IS605 family transposases (TnpA) are not related to the well known and best characterized DDE transposases (8) but are members of the large HUH (histidine–hydrophobic–histidine) endonuclease family that includes viral Rep proteins, conjugative plasmid relaxases and rolling circle replication initiator proteins (9). All use a catalytic tyrosine residue to attack the target phosphodiester bond creating a covalent 5′-phosphotyrosine enzyme–substrate intermediate (4,5,10).
As for other IS types, these reactions are carried out by a molecular machine, the transpososome or synaptic complex, a key controlling element in transposition (11) in which the IS ends are assembled into a complex with several transposase protomers. In a number of systems, strand cleavage can only occur once the transpososome is correctly assembled (12–14), ensuring that adventitious and potentially deleterious DNA cleavages do not occur before conditions for productive transposition are assured.
Our limited knowledge of transpososome architecture and behaviour has been obtained largely from transposable elements which use double-stranded (ds) DNA intermediates and employ DDE transposases (15–17). These transpososomes undergo a series of orchestrated transformations involving conformational changes leading to the positioning of transposon DNA strands for cleavage, elimination of flanking DNA, target DNA docking and DNA strand transfer. Tn10 and phage Mu transpososome assembly, for example, is highly ordered and the complexes become increasingly robust and refractory to denaturation as transposition progresses (18,19).
Little is known about assembly and behaviour of IS200/IS605 family ss transpososomes although structural studies have revealed large conformational changes on DNA binding (4).
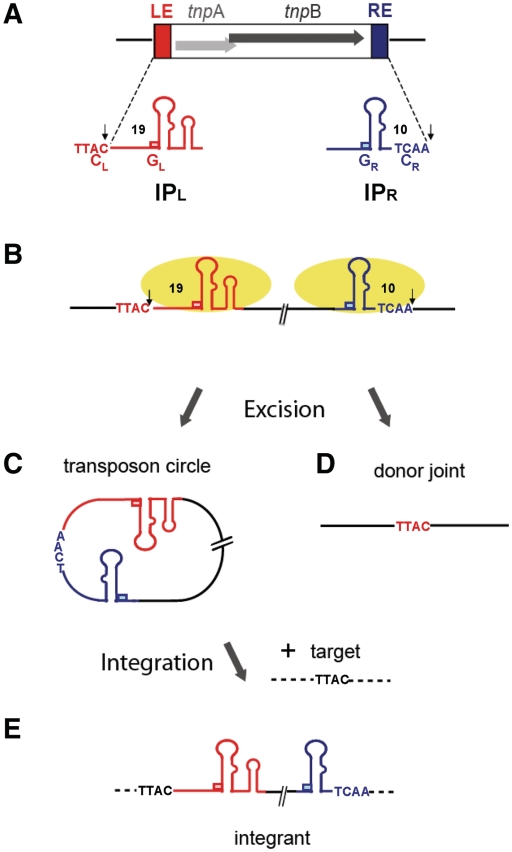
IS608 TnpA binds subterminal imperfect palindromes, IPL and IPR, at the left (LE) and right (RE) ends (Figure 1A and B) located some distance from the cleavage sites (4,20) (Figure 1B). Cleavage is strand specific occurring in the ‘top’ strand (Figure 1B). Strand transfer generates a circular ssDNA ‘top’ strand transposon intermediate with abutted RE and LE (the transposon or RE–LE junction) (Figure 1C). This inserts specifically into an ssDNA target 3′ to a TTAC tetranucleotide (Figure 1E) (3,7) also required for subsequent transposition (5). Strand transfer also joins the DNA originally flanking the excised strand to generate a donor joint and preserve the target TTAC without DNA gain or loss (5) (Figure 1D). The entire transposition cycle has been reconstituted in vitro with purified TnpA (3).
Figure 1.
IS608 and its transposition cycle. The left (LE) and right (RE) IS ends are shown in red and blue, respectively. (A) IS608 organization. Grey horizontal arrows: tnpA and tnpB; red and blue boxes, left (LE) and right (RE) ends (colour code retained throughout). The subterminal secondary structures IPL and IPR, the left (TTAC) and right (TCAA) cleavage (CL and CR) and guide (GL and GR) sequences and the distances in nucleotides between CL or CR and the foot of IPL or IPR are indicated. The cleavage sites are indicated by small vertical arrows. Flanking DNA is shown as black horizontal lines. (B) Binding and cleavage by TnpA. TnpA molecules are shown as yellow ellipses. The active top strand is shown in bold. Note that a 5′ phosphotyrosine bond is formed with LE and with the RE 5′ flank (arrows). (C) The IS608 ss circle intermediate. Excision of the IS608 ss circle intermediate with abutted left and right ends retaining the right cleavage site, TCAA. (D) The donor replicon after IS608 excision. Excision of the IS608 ‘top strand’ leads to rejoining of the left and right ends to form a transposon joint retaining the TTAC target sequence. (E) Integration. The circular IS recognizes the TTAC target sequence and inserts 3′ of this sequence.
TnpA itself is dimeric both in solution and in the X-ray crystal structure (4). The IPL and IPR binding sites recognize IP structural features and are located on one dimer face and the two active sites on the other (Supplementary Figure S3E) (4,20). Both active sites include amino acids from each monomer assembled by juxtaposition of an alpha helix (D) carrying the catalytic tyrosine (Y127) of one monomer and the two histidines of the HUH motif (that coordinate the catalytically essential divalent metal ion) from the other. In complexes with or without bound oligonucleotides (containing only IPL and IPR), the active sites are in an inactive configuration (4) but the ensemble undergoes a large conformational change if 4 nt 5′ to the foot of the IP are included. This permits divalent metal ion binding and places Y127 in an appropriate position for nucleophilic attack (Supplementary Figure S3E, right) (20).
IS608 shows important asymmetries in both its organization and in its transposition mechanism. Cleavage at LE occurs 3′ to a conserved TTAC located 19 nt 5′ from the foot of IPL whereas at RE, the cleavage site, TCAA, lies 10 nt and 3′ to the foot of IPR (Figure 1B). Astonishingly, these cleavage sites (CL and CR) are not recognized directly by the enzyme but via base pairing with four bases (guide sequences: GL and GR) that are located 5′ to the foot of IPL and IPR respectively (20,21). Furthermore, while strand cleavage creates 5′ phospho–tyrosine bonds between TnpA and substrates at both ends (4,5), this reaction occurs with the 5′-end of LE but with the 5′-end of the DNA flank at RE (Figure 1B).
The experiments reported here address questions central to the formation, organization and function of the ss transpososome involved in the excision of IS608 as an ss circular transposition intermediate. We reconstitute a biologically relevant complex including both LE and RE and TnpA and demonstrate that it is catalytically active. We identify DNA sequences within the ends required for transpososome assembly. Furthermore, we show that the guide sequences, GL and GR, and the LE and RE cleavage sites, CL and CR, together with a network of canonical (‘Watson and Crick’) and unusual non-canonical base interactions (reminiscent of certain catalytic RNA species (reminiscent of certain catalytic RNA species (22)) are necessary for assembly of a robust synaptic complex stable during gel electrophoresis. The results provide a detailed picture of the way in which the IS608 transpososome is assembled.
MATERIALS AND METHODS
Oligonucleotides and radioactivity
All the oligonucleotides used in this work were from Eurogentec (PAGE purified), except for 2-amino purine labelled oligonucleotides which were from Fidelity Systems. The oligonucleotide sequences are shown in Supplementary Table S1. Where necessary they were 5′ or 3′-end labelled by 32P.
TnpA-His6 purification
TnpA-His6 was purified according to (5).
Complex formation in vitro
32P-radiolabelled (10 000 cpm HIDEX liquid Scintillation Counter) or fluorolabelled oligonucleotide together with unlabeled (>100-fold in excess, 0.6 µM) oligonucleotide were incubated with TnpA-His6 in binding buffer containing 10 mM Tris pH 7.5, 200 mM NaCl, 0.5 mM EDTA, 15% glycerol, 3.5 mM DTT, 20 µg/ml BSA and 0.5 µg poly-dIdC competitor at 37°C for 45 min.
Electrophoretic mobility shift assay (EMSA)
Complexes were separated in an 8% native polyacrylamide gel in TGE buffer (25 mM Tris, 200 mM glycine and 1 mM EDTA) at 170 V for 3 h at 4°C.
In gel crosslinking
The preparative electrophoretic mobility shift assay (EMSA) gel was analysed by Gel doc 2000 (Biorad) and the species of interest complex carrying fluorescein-labelled oligo were excised and rinsed with cross-link buffer (10 mM HEPES pH 7.5, 1 mM EDTA and 10% glycerol), and then were incubated at 37°C for 1 h in 400 µl crosslink buffer containing 0.5 mM cross-linker BMH [bis(maleimido)hexane]. The solution was then replaced with 400 µl of 30 mM DTT for 15 min. The gel slice was rinsed with 500 µl of 10 mM HEPES pH 7.5, 1 mM EDTA and 1% SDS. An amount of 30 µl of 1× SDS loading dye was added to the gel slice and heated at 70°C for 2 min. The sample was loaded on 16% SDS–PAGE gel containing 0.2% SDS and electrophoresis was in Tris–glycine buffer containing 0.2% SDS. TnpA-His6 was detected by western blot using 6His-HRP antibody (Clontech).
In vitro cleavage
The EMSA gel was exposed to X-ray film (Kodak), and the species of interest carrying 32P-radiolabelled oligonucleotides were excised then soaked in a cleavage buffer with Mg2+ (10 mM Tris pH 7.5, 200 mM NaCl, 0.5 mM EDTA, 15% glycerol, 3.5 mM DTT, 20 µg/ml BSA and 5 mM MgCl2) for 3 h. DNA was eluted in elution buffer (10 mM Tris pH 8, 1 mM EDTA, 0.2% SDS and 300 mM NaCl) overnight at 37°C, the radioactivity quantified using a HIDEX liquid Scintillation Counter and equal amounts of radioactivity were loaded and separated on a 9% denatured sequencing gel.
RESULTS
Composition of the IS608 transpososome
TnpA generates two nucleoprotein complexes
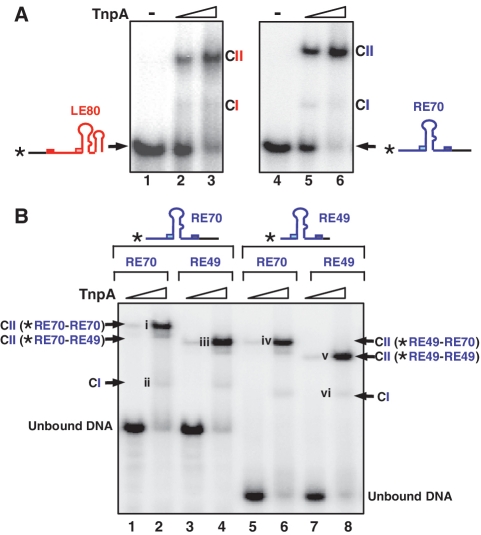
TnpA binds the ‘top’ strand of both RE and LE when they are in ssDNA form (5). EMSA with either LE80 (an oligonucleotide including LE and 18 nt of flanking DNA including the cleavage site) or RE70 (including RE and 17 nt of flanking DNA) (Supplementary Table S1) and TnpA revealed a major and a minor complex, CII and CI, respectively (Figure 2A).
Figure 2.
Synaptic complexes. (A) EMSA analysis of complexes formed between TnpA and LE (left panel) and TnpA and RE (right panel). The experiments were performed in the absence of Mg2+. The nature of the two complexes CI and CII are described in the text. LE80 and RE70 indicate the length of the oligonucleotides used (Supplementary Table S1). Asterisk indicates the 5′ label and the light coloured boxes represent the guide sequences GR (pale blue) and GL (pink). The full boxes represent the cleavage sequences CL (red) and CR (blue). ‘–’ indicates absence of TnpA-His6. Increasing TnpA-His6 concentrations (0.5 and 2.5 µM) are indicated by the triangle. Complexes were separated in 8% polyacrylamide native gels. (B) EMSA analysis of DNA content of CI and CII. Lanes 1–4: 5′ end-labelled RE70 with excess unlabelled RE70 (lanes 1 and 2); with excess unlabelled RE49 (lanes 3 and 4). Lanes 5–8: 5′ end-labelled RE49 with excess unlabelled RE70 (lanes 5 and 6); with excess unlabelled RE49 (lanes 7 and 8). Triangle presents different TnpA-His6 concentration (0.1 and 2.5 µM). The bands indicated by roman numerals were cut from the gel and used for the analysis of catalytic activity presented in Figure 3C.
To determine DNA stoichiometry in these complexes, we used a mixture of two RE ends with distinguishable sizes (RE49 and RE70; Supplementary Table S1). In these experiments, a mixture of a labelled IS end and a 100-fold excess of unlabelled end was used. This assures that the labelled end is accompanied by an unlabelled partner in the complex. The 5′-end-labelled RE70 alone (Figure 2B, lane 2) generated predominantly CII (i) and little CI (ii). Inclusion of the unlabelled but shorter RE49 generated a slightly faster migrating CII species (lane 4; iii). An identically migrating species was also formed using 5′-end-labelled RE49 and unlabelled RE70 (lane 6; iv). Both CII species, therefore, carry two ends: one RE70 and one RE49 copy. Note that the position of the minor CI depends only on the size of the end-labelled RE used. This is consistent with the notion that CI contains a single RE.
Assembly of a biologically relevant synaptic complex containing left and right ends
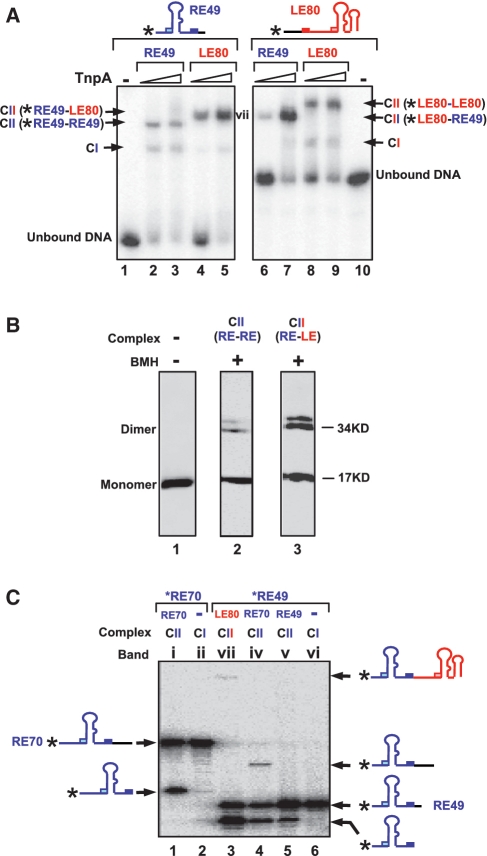
Synapsis during transposition must include both LE and RE. However, the complexes above are composed of only one type of IS608 end, either RE or LE. To assemble complexes containing both ends, we used a mixture of 5′-end-labelled RE49 and a longer unlabelled LE80 (Supplementary Table S1) in excess. The results (Figure 3A) show that RE49 alone (lane 1) generates both CI and CII species (lanes 2 and 3). Inclusion of unlabelled LE80 leaves some CI complex intact but all the CII observed with RE49 alone chases into a slower migrating complex presumably carrying single copies of labelled RE49 and unlabelled LE80 (lanes 4 and 5, vii). Reversal of the labelling regime revealed that end-labelled LE80 (lane 10) generated both CI and CII species (lanes 8 and 9) and addition of excess of unlabelled RE49 chased the more slowly migrating CII into a faster migrating species (lanes 6 and 7).
Figure 3.
Identification of the IS608 transpososome. (A) Identification of complexes including both RE and LE. The 5′-end-labelled RE49 (lanes 1–5); alone (lane 1); with TnpA and excess unlabelled RE49 (lanes 2 and 3); with TnpA and excess unlabelled LE80 (lanes 4 and 5); 5′-end-labelled LE80 (lanes 6–10) alone (lane 10); with TnpA and excess unlabelled LE80 (lanes 8 and 9); with TnpA and excess unlabelled RE49 (lanes 6 and 7). Asterisk indicates the 5′ label. Triangle presents different TnpA-His6 concentration (0.5 and 2.5 µM). The band indicated by vii was cut from the gel and used for the analysis of catalytic activity presented in Figure 3C. (B) Identification of crosslinked TnpA. CII composed of two REs (RE49F) or one RE and one LE (LE63) were excised from an EMSA gel and treated with the crosslinking agent BMH. They were loaded onto an SDS–PAGE gel and identified by western-blot using a 6His-HRP antibody. Lane 1: TnpA-His6 alone; lane 2: CII carrying two RE49F treated with BMH (the subscript F indicates that the oligonucleotides used were labelled with a 3′ fluorescein to identify the synaptic complexes to be isolated from the initial EMSA gel; see oligonucleotide list, Supplementary Table S1); lane 3: CII carrying one RE49F and one LE63 treated with BMH. (C) Activities of CI and CII. Bands labelled with roman numerals in Figures 2B and 3A were excised from the gels and soaked in a cleavage buffer containing Mg2+ for 3 h. DNA was eluted and analysed on a 9% denatured sequencing gel. Lane 2 and lane 6: CI composed of TnpA-His6 carrying only one DNA end RE70 or RE49; lane 1 and lane 5: CII composed of TnpA-His6 carrying two same DNA ends RE70 or RE49; lane 4: CII composed of TnpA-His6 carrying RE49 and RE70; lane 3: CII composed of TnpA-His6 carrying RE49 and LE80.
To determine protein stoichiometry, CII complexes containing both LE and RE or RE alone were isolated from the gel and cross-linked using BMH (see ‘Materials and Methods’ section). Two closely migrating complexes were observed at a position expected for a dimer (Figure 3B) in SDS–PAGE. These presumably represent dimers that are cross-linked in different ways. Thus CII is a TnpA dimer containing two IS608 ends.
Activity and stability of the IS608 transpososome
Complex CII but not CI is active in strand cleavage and transfer
To determine if the complexes observed in EMSA have catalytic activity, bands were cut from the gels, incubated with buffer containing Mg2+ and the reaction products were eluted and separated on a denaturing gel (Figure 3C). Comparable amounts of radioactivity were used in each sample to compensate for the different amounts of CI and CII formed. CII [Figure 2B(i), (iv), (v) and Figure 3A (vii)] clearly catalysed Mg2+-dependent cleavage (Figure 3C, lanes 1, 3, 4 and 5). Furthermore, CII formed with RE49 and RE70 exhibited a band consistent with strand transfer corresponding to exchange between the RE49 and RE70 (3) (lane 4). CII formed with LE80 and RE49 also generated a strand transfer product with a size expected for the RE–LE junction (Figure 3C, lane 3). Note that in lanes 1 and 5 because the labelled and unlabelled DNA substrates in these reactions were identical in length. Strand transfer products could not be distinguished because they would be of the same length as the initial substrates. In contrast, CI generated by RE70 [Figure 2B (ii)] or RE49 [Figure 2B (vi)] appeared catalytically inactive (Figure 3C, lanes 2 and 6).
DNA sequence of RE required for complex stability
Although complexes between TnpA and the hairpins, IPL and IPR, can be readily detected in solution by gel filtration (4) and by fluorescence techniques (23), we were unable to identify them using EMSA, presumably because they are unstable and are lost during electrophoresis (Supplementary Figure S1A). However, as shown above, complexes can be clearly identified by EMSA when longer DNA substrates are used suggesting that DNA in addition to IPL and IPR is necessary to generate a robust complex.
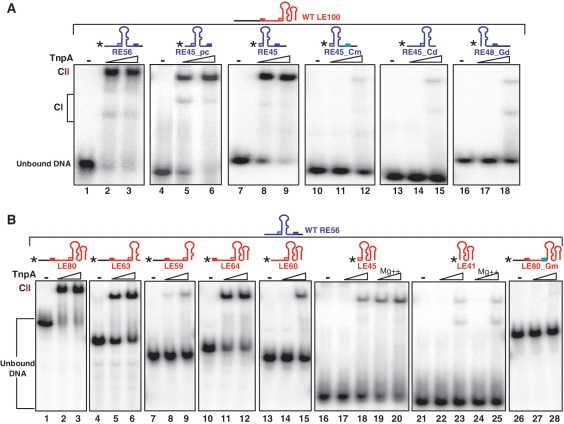
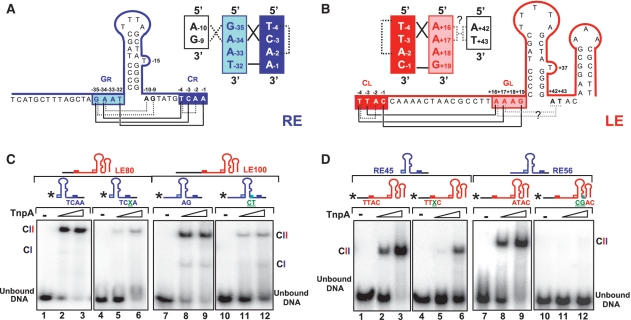
To identify the RE sequences involved, a series of RE derivatives were 5′-end labelled and used in conjunction with an excess of unlabelled full length LE100 (Supplementary Table S1) to visualize CII complexes carrying both LE and RE. Full-length RE generated a robust CII species (Figure 4A, RE56, lanes 2 and 3) as did RE derivatives without the 3′ flank of CR (Figure 4A, RE45_pc, lanes 5 and 6) or the sequence 5′ of GR (Figure 4A, RE45, lanes 8 and 9). However, almost no CII could be detected with a derivative in which the CR had been either mutated (TCAA>AGAG; Figure 4A, RE45_Cm, lanes 11 and 12) or deleted (Figure 4A, RE45_Cd, lanes 14 and 15). Moreover, removal of GR also prevented robust CII formation (Figure 4A, RE48_Gd, lanes 17 and 18). These results suggest that the interaction between GR and CR is important in generating CII.
Figure 4.
DNA sequences necessary for transpososome stability. (A) Sequences required in RE. EMSA of excess unlabelled LE100 mixed with 5′-end-labelled RE derivatives performed with 0, 0.5 and 2.5 µM TnpA-His6. Two complexes with different mobility are indicated as CI and CII. Lanes 1–3, wt RE56; lanes 4–6, precleaved RE45; lanes 7–9, RE45 deleted for DNA 5′ to GR; lanes 10–12, RE45 deleted for DNA 5′ to GR and mutated in CR (TCAA to AGAG); lanes 13–15, RE45 deleted for CR; lanes 16–18, RE48 deleted for GR. (B) Sequences required in LE. EMSA of excess unlabelled RE56 mixed with 5′-end-labelled LE derivatives performed with 0, 0.5 and 2.5 µM TnpA-His6. Lanes 1–3, wild-type LE80; lanes 4–6, LE63, deleted for the 3′ potential secondary structure; lanes 7–9, LE59 deleted for the ATAC tetranucleotide 3′ to the foot of IPL; lane 10–12, LE64, lacking the 5′ left flanking DNA but retaining CL; lanes 13–15, LE60, precleaved LE lacking the 5′ left flanking DNA and CL; lanes 16–20, LE45 retaining only GL 5′ to IPL and the region 3′ to IPL without (lanes 17 and 18) or with (lanes 19 and 20) Mg2+; lanes 21–25, LE41 deleted for GL 5′ to IPL but retaining the region 3′ to IPL without (lanes 22 and 23) or with (lanes 24 and 25) Mg2+; lanes 26–28, LE80 mutated in GL (AAAG to GGAA) ‘*’ indicates 5′ label.
DNA sequence of LE required for robust complexes
In a similar manner, we investigated the role of various LE interactions in generating robust CII complexes.
A series of 5′-end-labelled LE derivatives were used in conjunction with an excess of unlabelled full-length RE56 to visualize CII. Full-length LE generated a robust CII species with RE56 (Figure 4B, LE80, lanes 2 and 3). In addition to IPL, LE includes a second 3′ hairpin. Deletion of this reduced complex levels as judged by the presence of uncomplexed DNA (Figure 4B, LE63, lanes 5 and 6). Surprisingly, deletion of the 4 nt 3′ to IPL (which separate the two hairpins) significantly reduced CII levels in spite of the presence of CL and GL (Figure 4B, LE59, lanes 8 and 9). The absence of the DNA flank 5′ to CL decreased complex stability slightly as judged by the level of uncomplexed DNA (Figure 4B, LE64, lanes 11 and 12). Deletion of CL with or without the DNA between CL and GL significantly reduced, but did not eliminate, CII (Figure 4B, LE60, lanes 14 and 15 and LE45, lanes 17 and 18). Almost no complex could be detected if GL was also deleted (Figure 4B lanes, LE41, 22 and 23). Mutation of GL alone (AAAG to GGAA) in full-length LE also eliminated CII (Figure 4B, LE80_Gm, lanes 27 and 28).
Mg2+ can increase the robustness of CII
As a metal ion is required for catalysis, we wondered if divalent cations might stabilize the TnpA–LE complex even in the absence of the CL/GL interactions. Comparison of LE45 and LE41 showed this to be the case. Addition of Mg2+ slightly increased the level of CII if GL was present (LE45; Figure 4B, compare lanes 19 and 20 with 17 and 18) but not if it was absent (LE41; compare Figure 4B lanes 24 and 25 with 22 and 23). Stabilization, therefore, requires GL suggesting that activation of TnpA by GL (20) is involved.
Effect of non-canonical base pairing on complex stability
Structural analysis previously revealed a complex network of base interactions within LE and within RE [Figure 5; (20)] which could increase the robustness of CII by facilitating correct folding of the ssDNA. Indeed, interactions between GL and CL are important in determining the tetranucleotide sequence specificity in integration of the IS (21).
Figure 5.
Non-canonical base interactions. (A) Schematic of the canonical and non-canonical base interactions in RE as identified from the crystal structure. Cleavage sequence CR is shown in dark blue, guide sequence GR in pale blue. The left hand section shows the nucleotide sequence of RE and the base paring within IPR. The extra-helical T is also indicated. The filled lines beneath indicate base canonical interactions between CR and GR. The dotted lines indicate additional non-canonical base interactions. These are schematized in the cartoon on the right. The nucleotide coordinates are those from (20). (B) Schematic of the canonical and non-canonical base interactions in LE as identified from the crystal structure. CL is shown in red, GL in pink. The right hand section shows the nucleotide sequence of LE and the base paring within IPL. Base interactions are marked as for RE. It should be noted that the base triplets indicated between the AT of the ATAC tetranucleotide have not been demonstrated by crystallography but are implied from the results obtained here. (C) Effect of mutations in non-canonical base interactions in RE. EMSA of 5′ labelled WT RE45 (lanes 1–3) or 2-aminopurine modified RE45 (lanes 4–6) and unlabelled LE80 with 0, 0.5 and 2.5 µM TnpA-His6. EMSA of 5′ labelled WT RE56 (lanes 7–9) or A−10G−9 mutated RE (lanes 10–12) and unlabelled LE100 with 0, 0.5 and 2.5 µM TnpA-His6. Two complexes with different mobility are indicated as CI and CII. X is 2-amino purine (2-AP). (D) Effect of mutations in non-canonical base interactions in LE. EMSA of 5′-labelled WT LE80 (lanes 1–3) or 2-aminopurine modified LE80 (lanes 4–6) and unlabelled RE45 with 0, 0.5 and 2.5 µM TnpA-His6. EMSA of 5′ labelled WT LE80 (lanes 7–9) or A+42T+43 mutated LE (lanes 10–12) and unlabelled RE56 with 0, 0.5 and 2.5 µM TnpA-His6. X is 2-amino purine (2-AP) ‘*’ indicates 5′ label.
Not only do bases in GL/R form canonical base pairs with CL/R in both RE and LE (Figure 5A and B, continuous lines; and Supplementary Figure S3A–D), but at RE, the two bases, A−10 and G−9, 3′ to the foot of IPR form base triplets with the first two bases, G−35 and A−34, of GR (Figure 5A, dotted lines). It is not known whether equivalent base triplets occur at LE. Additional base interactions were observed within both CR and CL. These occur between T−4 and A−2 in CR and between T−4 and A−2 in CL via a hydrogen bond between the 2-oxygen group of the first nucleotide T−4 and the 6-amino group of the third unpaired nucleotide A−2 of CL and of CR (dotted lines in Figure 5A and 5B, S2A and S2C). Note that A−2 has no other base interactions. We have examined the effects of different components of this interaction network.
We first asked whether the internal nucleotide interactions within CR and CL assist in stabilizing CII. Replacing the A−2 by its analogue 2-aminopurine (2-AP), thus moving the 6-amino group to position 2 to eliminate this hydrogen bond (Supplementary Figure S2B and D), significantly decreased complex robustness. A 2-AP-modified RE formed much less stable CII with LE80 (Figure 5C, lanes 5 and 6) than did a wild-type RE (Figure 5C, lanes 2 and 3). Similarly, the equivalent 2-AP derivative of LE showed lower CII levels (Figure 5D, lanes 5 and 6) with RE45 compared to wild-type LE (Figure 5D, lanes 2 and 3). This supports the idea that the T−4–A−2 hydrogen bond in the network plays a role in increasing CII robustness although it does not rule out that displacement of the amino group has additional effects which in turn lead to reduced stability.
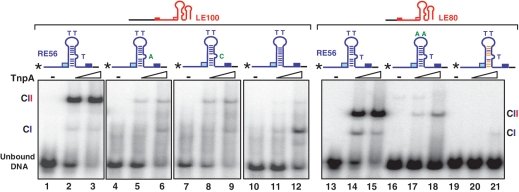
We next examined the effect of base triplet interactions between the first two bases of GR and two bases directly 3′ of IPR (C−3G−35*G−9 and T−4A−34*A−10, where asterisk indicates the non-canonical base pairing, Figure 5A and S2E and S2F) (20). If these interactions are important, mutating both purines (A−10/G−9) 3′ of IPR to pyrimidines (C/T) should destabilize the triplet structure. Indeed, mutation of RE led to a much lower CII level with LE100 (Figure 5C lanes 11 and 12) compared to wild-type RE (Figure 5C, lanes 8 and 9).
By analogy to RE, it seemed possible that LE might also possess a similar triplet structure, yet direct structural evidence for this is not available. It is also not clear which region of LE might contribute the ‘missing’ nucleotide of the triplets. However, deletion analysis indicated that the 4 nt (A+42T+43A+44C+45) localized 3′ to IPL affect CII robustness (Figure 4B, lanes 7–9). The A+42T+43 dinucleotide is located in the equivalent position in LE (Figure 5B) to the triplet-forming nucleotides in RE (Figure 5A). By symmetry with RE, the equivalent GL and CL bases involved would be A+16A+17 and T−4T−3, respectively (Figure 5B). Note that in triplet formation, such T.A pairs prefer A or T as the third base (24). Mutation of A+42 to C led to a large decrease in CII (Supplementary Figure S1B, lanes 2 and 3). While mutation of T+43 to G slightly reduced CII (Supplementary Figure S1B, lanes 5 and 6), the double mutation A+42T+43 to CG completely abolished CII (Figure 5D, lanes 11and 12). The influence of the mutation of A+44 to C was much less important (Supplementary Figure S1B, lanes 8 and 9). Thus A+42 T+43 might be involved in TA*A and TA*T triplet formation (Supplementary Figure S2G and S2H) by interacting with A+16A+17 in GL (indicated as dotted line in Figure 5B).
Bases within IP which are important for complex stability
Although TnpA does not use a classical DNA sequence readout for recognition, it does make specific key base interactions. In addition to the base interactions within LE and RE, it is also likely that the limited number of TnpA–base interactions are involved in CII assembly.
One example is the extra-helical T in both IPL and IPR (Supplementary Figure S3). In IPR, this (T−15) is positioned in a pocket of one TnpA monomer and stacked between the benzene ring of Phe75 and the guanidinium moiety of Arg52 of the second monomer (4). Moreover, the carbonyl oxygen of Leu51 forms a short hydrogen bond with the N3 of T−15 (Supplementary Figure S2I). Deleting or replacing T−15 by a purine (A) or a pyrimidine (C) significantly decreased CII stability (Figure 6, lanes 4–12). The similar increase in disassociation constant of IPL and TnpA observed when replacing T+37 by A, which was explained by space limitation to accommodate a purine base (23). Here, the decrease in CII robustness cannot simply due to a limitation by the available space in the surrounding TnpA pocket because the mutation to C does not increase the space required to accommodate them. However, mutation eliminates a hydrogen bond, between the N3 position of T and the carbonyl oxygen of Leu51 (Supplementary Figure S2J and S2K). Also when T−15 is deleted to generate a perfect palindrome, several interactions between T−15 of RE bound to one monomer and the other monomer are abolished. Similar results were obtained with mutants of LE (data not shown). This suggests that these DNA-mediated ‘crossed’ interactions between monomers contribute to the robustness of the complex.
Figure 6.
IP shape and synaptic complex stability. Role of extra-helical T in RE (lanes 1–12). EMSA of excess unlabeled LE100 with 5′ labelled WT RE56 or RE mutant in which T−15 was mutated to A or C or deleted performed with 0, 0.5 and 2.5 µM TnpA-His6. Two complexes with different mobility are indicated as CI and CII. Role of tip Ts and IP shape of RE (lanes 13–21). EMSA of excess unlabeled LE80 with 5′ labelled WT RE56 or RE mutants in which T−21T−22 was mutated to AA or the bases of IPR stem which do not interact directly with the protein was reversed performed with 0, 0.5 and 2.5 µM TnpA-His6. Two complexes with different mobility are indicated as CI and CII. The blue lines inside the IP indicate the original base pairing while the orange lines show pairing of the reversed bases (Supplementary Table S1) ‘*’ indicates 5′ label.
Another example occurs in the tip of IPL and IPR. Previous studies showed a significant decrease in TnpA binding to a minimal IPL substrate when all three Ts at the tip were mutated to A (23). To confirm the importance of these bases in the context of the active transpososome (CII complexes containing LE and RE), we analysed comparable mutations within IPR by EMSA. The results (Figure 6, lanes 17 and 18) show that mutation of T−21T−22 to AA indeed reduces CII stability. The decreased CII stability could also be explained by specific base interactions with TnpA since T−22 also interacts with Gly86 and Arg87 (4).
We have also investigated the importance of sequence of the IPR stem by changing those bases which do not appear to interact directly with the protein in the crystal structures (4). We exchanged C−31 and G−11, C−30 and G−12, C−29 and G−13, T−27 and A−16, A−26 and T−17, C−24 and G−19, T−23 and A−20 (‘HP_m’ in Supplementary Table S1). CII formation was abolished using this mutant (Figure 6, lanes 20 and 21). In the absence of specific TnpA–base interactions at these positions, it seems probable that this is due to a change in shape of the non-B form IP stem.
These results, therefore, suggest that both the specific TnpA–base interactions and the surface shape of IPs are important for binding TnpA and CII formation.
DISCUSSION
Learning from structure
ss transposition is a recently identified mechanism adopted by members of the widespread bacterial and archeal IS200/IS605 family (3). Several structures of the IS200/IS605 ss transpososome, the protein–DNA machinery that accomplishes this, have been solved and have provided key information concerning its central role in orchestrating the catalytic steps of transposition. The structures of the IS200/IS605 family paradigms, IS608 and ISDra2 (20); (23) with and without their minimal DNA binding sequences (subterminal ssDNA hairpin structures, IPL and IPR) have revealed a TnpA dimer with two DNA binding sites. Unusually, TnpA recognizes transposon DNA through the subterminal IPL and IPR rather than by classical DNA sequence-specific readout. At the left end, LE, cleavage requires a set of base interactions between a tetranucleotide cleavage site (CL) located 19 nt 5′ to IPL, and not part of the IS, and a tetranucleotide guide sequence (GL) located 5′ to the foot of IPL (Figure 5). A similar arrangement occurs at the right end, RE, but the cleavage site (CR) is located within the IS only 10 nt from the foot of IPR and to its 3′ side. In addition, a single A residue within GL and GR is inserted into the protein resulting in a large change in its structure and activation of the catalytic site. The crystal structure has also revealed a network of additional non-canonical base interactions.
However, in spite of this information, little is known about the biologically relevant transpososome containing both left and right IS ends. Here, we have identified this complex and present a detailed and extended picture of its assembly, activity and the interactions involved in its stability in solution.
Identification of the IS608 transpososome
EMSA analysis using either LE or RE identified two TnpA DNA complexes, CI and CII. CII contained two IS608 ends and, unlike CI, was catalytically active. The minor CI complex may carry only a single DNA molecule. Using an end-labelled LE or RE oligonucleotide together with an excess of the opposite, unlabelled, end we have also identified biologically relevant CII species containing both LE and RE (a simulation of this complex is shown in Supplementary Figure S3F). Again, these complexes were catalytically active in cleavage and recombination.
The relationship between CI and CII is at present unclear. CI is generally present only at very low levels at all TnpA concentrations used. While this might suggest that it is a precursor of CII, it may simply indicate that CI is a non-productive complex either formed independently of CII or resulting from partial disassembly of CII.
However, binding leading to CII formation is clearly strongly cooperative and might have important consequences for transpososome assembly within the cell. Cooperativity would presumably favour the use of two closely spaced ends. It might also impose a constraint that both IS ends be in their active ss form before the assembly process can be initiated. This provides a regulatory mechanism which would prevent adventitious activation of TnpA-mediated cleavage on single IS ends.
Formation of a robust transpososome
Gel filtration had identified complexes between a TnpA dimer and an oligonucleotide including the 22-nt IPL or 21-nt IPR (4) and derivatives carrying the tetranucleotide GL or GR. These were not detected by EMSA (Supplementary Figure S1A). However, robust complexes able to withstand the conditions of gel electrophoresis were observed with longer LE or RE oligonucleotides (5). We show that robust complexes require CL, CR, GL and GR. Moreover, in addition to the canonical CL/R–GL/R base interactions, the network of non-canonical base interactions identified from the co-crystal structures with RE and with LE alone [Supplementary Figure S3 (20)] are necessary for synaptic complex formation. In RE, these include interactions between bases 3′ to IPR and GR which form base triplets, and between T−4 and A−2 in CR. Similar requirements were shown in LE between T−4 and A−2 in CL. Moreover, although for technical reasons we did not identify base triplets in the LE co-crystal structures, the results presented here suggest that such interactions indeed exist: A+42 and T+43 (at equivalent positions to triplet forming bases A−10 and G−9 in RE) are required for robust complex formation.
Our data also suggest that the changes in TnpA conformation on activation by A+18 in GL and A−34 in GR which create a catalytic pocket with the correct architecture for binding of the essential metal ion, Mg2+ also leads to a more robust transpososome and that addition of Mg2+ can increase complex stability in compromised complexes lacking CL or CR.
Implications for target choice
Base changes in GL (but not in GR) lead to predictable changes in the tetranucleotide used as a target site for insertion (21). This is consistent with the fact that the natural target tetranucleotide, TTAC (CT), is also the same as the left cleavage site CL and is recognized in a similar way by GL. Although changing GL resulted in predictable changes in the target tetranucleotide, we observed large differences in insertion frequency. The influence of the presumed non-canonical interactions between bases in LE would provide an explanation for this variability since these were not taken into account in our choice of GL sequence. Thus although target choice depends critically on the GL sequence, it may be ‘fine-tuned’ by the additional non-canonical base interactions. These interactions would provide important constraints for maintaining target choice. Simple mutation of bases within GL would not, on their own, be expected to lead to the efficient use of alternative target sequences in vivo. From the point of interactions of the IS with its host genome, this means that spontaneous changes in the target sequence resulting from random mutations in GL would be limited by the absence of the corresponding changes required to maintain the network of base interactions necessary for transpososome stability.
These data provide a solid basis for understanding the forces involved in IS608 transpososome formation. They lead directly to questions such as how LE and RE are initially recognized (e.g. whether they form the fold-back structures prior to binding or by subsequent interaction with TnpA) and how transposition activity is mechanistically coupled to replication forks. Initial low energy circular dichroism (25) and fluorescence studies suggest that the IP structures exist in solution in the absence of TnpA (Susu He, Bao Ton Hoang, Michael Chandler and Neil P. Johnson, unpublished data), and further preliminary studies suggest that TnpA preferentially binds forked DNA structures in vitro and targets replication forks in vivo (Laure Lavatine, Bao Ton Hoang and Michael Chandler, unpublished data).
Relationship with other mobile genetic elements and RNA
The ss transpososome is a relatively simple and interesting example of DNA recognition using a structural rather than classical sequence-specific readout. Very few examples of this type are known. Integrons and certain ss phage are known to use ss intermediates for integration into and excision from their host chromosomes (26,27) involving sequence independent structural recognition. Non-canonical base interactions are often observed in RNA structures but rarely in DNA structures. IS608 is therefore somewhat analogous to certain ribonucleoproteins in which the nucleic acid itself is involved in establishing the active complex and, in this case, providing the necessary recognition signals for catalysis. In addition TnpA has adopted a strategy of base interactions similar to the common use of guide RNA for sequence recognition in RNA binding proteins (22). This suggests that members of this IS family may be more closely related to the ‘RNA world’ than are other ISs and perhaps of more ancient origin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (France), by Agence National de Recherche (France) grant Mobigen (M.C. and N.P.J.); the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (F.D.). Funding for open access charge: Agence National de Recherche (France) (Grant MOBIGEN to M.C. and N.P.J.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank C. Guynet, L. Lavatine, G. Duval-Valentin and B. Hallet for discussions.
REFERENCES
- 1.Siguier P, Filee J, Chandler M. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006;9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B. In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol. Cell. 2008;29:302–312. doi: 10.1016/j.molcel.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell. 2005;20:143–154. doi: 10.1016/j.molcel.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 5.Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 2005;24:3325–3338. doi: 10.1038/sj.emboj.7600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ton-Hoang B, Pasternak C, Siguier P, Guynet C, Hickman AB, Dyda F, Sommer S, Chandler M. Single-stranded DNA transposition is coupled to host replication. Cell. 2010;142:398–408. doi: 10.1016/j.cell.2010.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersulyte D, Velapatino B, Dailide G, Mukhopadhyay AK, Ito Y, Cahuayme L, Parkinson AJ, Gilman RH, Berg DE. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 2002;184:992–1002. doi: 10.1128/jb.184.4.992-1002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman AB, Chandler M, Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochemi. Mol. Biol. 2010;45:50–69. doi: 10.3109/10409230903505596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin EV, Ilyina TV. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 10.Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 11.Gueguen E, Rousseau P, Duval-Valentin G, Chandler M. The transpososome: control of transposition at the level of catalysis. Trends Microbiol. 2005;13:543–549. doi: 10.1016/j.tim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Savilahti H, Mizuuchi K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 13.Savilahti H, Rice PA, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naumann TA, Reznikoff WS. Trans catalysis in Tn5 transposition. Proc. Natl Acad. Sci. USA. 2000;97:8944–8949. doi: 10.1073/pnas.160107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 2010;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 17.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haniford DB, Benjamin HW, Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991;64:171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- 19.Surette MG, Buch SJ, Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- 20.Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell. 2008;132:208–220. doi: 10.1016/j.cell.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guynet C, Achard A, Hoang BT, Barabas O, Hickman AB, Dyda F, Chandler M. Resetting the site: redirecting integration of an insertion sequence in a predictable way. Mol. Cell. 2009;34:612–619. doi: 10.1016/j.molcel.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickman AB, James JA, Barabas O, Pasternak C, Ton-Hoang B, Chandler M, Sommer S, Dyda F. DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. EMBO J. 2010;29:3840–3852. doi: 10.1038/emboj.2010.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beal PA, Dervan PB. The influence of single base triplet changes on the stability of a pur.pur.pyr triple helix determined by affinity cleaving. Nucleic Acids Res. 1992;20:2773–2776. doi: 10.1093/nar/20.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson NP, Baase WA, von Hippel PH. Investigating local conformations of double-stranded DNA by low-energy circular dichroism of pyrrolo-cytosine. Proc. Natl Acad. Sci. USA. 2005;102:7169–7173. doi: 10.1073/pnas.0502359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.