Abstract
We have developed a novel technique of using fluorescent tRNA for translation monitoring (FtTM). FtTM enables the identification and monitoring of active protein synthesis sites within live cells at submicron resolution through quantitative microscopy of transfected bulk uncharged tRNA, fluorescently labeled in the D-loop (fl-tRNA). The localization of fl-tRNA to active translation sites was confirmed through its co-localization with cellular factors and its dynamic alterations upon inhibition of protein synthesis. Moreover, fluorescence resonance energy transfer (FRET) signals, generated when fl-tRNAs, separately labeled as a FRET pair occupy adjacent sites on the ribosome, quantitatively reflect levels of protein synthesis in defined cellular regions. In addition, FRET signals enable detection of intra-populational variability in protein synthesis activity. We demonstrate that FtTM allows quantitative comparison of protein synthesis between different cell types, monitoring effects of antibiotics and stress agents, and characterization of changes in spatial compartmentalization of protein synthesis upon viral infection.
INTRODUCTION
Dynamic spatial compartmentalization of different constituents of the protein translation apparatus plays a significant role in the regulation of this essential cellular function (1–3). Modern microscopy-based techniques enable the characterization of the dynamics of fluorescent molecules at high temporal/spatial resolutions. Genetically encoded fluorescent protein fusion constructs are a principal experimental approach for the study of protein function in intact cells (4). A complementary approach involves the exogenous introduction of labeled molecules, such as antibodies or ligands, allowing the observation of in-vivo dynamics of different classes of macromolecules, in cases where the expression of genetically modified proteins is either impossible or undesired.
The measurement of protein levels, even when carried out in single cells with high temporal and spatial resolution, generally reflects their steady state levels and fails to distinguish between contributions of different processes such as synthesis and degradation. In particular, this approach measures the products of translation, and not the translation process itself, and hence suffers in terms of temporal and spatial resolutions. Alterations or impairments to cellular functions such as differentiation or viral infection may induce, involve or depend on modulation of the protein synthesis apparatus. For example, the usurpation of cellular resources by viruses may involve extensive modulations to the spatial/temporal parameters of mRNA translation. Specifically, the replication cycle of reoviruses involves the formation of viral factories, a compartmentalization of various cellular components aimed at maximizing replication efficiency and neutralizing host defenses (5,6). The molecular mechanisms of the induction and the execution of these processes is currently the object of intense study in various viral model systems (7–9). The extent and means by which Reoviridae such as epizootic hemorrhagic disease virus-2 (EHDV2) (Ibaraki strain, EHDV2-IBAV) (10,11) manipulate and alter the cellular protein synthesis apparatus is currently unknown.
Here, we introduce and apply a novel technique, termed fluorescent tRNA for translation monitoring (FtTM) to identify and monitor intracellular sites of active protein synthesis. FtTM utilizes quantitative fluorescence microscopy and transfected bulk uncharged tRNA, labeled with fluorescent probes [which can also function as a fluorescence resonance energy transfer (FRET) donor/acceptor pair]. With FtTM, two modes of protein synthesis assessment are possible: a high sensitivity, lower resolution mode in which fl-tRNA functions as a reporter for protein synthesis site in single live cells, and a higher resolution FRET-based method (characterized by a lower sensitivity) that generates a quantitative report, at submicron level, of translation events. Employing FtTM, we quantitatively compared protein synthesis in activated and resting primary brain astrocytes, and provide evidence for the spatial compartmentalization of protein synthesis in EHDV2–IBAV-infected cells.
MATERIALS AND METHODS
Cell lines and culture conditions
Chinese hamster ovary cells (CHO) were maintained in nutrient mixture F-12 (Ham) supplemented with 10% (v/v) fetal calf serum, 5 mM l-glutamine and antibiotics (all from Biological Industries Ltd). Astroglial primary cell culture was produced from the cerebral cortex of 10 newborn (P1–P3) C57BL mice. For activated astrocytes, cells were treated with final concentrations of 2 µg/ml LPS (Sigma) and 3 ng/ml INFγ (Roche Pharmaceuticals Ltd) for 24 h.
Virus
The Ibaraki Virus was obtained from Dr T. Tsuda, of the Kyushu Research Station, National Institute of Animal Health, Chuzan, Kagoshima, Japan and donated to this study by Dr Hagai Yadin of the Kimron Veterinary Institute, Beit Dagan, Israel. The virus (denoted strain 2) was isolated in September 1959 in Ibaraki, Japan. The virus was passaged 19 times (8 passages in bovine embryo kidney cells, 3 passages in bovine kidney cells and 8 passages in Hamster lung cells).
tRNA
Commercial preparations of total yeast (Sigma, Invitrogen or Ambion) or bovine (Sigma) tRNA were labeled at dihydrouridine positions with either rhodamine 110 or Cy3 as described (8,12). In some cases, denaturing urea–PAGE analysis indicated the need for purification to remove tRNA fragments and high-molecular weight species, most likely contaminating DNA, prior to labeling. Such purification involved two extractions with phenol/chloroform and ethanol precipitation, followed by Qiagen-tip-500-gravity flow column chromatography (anion exchange) and RP-HPLC (C18). Labeled tRNAs typically contained 0.6–0.8 dye/tRNA. Yeast bulk tRNA: 0.75 Cy3, 0.76 Rho110; bovine bulk tRNA: 0.63 Cy3, 0.65 Rho110.
Virus purification
EHDV2–IBAV-infected OK cells were collected 48 h post infection and pelleted at 4°C. The pellet was resuspended in 6 ml TNET Buffer (50 mM Tris–HCl pH 8.0, 0.2 M NaCl, 5 mM EDTA, 0.5% Triton X-100) and homogenized (10 strokes) using a Glass homogenizer (7 ml). The homogenate was layered onto a sucrose cushion comprised of 66% and 40% sucrose each prepared in 0.2 M Tris. Samples were centrifuged in a Beckman Ultracentrifuge using a SW41 Rotor, at 23 000 r.p.m., 4°C for 3 h. Purified virus was extracted from the interface of the sucrose cushions and 10 mM dithiothreitol (DTT) was added to prevent virus aggregation.
Virus infection
CHO cells, seeded at 1 × 105 cells/well in 24-well plates on coverslips, were infected with semi-purified Ibaraki virus (MOI = 1) for 24 h, and transfected with labeled yeast tRNA (see under Transfection protocol and FRET assay protocol). For protein synthesis assay, cells were seeded at 2 × 105 cells/well in 6-well plates, infected the next day and radio-labeled 30 h post infection.
In vitro translation assays
Two systems were used: wheat germ and rabbit reticulocyte lysate (Promega). Different concentrations (0–50 µM) of labeled or unlabeled tRNA (Sigma) were added to the in vitro translation mixes supplemented with luciferase RNA and luciferase activity was determined by using Veritas luminometer according to manufacture protocol.
Antibodies
Monoclonal mouse anti-non-structural protein 2 (NS2) antibodies were specially generated for the present study. Mice were immunized with the following peptide: n′–PEPKGYVLEISEVGSYRIQDG–c′ [corresponding to amino acids 51–71 of EHDV2-IBAV NS2, GeneScript Corporation (NJ, USA)].
Immunostaining
CHO cells pre- or post-viral infection and/or transfection, were fixed (4% PFA, 20 min, room temperature), permeabilized (0.5% Triton-X, 10 min), extensively washed with PBS, blocked (4% BSA and 5% normal goat serum in PBS (PBS/BSA), 1 h) and stained. The following primary antibodies were employed (typically at 1:500 dilution of the commercially supplied sample, in PBS/BSA, 4°C, overnight): arginyl-tRNA synthetase (Abcam), eEF-1A (Cell Signaling), calnexin (Sigma), rpS6 (Cell Signaling), clathrin (Novus) and anti-NS2 (generated in the course of this study). Secondary antibodies were AlexaFluor-488 IgG goat anti-rabbit and/or AlexaFluor-555 IgG goat anti-mouse (Invitrogen; 1:500 dilution, PBS/BSA, 2 h, room temperature). Coverslips were mounted with Fluoromount (Sigma).
Transfection
Cells were seeded in the day preceding the transfection (1.5 × 105 cells/well, 13 mm coverslips, in 24-well plates). Transfection was with 3 µg of yeast labeled—bulk tRNA. In all cases, transfections were performed according to manufacture's instructions. For the majority of experiments, Interferin™ (Polyplus Transfection) of Polyplus transfection was employed. For DNA transfection (pSuper-retro-GFP), 9 × 104 cells/well were transfected with 1 µg supercoil plasmid DNA (TransIT-LT1 transfection reagent (Mirus) according to manufacture's protocol). The following day, cells were re-transfected with labeled tRNA as described above.
Quantitative image analysis
For quantitative analysis of spatial distribution of the labeled tRNA and additional cellular markers cells were imaged with spinning disk confocal microscope [Yokogawa CSU-22 Confocal Head; Axiovert 200M, Zeiss; under the control of SlideBook™ (Intelligent Imaging Innovations) and coupled to a CoolSnap HQ2 camera (Photometrics)]. Typically, the entire cell volume was captured (0.15 µm between planes of the z-stack) with a 100× lens (1.45 NA, Zeiss) and 1 × 1 ‘on chip binning’ yielding a pixel size of 65 × 65 nm; and deconvolved with the constrained iterative algorithm of Slidebook™ which employs an experimentally measured point spread function (PSF). Images, segmented by the intensity of the specific fluorescence signal (determined experimentally through non-specific staining controls), and served as a basis for the calculation of the percentage of overlapping pixels (denominated CL, co-localization). The Pearson's correlation coefficient (CCF) and van Steensel's cross-correlation (CCF, dx) were calculated with the JaCoP plugin of the ImageJ software (13).
FRET assay
Cells transfected with bulk yeast or bovine tRNA labeled with Rho110- (donor) and/or Cy3- (acceptor) were fixed 7 h post transfection, mounted and imaged with a spinning disc confocal microscope. A raw FRET signal consisting of the recorded emission above 570 nm in response to illumination at 473 nm served as a basis for the calculated FRET signal [FRETc; (raw FRET signal after the elimination of background and of the non-specific contribution originating from the experimentally measured donor bleedthrough and direct excitation of the acceptor under the FRET illumination)]. Images were acquired with 2 × 2 binning, yielding a pixel size of 260 × 260 nm.
Live cell imaging
Cells were grown in glass-bottomed 35 mm dishes and transfected as described. Seven hours post transfection, cells were extensively washed with Hepes-buffered media (pH 7.4) and imaged at 37°C with a inverted fluorescence microscope. Time-lapse sequences were acquired with a 30-s interval between frames for ~90 min. Images were deconvolved with the NoNeighbors algorithm of Slidebook™.
Protein synthesis and western blot assays
Cells, transfected or not with labeled tRNA, and under different treatments [(puromycin, 1 mM, 30 min, Mercury), (cycloheximide 0.1 mg/ml, 10 min, Sigma) or (thapsigargin 1 or 2 µM, 15 min, Sigma) were labeled (30 min, in 0.3 ml of methionine–cysteine-free Dulbecco's modified Eagle's medium (DMEM), supplemented with 2 mM l-glutamine, 10% dialyzed fetal calf serum (Sigma), 20 μCi/ml of l-[35S] methionine and l-[35S] cysteine). Cells were harvested following addition of 0.5 ml of cold PBS containing 100 μg/ml CHX and washed twice with cold PBS. The global protein synthesis rate was determined as described (14). Virally infected or uninfected control cells were labeled with (15 μCi/ml of l-[35S] methionine–cysteine, 10 min). For total protein synthesis rates, 5 µg of total proteins were separated by 12% SDS–PAGE transferred to nitrocellulose membrane and [35S]-Met/Cys incorporation was determined by Phosphor-Imager analysis. The same membrane was also washed, blocked and incubated with anti-NS2 antibody, which was visualized using ECL (Amersham).
Electron microscopy
Cells were fixed in 2.5% glutaraldehyde in PBS overnight at 4°C. After several washings in PBS the cells were post-fixed in 1% OsO4 in PBS for 2 h at 4°C. Dehydration was carried out in graded ethanol followed by embedding in glycid ether. Thin sections were mounted on Formvar/Carbon coated grids. Sections were stained with uranyl acetate and lead citrate and analyzed using Jeol 1200EX transmission electron microscope (Jeol, Japan).
RESULTS
Fluorescent labeled tRNA is functional
In FtTM, we transfect cells with uncharged, fluorescent labeled tRNA (fl-tRNA) and analyze the transfected cells by quantitative fluorescence microscopy. We used bulk uncharged yeast tRNA, labeled with either Cy3 hydrazide (8) or Rhodamine 110 (Rho110) (12) in the D-loop (fl-tRNAs). Initially, we tested the biological activity of either unlabeled or fl-tRNA in wheat germ (unlabeled) and rabbit reticulocyte (labeled and unlabeled) cell-free translation systems (Supplementary Figure S1). In both systems, and similarly to the unlabeled tRNA control, addition of 0.15 μM fl-tRNA led to a significant increase in luciferase synthesis (Supplementary Figure S1). Of note, this increase was saturable, as higher concentrations did not add to the observed effect. We concluded that yeast uncharged fl-tRNA is functional, and can mediate protein synthesis in higher eukaryotic translation machineries. Next, we studied conditions for fl-tRNA transfection into live CHO cells. An array of commercial reagents (Supplementary Table S1) was screened and classified according to: (i) degree of delivery (percentage of cells presenting a specific intracellular fluorescence signal); (ii) specificity of intracellular distribution (presence/absence of extra-cellularly localized aggregates, diffuse versus reticulate staining); and (iii) toxicity. Optimal conditions were specified as delivery into ~100% of the cells, of which over 90% show reticulate pattern of biologically relevant (see below), quasi-uniform cytoplasmic fluorescence distributions. Transfection of CHO cells with fl-tRNA consistently and significantly enhanced 35[S]-Met/Cys incorporation (by ~8%, P = 0.003, Supplementary Figure S2), consistent with the effect we observed in reticulocyte lysates in vitro (Supplementary Figure S1). As expected, the translational inhibitors puromycin (1 mM, 30 min) and cycloheximide (0.1 mg/ml, 10 min) reduced 35[S]-Met/Cys incorporation by more than 90%. Interestingly, whereas the degree of reduction by cycloheximide was insensitive to fl-tRNA transfection, the residual protein synthesis in puromycin treated cells was enhanced by ~20% by fl-tRNA transfection (Supplementary Figure S2), in accord with the structural similarity of puromycin and the 3′-terminus of aminoacyl-tRNA. This observation suggests that puromycin and fl-tRNA compete for binding to the ribosome A-site.
Transfected fl-tRNA co-localizes with translation machinery components
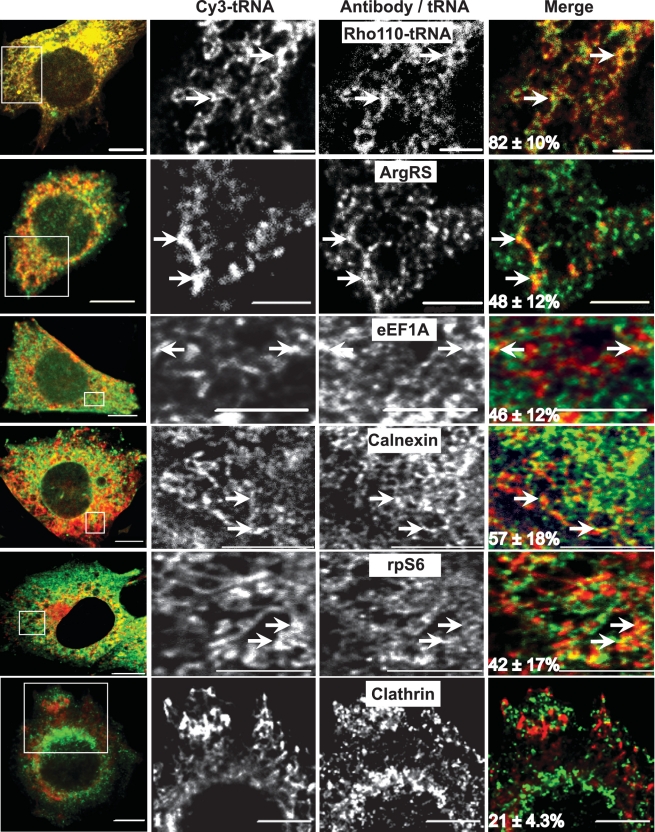
For quantitative analysis of the spatial distribution of the transfected fl-tRNA, the entire cell volume was imaged with a spinning disk confocal microscope. For optimal axial resolution, images were deconvolved with the Constrained Iterative deconvolution algorithm (Slidebook™) which employs an experimentally measured point spread function. The fluorescence signal of transfected fl-tRNAs was unevenly distributed throughout the cell interior, supporting the notion that the transfected fl-tRNA associates with definite subcellular components (Figure 1). Importantly, transfection of Cy3-tRNA into cells expressing green fluorescent protein (GFP) yielded reticulate-like structures in the cytoplasm that often did not co-localize with GFP, which characteristically occupies extensive portions of the cytoplasm and also localizes to the nucleus (Supplementary Figure S4A). Here, as well as in the image analyses described below, two parameters were measured: (i) co-localization (CL) and (ii) Pearson's correlation coefficient [CCF, calculated with the JaCop plugin of the imageJ software (13)]. Transfected Cy3-tRNA yielded intermediate CL and low CCF with GFP (59 ± 10% and 0.095 ± 0.06, respectively; Supplementary Figure S4A). In contrast, the signals produced by co-transfection of Cy3-tRNA and Rho110-tRNA presented both high CL and CCF (82 ± 10% and 0.82 ± 0.02%, respectively; Figure 1, top panel and Supplementary Figure S3), demonstrating that fluorophore identity is not a determinant of fl-tRNA intracellular localization of transfected fl-tRNA. To examine whether transfected fl-tRNA gains access to the translation machinery, we measured by confocal microscopy the degree of overlap and correlation of its fluorescence with the fluorescence signal obtained through the immunostaining of cellular components related to the protein synthesis apparatus. Figure 1 shows representative images (and the average CL) of transfected Cy3-tRNA and endogenous immunostained arginyl-tRNA synthetase (ArgRS), translation elongation factor 1A (eEF1A), ribosomal protein S6 (rpS6), calnexin [a marker of the endoplasmic reticulum (ER), a main site of protein synthesis (15)] and clathrin (a non-ER marker of endomembranes). The positive, albeit moderate levels of CL (Figure 1) and CCF (Supplementary Figure S3), are indicative of a partial co-localization with the different cellular factors and in accord with the various functional interactions of tRNA. Next, we calculated the van-Steensel's cross-correlation of the Pearson's coefficient with distance (CCF, dx; Supplementary Figure S3A and F) and the degree of ‘enrichment’ of co-localized pixels (relative to random co-localization stemming from signal abundance, Supplementary Figure S3G). The observed enrichment and the observance of the maximal CCF at dx = 0 pixels support the notion of the specificity of the co-localization, in spite of its partiality. In summary, the near-complete colocalization of Cy3-tRNA and Rho110-tRNA, and the partial and specific co-localization of Cy3-tRNA with different components of the translation machinery (ribosomes, tRNA synthetases, ER membrane), reflect the targeting of fl-tRNA to the multiple intracellular locations in which charging and/or translation occurs. To further establish a correlation between the intracellular distribution pattern of transfected fl-tRNA and its proposed function in protein synthesis, we followed by live cell microscopy the alteration in this pattern upon inhibition of protein synthesis with puromycin (1 mM, 90 min; Figure 2 and Supplementary Movie S1). In a time-dependent manner, puromycin abrogated the reticulate ER-like fluorescence pattern of Cy3-tRNA, while concomitantly inducing a concentration of this signal to the cell nucleus (Figure 2A and B). Quantification of this phenomenon further established that puromycin (1 mM, 30 min) led to a ~2-fold increase in co-localization of 4′-6-Diamidino-2-phenylindole (DAPI) and fl-tRNA (Figure 2B and C) while abrogating the co-localization of fl-tRNA and calnexin (Figure 2B). Taken together, these results establish the accessibility of transfected fl-tRNA to sites associated with protein synthesis, and support the concept of their functional participation in mRNA translation.
Figure 1.
Transfected Cy3-labeled yeast tRNA shows partial co-localization with cellular factors involved in protein synthesis. CHO cells were transfected with bulk Rho110-labeled and/or Cy3-labeled yeast tRNA, fixed 7 h post transfection and immunostained for ArgRS, eEF1A, calnexin, ribosomal S6 (rpS6) or clathrin as indicated. Panels depict a single middle plane of representative cells imaged with a spinning disc confocal microscope (prior to or following deconvolution employing the constrained iterative algorithm of Slidebook™). Pictures of randomly selected cells (n = 30 for each condition) were employed for the calculation of the co-localization and Pearson's correlation coefficient of the fluorescent signals obtained with the different wavelengths (CL and CCF). The percentage of Cy3-fl-tRNA co-localizing with the different cellular components (or Rho110-tRNA) appears at the lower left hand corner of the merged images. CCF values are detailed in Supplementary Figure S3. Bars are 5 μm.
Figure 2.
Puromycin alters the intracellular distribution of transfected fl-tRNA. (A) Live cell imaging of puromycin treatment. CHO cells, transfected with Cy3-labeled yeast tRNA and treated with puromycin (1 mM, 7 h post transfection, up to 90 min treatment) were imaged by fluorescence time-lapse microscopy (90 min, 30 s between frames). Time-lapse sequence (shown in its entirety in Supplementary Movie S1) was deconvolved with the No Neighbors algorithm and submitted to a Gaussian filter with Slidebook™. Upper panels depict the first and last frames of the sequence. Lower panels are the ‘close-up’ of the indicated inset. Bars are 5 μm. (B and C) Puromycin increases the nucleus-to-cytoplasmic ratio of fl-tRNA fluorescence. CHO cells, transfected as in Figure 1, were treated with puromycin (1 mM, 30 min) prior to fixation, permeabilization and staining against DNA and calnexin. Cells were imaged by confocal microscopy (n = 50) and the percentage of Cy3-tRNA co-localizing with the DAPI signal was calculated by intensity-based segmentation with Slidebook™. Graph depicts the average ± SD of the percentage of Cy3-tRNA showing nuclear localization. *P < 10−27.
FRET-based measurements detect high local levels of protein synthesis
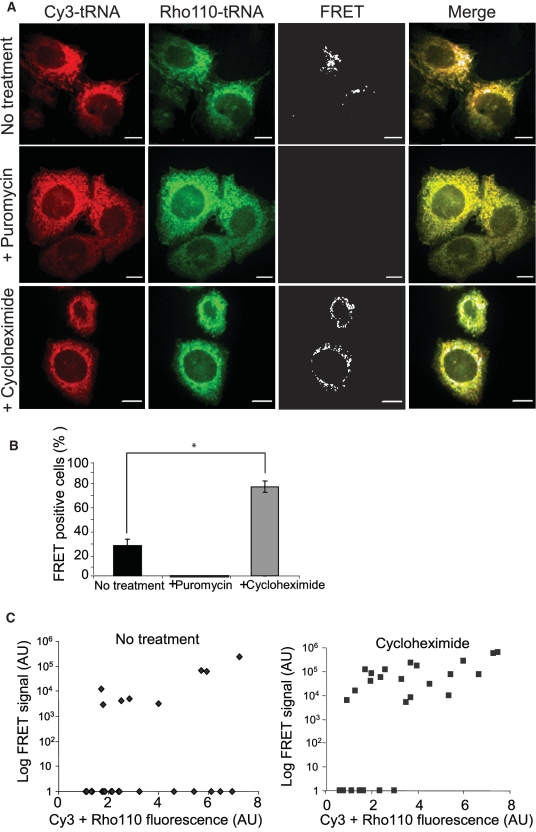
To ascertain the functionality of fl-tRNA in protein synthesis, we looked for the FRET signal expected to be generated by Rho110-tRNA and Cy3-tRNA when occupying adjacent sites on the ribosome (8,16). We imaged CHO cells co-transfected with Rho-110-tRNA and Cy3-tRNA under three different excitation/emission conditions, aimed at detecting signals of Rho110-tRNA and Cy3-tRNA through their direct excitation, as well as the sensitized emission, or FRET signals arising from their juxtaposition (see ‘Materials and Methods’). To ensure specificity, we performed a pixel-by-pixel signal correction by experimentally measuring and subtracting the contributions of the direct excitation of the FRET-acceptor and the bleed-through of the FRET-donor to the intensity of the obtained FRET signal. The corrected FRET signal (FRETc), calculated with Slidebook™ software and employed in all further measurements and calculations, is referred to as FRET throughout the text for the sake of simplicity. Imaging of a single confocal midplane yielded a detectable FRET signal in 25 ± 5% of the transfected cells (representative pictures in Figure 3A, top panel). In the majority of cases, the FRET signal exhibited peri-nuclear localization and a reticulate pattern, consistent with protein synthesis on the ER membrane. In contrast, Cy3-tRNA did not yield any detectable FRET signal in cells highly expressing evenly distributed GFP (Supplementary Figure S4B). To expand on the above-presented results and verify that tRNA originating from higher eukaryotes can also be employed in a similar experimental setup, we performed analogous FRET measurements (in fixated transfected CHO cells, at 6 h post transfection) with labeled bovine bulk tRNA (Supplementary Figure S5). Importantly, in this setting, we confirmed the ‘excitation’ FRET through measurements of the increase in Rho-110-tRNA fluorescence upon the bleaching of Cy3-tRNA (‘acceptor-bleaching’). To verify that the FRET signal originated from the proximity of the tRNA FRET pairs on translating ribosomes, we measured FRET under conditions of protein synthesis arrest, achieved by either puromycin or cycloheximide treatments. Puromycin (1 mM for 10 min, indicative of the initial stages of the inhibition process) completely abrogated the FRET signal (representative pictures in Figure 3A, middle panel), in accord with its binding to the ribosomal A-site, and with abortion of protein synthesis and polysome breakdown which it induces (17,18). In contrast, cycloheximide (0.1 mM, 10 min) enhanced both the number of FRET positive cells as well as the specific FRET intensity per cell (total FRET signal divided by the fluorescent signal obtained through the direct excitation of the fluorophores) (representative pictures in Figure 3A, bottom panel, quantifications in Figure 3B and C), in accord with its inhibitory effect on ribosomal translocation and translational arrest at the elongation level (19), resulting in the freezing of bound tRNAs on the ribosome. Importantly, the FRET signals obtained under cycloheximide treatment were also peri-nuclear and reticulate, in accord with localization to the ER membrane.
Figure 3.
FRET assay involving Cy3- and Rho110-labeled tRNAs identifies protein synthesis sites. (A) FRET signals are sensitive to protein synthesis inhibitors. Cells were co-transfected with bulk Cy3- and Rho110-labeled yeast tRNAs, at 7 h post transfection, were treated or not with puromycin or cycloheximide prior to fixation and imaging. Panels show representative cells. Bars are 5 μm. (B) Percentage of cells showing a specific FRET signal. Graph depicts the average ±SD of the percentage of FRET positive cells in 15 fields per condition, from two independent experiments. *P < 0.05. (C) Cycloheximide enhances FRET signal intensity. Graphs depict the correlation of the FRET and total fluorescence signal of single cells in untreated and cycloheximide-treated conditions (n = 30). Note: the arbitrary units (AU) are not identical between panels.
Taken together, these results show that FtTM provides two modes of analyses: (i) A highly sensitive co-localization detection method to assess alterations to the topography of protein synthesis involving tRNA mislocalization; and (ii) FRET-based measurements for the detection of high local levels of protein synthesis. The FRET-mode of FtTM also reports the action of chemicals, which block protein synthesis while leaving the tRNA associated with ribosomes.
FtTM provides quantitative assessment of protein synthesis in single mammalian cells at submicron resolution
Next, we wished to test if our newly developed methods can faithfully reflect reduction in protein synthesis in response to agents that induce cellular stress. To this end, we treated cells with thapsigargin, an inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase, in order to induce ER-stress-mediated translation inhibition (20). Indeed, thapsigargin reduced 35[S]-Met/Cys incorporation in fl-tRNA-transfected CHO cells in a concentration dependent manner (Figure 4A). Specifically, 2 μM thapsigargin significantly reduced both the percentage of FRET positive cells (by 45 ± 6%, Figure 4B) and the specific FRET intensity of single FRET positive cells (Figure 4C). These results encouraged us to test the use of FtTM in comparing protein synthesis levels between cells in different physiological states. To this end, we compared FRET signals of primary brain astrocytes prior to, and following, activation with bacterial lipopolysaccharides (LPS) and interferon-γ. In resting primary brain astrocytes transfected with fl-tRNA, the FRET signal was below detection level (Figure 5A), whereas upon their activation both the percentage of FRET-positive astrocytes and the specific FRET intensity per cell were enhanced (Figure 5A and B). Importantly, increases in both FRET parameters upon astrocytic activation correlated qualitatively with similar increases in 35[S]-Met/Cys incorporation (Figure 5C) (albeit quantitatively distinct due to the different sensitivities of the FtTM and metabolic labeling assays), reinforcing the ability of FtTM, in its current form, to provide at least semi-quantitative assessment of protein synthesis in single mammalian cells at submicron resolution. Interestingly, an additional correlation could be established between the degree of FRET positive cells and the degree of 35[S]-Met/Cys incorporation (astrocytes versus CHO cells, Figure 5C).
Figure 4.
Thapsigargin (Tg) reduces protein synthesis and tRNA-mediated FRET. (A) CHO cells at 7 h post transfection with Cy3-labeled tRNA were treated with Tg (0, 1 or 2 μM, 15 min), prior to incubation with 35[S]-Met/Cys (30 min, 20 µCi/ml). Cells were subsequently lysed and 35[S]-Met/Cys incorporation was measured as previously described (14). The graph depicts average ±SD of three independent experiments done in triplicates. *P < 0.0002. (B) Cells transfected with both Cy3- and Rho110-labeled yeast tRNAs, were treated or not with 2 μM Tg for 15 min and subsequently fixed and imaged. The specific FRET signal (FRETc) was calculated as described in ‘Materials and Methods’. Graph depicts the percentage of FRET-positive cells in 15 randomly selected fields per condition from three independent experiments. *P < 0.03. (C) Tg reduces FRET signal intensity. Graphs show correlation of FRET and of total fluorescence signals (direct measurements of Cy3 and Rho110) of single cells (n = 50) in untreated and Tg treated condition. Note: the arbitrary units (AU) are not identical between panels.
Figure 5.
FtTM measures differences in protein synthesis between resting and activated astrocytes. (A) Mouse primary astrocytes prior to or following activation with 2 µg/ml LPS and 3 ng/ml INFγ for 24 h, were assayed by FtTM. Panels show representative cells. Normalization of the fluorescence signal was identical to allow visual comparison. Bars are 10 μm. (B) Quantification of the percentage of cells which show specific FRET signal. Graph depicts the percentage of FRET-positive cells in randomly selected fields. n = 30 cells/experiment/condition), two independent experiments. *P < 0.03. (C) Levels of 35[S]-Met/Cys incorporation directly correlate with FtTM measurements. Transfected astrocytes (resting or activated) and CHO cells were labeled with 35[S]-Met/Cys (30 min, 20 µCi/ml). Cells were subsequently lysed and 35[S]-Met/Cys incorporation was measured as previously described (14). Graph depicts the average ±SD of three independent experiments done in duplicates.*P < 5E-7; **P < 1E-12.
FtTM characterizes changes in spatial compartmentalization of protein synthesis upon viral infection
Next, we set to measure levels of viral protein synthesis and visualize its subcellular localization patterns in a population of infected host cells. Genome replication and assembly of viruses often take place in specific intracellular compartments where viral components concentrate. For a number of viruses, the formation of ‘viral factories’ or ‘viroplasm’ has been described, which consist of peri-nuclear or cytoplasmic foci that may exclude host proteins to a significant extent (5,6,21). We focused on the EHDV (serotype 2, strain Ibaraki) (10,11), an orbivirus (arbovirus of the Reoviridae family), which is closely related to the Bluetongue Virus (BTV) (22,23). The intracellular localization of active protein synthesis complexes in EHDV2-IBAV-infected cells as well as the correlation of this localization with viral factories has not been established yet. Infection of CHO cells (30 h, at multiplicity of infection of one plaque forming unit per cell) induced a reduction of 30% in total protein synthesis (measured through 35[S]-Met/Cys incorporation and quantification of the intensity of the signal of the entire SDS–PAGE lane of labeled whole cell lysates; Figure 6A). The partial shut-off of host protein synthesis was accompanied by the appearance of a number of specific bands exclusively in the infected cells, most likely representing viral proteins (Figure 6A, arrows). To identify and visualize the main protein component of viroplasms induced by orbiviruses, we raised peptide-specific monoclonal antibodies against the virally encoded non-structural protein 2 (NS2). Indeed, staining the radioactive SDS–PAGE blot presented in Figure 6A with anti-NS2 antibodies resulted in clear and specific bands. The main band had the expected molecular weight of 41 KDa, and thus corresponds to EHDV2-IBAV-NS2 protein, while the weaker and lower weight bands may reflect proteolytic fragments of NS2, and will be the object of future study. Immunofluorescence staining of EHDV2-IBAV-infected CHO cells with anti-NS2 antibodies showed a characteristic distribution of NS2 within discrete cytosolically localized puncta of varying size, in addition to a more diffuse staining observed in the cytoplasm and cell nucleus. Importantly, this same distribution was observed in cells infected with EHDV2-IBAV and transfected with fl-tRNA (30 h infection followed by 7 h transfection; Figure 6B and C, arrows), in accord with its localization to viral factories. Strikingly, fl-tRNA exhibited high and significant CL as well as CCF, in both the discrete cytoplasmic punctae and the cell nucleus (Figure 6B). Moreover, in EHDV2-IBAV-infected cells co-transfected with Rho110-tRNA and Cy3-tRNA and labeled for NS2, FRET signals could also be observed in the proximity/overlap of viral factories (Figure 6C), indicating the virally induced compartmentalization of active protein synthesis. Electron microscopy of infected CHO cells at 36 h post infection revealed unique structures in different localizations in the cytosol, consisting of an electron-dense matrix interspersed with icosahedral structured virions and interpreted by us as viral factories (Figure 6D, arrow). Typically, these viral factories were surrounded with multiple layers of electron-dense spots, considerably smaller than the virions. To directly probe for the presence of ribosomes in the vicinities of factories, infected CHO cells were processed for immunofluorescence and concomitantly stained for NS2 and for rpS6. A considerable amount of NS2 and rpS6 co-localized (70% and 28% of NS2 and rpS6, respectively, Figure 6E) yielding a CCF of 0.416 ± 0.05, in accord with the FRET measurements and the EM analysis.
Figure 6.
Co-localization of EHDV2-IBAV NS2 protein and fl-tRNA and rpS6 reveals infection-dependent compartmentalization of protein synthesis. (A) CHO cells, infected with EHDV2-IBAV (MOI = 1; 30 h), labeled with 35[S]-Met/Cys (15 μCi/ml, 10 min, 37°C), were lysed and processed by SDS–PAGE. Nitrocellulose blots were either visualized with a phosphor-imager (left panel) or immunoblotted with monoclonal anti-NS2 antibodies (right panel). Arrows indicate putative NS2 bands in the radioactive blot. (B) CHO cells were infected as in (A) and transfected at 30 h post infection with Cy3 fl-tRNA. At 7 h post transfection, cells were fixed, immunostained (anti-NS2/Alexa-488 goat-anti-mouse) and imaged (CCF, lower left hand corner of merge image; arrows point to co-localizations). (C) CHO cells, infected as in (A) and co-transfected with Cy3 and Rho110 fl-tRNA were immunostained (anti-NS2/Alexa 648 goat-anti-mouse). Arrows point to typical triple co-localizations. Note: due to imaging constraints the NS2 image of the same field was acquired through a different microscope port. (D) CHO cells, infected as above, were imaged by electron microscopy. Micrographs show: entire cell (left, arrows indicate factories), a close-up of the viral factory with interspersed virions (middle), accumulation of electron dense spots in the factory vicinity (elipse points to typical accumulations, interpreted as ribosomes). (E) CHO cells, infected as above, were stained with anti-NS2 and anti-rpS6 antibodies and imaged by confocal microscopy (arrows point to typical instances of co-localization, CCF at the lower left hand corner of merged picture). Bars in B, C and E are 10 μm and in D are 2 µm, 1 µm and 500 nm (left to right, respectively).
DISCUSSION
FtTM represents a novel addition to the existing roster of methods for monitoring and localizing protein synthesis within cells (24–27). This currently include the use of fluorescent derivatives of cycloheximide to identify a specific site of protein synthesis (25) transfection of genetically engineered vectors for the expression of tagged mRNAs to visualize growing nascent peptide at sites of specific mRNA localization (26); expression of tagged translation initiation factors that serve as FRET pairs to specifically monitor translation initiation (24); and in situ spectral monitoring using the ratio between the peak areas of RNA and proteins as a measure of mRNA translation (28). The results presented in the current study demonstrate the ability of FtTM to quantitatively measure the process of translation and delineate the intracellular topography of protein synthesis activity under normal physiological conditions, in single cells and at subcellular resolution. FtTM achieves this by following the distribution and dynamics of fl-tRNA and by the direct monitoring of the activity of translating ribosomes. Application of FtTM to monitoring protein synthesis revealed, for the first time, differential translation activity in resting versus activated primary astrocytes and in addition enabled, for the first time, the visualization of actively translating ribosomes in the vicinity of viral factories in EHDV2-IBAV-infected CHO cells. This latter result demonstrates a clear example of protein synthesis compartmentalization linked to the usurpation of the translation machinery during the course of infection. In its current mode, employing bulk fl-tRNAs, FtTM is useful for monitoring overall protein synthesis in many different kinds of fixated and living cells under a variety of growth conditions. The limited percentage of cells showing a detectable FRET signal in the transfected CHO cell population, and the restricted percentage of FRET positive pixels among the ER-distributed fl-tRNA in those cells, are both indicative of the limited sensitivity of the FRET measurements using the current probes. Nevertheless, the quantitative nature of fl-tRNA-FRET, enables the detection of intra-populational variability and quantifies protein synthesis in defined cellular regions. Moreover, the ability to monitor alterations in the intracellular distribution of fl-tRNA in live cells enables the analysis of the kinetics of perturbations to protein synthesis (Figure 2). Specifically, the observed accumulation of fl-tRNA in the nuclei of puromycin-treated cells may reflect the spatial compartmentalization of an important cellular resource under conditions interpreted by the cell as stressful.
Labeling of tRNA in the D-loop region leads to the introduction of a bulky fluorophore which may interfere with aminoacylation of some labeled tRNAs by their cognate aminoacyl-tRNA synthetases, thus preventing their participation in translation. Earlier we showed that charged bacterial tRNAs labeled with Rho110 or Cy3 dyes in their D-loops were functional in in vitro assays of ribosome function (8,12). Here, we show that similarly labeled, but uncharged, yeast tRNAs are functional in protein synthesis when transfected into mammalian cells. The evidence for this functionality is based on four observations: (i) localization of the labeled tRNA with components of the protein synthesis machinery (including Arg-synthetase, Figure 1); (ii) alteration of the localization of tRNA upon inhibition of protein synthesis by puromycin (Figure 2); (iii) similar enhancements of luciferase protein synthesis levels upon addition of bulk-labeled or bulk-unlabeled uncharged tRNA, suggesting that both types of tRNA are charged in the rabbit reticulocyte lysate; and (iv) measured FRET between Rho110- and Cy3-labeled tRNAs, direct evidence for their co-localization to the ribosome, after charging. These results demonstrate that exogenous uncharged tRNA are aminoacylated shortly after their entry into CHO cells, consistent with earlier reports (29) and further demonstrate that following longer incubation time they are channeled to the tRNA cycle and repeatedly delivered to translating ribosomes while being protected from RNase digestion (30).
Future applications, currently under development, include applying FtTM to measure translation dynamics and synthesis patterns in normal and diseased cells under various physiological, pathological and environmental conditions and in response to drugs. In addition, it should prove possible to monitor the synthesis rates of a specific protein using specifically selected fl-tRNA pairs that are used at high frequency during the synthesis of that protein. This capability would permit the visualization and quantification of the expression of specific genes in real time as recently described in vitro for single translating ribosomes and synthetic polypetides (31).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Anima Cell Metrology research grant (to O.E.S. and M.E.), National Institutes of Health (grants No. GM090404-01 and GM-071014 to B.C.), BARD (grant No. 15-4192-09 to M.E.) and the Legacy Heritage Biomedical Science Partnership Program of the Israel Science Foundation (grant No. 1911/08 to O.E.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEGEMENTS
The authors thank Yale Goldman for numerous helpful discussions during the course of this work; Diana Cabral for early preparations of bulk fl-tRNAs; Hagai Yadin and Valizar Bombarov for the original viral isolate; Galit Horn and Naomi Ziv for early work on the anti-NS2 antibody; Ben Shai for virus purification; Yuval Cabili for astrocytes cell isolation and Vered Holdengreber for electron microscopy.
REFERENCES
- 1.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 2.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr. Opin. Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castello A, Quintas A, Sánchez EG, Sabina P, Nogal M, Carrasco L, Revilla Y. Regulation of host translational machinery by African swine fever virus. PLoS Pathog. 2009;5:e1000562. doi: 10.1371/journal.ppat.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan D, Qin H, Cooperman BS. Synthesis and functional activity of tRNAs labeled with fluorescent hydrazides in the D-loop. RNA. 2009;15:346–354. doi: 10.1261/rna.1257509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matumoto M, Inaba Y, Tanaka Y, Morimoto T, Omori T. Ibaraki virus, an agent of epizootic disease of cattle resembling bluetongue. 3. Experimental inoculation of cell cultures, embryonated eggs and laboratory animals. Jpn. J. Microbiol. 1970;14:99–109. doi: 10.1111/j.1348-0421.1970.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 11.Omori T, Inaba Y, Morimoto T, Tanaka Y, Kono M. Ibaraki virus, an agent of epizootic disease of cattle resembling bluetongue. II. Isolation of the virus in bovine cell culture. Jpn. J. Microbiol. 1969;13:159–168. doi: 10.1111/j.1348-0421.1969.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 12.Betteridge T, Liu H, Gamper H, Kirillov S, Cooperman BS, Hou YM. Fluorescent labeling of tRNAs for dynamics experiments. RNA. 2007;13:1594–1601. doi: 10.1261/rna.475407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Steensel B, van Binnendijk EP, Hornsby CD, van der Voort HT, Krozowski ZS, de Kloet ER, van Driel R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 1996;109:787–792. doi: 10.1242/jcs.109.4.787. [DOI] [PubMed] [Google Scholar]
- 14.Gerlitz G, Jagus R, Elroy-Stein O. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 2002;269:2810–2819. doi: 10.1046/j.1432-1033.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- 15.Stephens SB, Nicchitta CV. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol. Biol. Cell. 2008;19:623–632. doi: 10.1091/mbc.E07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl Acad. Sci. USA. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathans D. Puromycin Inhibition of Protein Synthesis: Incorporation of Puromycin into Peptide Chains. Proc. Natl Acad. Sci. USA. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munro HN, Baliga BS, Pronczuk AW. In vitro inhibition of peptide synthesis and GTP hydrolysis by cycloheximide and reversal of inhibition by glutathione. Nature. 1968;219:944–946. doi: 10.1038/219944a0. [DOI] [PubMed] [Google Scholar]
- 20.Wong WL, Brostrom MA, Kuznetsov G, Gmitter-Yellen D, Brostrom CO. Inhibition of protein synthesis and early protein processing by thapsigargin in cultured cells. Biochem. J. 289(Pt 1):71–79. doi: 10.1042/bj2890071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anthony SJ, Maan N, Maan S, Sutton G, Attoui H, Mertens PP. Genetic and phylogenetic analysis of the non-structural proteins NS1, NS2 and NS3 of epizootic haemorrhagic disease virus (EHDV) Virus Res. 2009;145:211–219. doi: 10.1016/j.virusres.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Roy P. Functional mapping of bluetongue virus proteins and their interactions with host proteins during virus replication. Cell Biochem. Biophys. 2008;50:143–157. doi: 10.1007/s12013-008-9009-4. [DOI] [PubMed] [Google Scholar]
- 24.Miyata S, Mori Y, Fujiwara T, Ikenaka K, Matsuzaki S, Oono K, Katayama T, Tohyama M. Local protein synthesis by BDNF is potentiated in hippocampal neurons exposed to ephrins. Brain Res. Mol. Brain Res. 2005;134:333–337. doi: 10.1016/j.molbrainres.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 25.Paoletti F, Ainger K, Donati I, Scardigli R, Vetere A, Cattaneo A, Campa C. Novel fluorescent cycloheximide derivatives for the imaging of protein synthesis. Biochem. Biophys. Res. Commun. 2010;28:258–264. doi: 10.1016/j.bbrc.2010.04.075. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J. Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shav-Tal Y, Singer RH, Darzacq X. Imaging gene expression in single living cells. Nat. Rev. Mol. Cell Biol. 2004;5:855–861. doi: 10.1038/nrm1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notingher I, Bisson I, Bishop AE, Randle WL, Polak JM, Hench LL. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal. Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- 29.Stapulionis R, Deutscher MP. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl Acad. Sci. USA. 1995;92:7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Negrutskii BS, Deutscher MP. A sequestered pool of aminoacyl-tRNA in mammalian cells. Proc. Natl Acad. Sci. USA. 1992;89:3601–3604. doi: 10.1073/pnas.89.8.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.