Abstract
Riboswitches are RNA-based regulatory devices that mediate ligand-dependent control of gene expression. However, there has been limited success in rationally designing riboswitches. Moreover, most previous riboswitches are confined to a particular gene and only perform one-way regulation. Here, we used a library screening strategy for efficient creation of ON and OFF riboswitches of lacI on the chromosome of Escherichia coli. We then engineered a riboswitch-LacP hybrid device to achieve portable gene control in response to theophylline and IPTG. Moreover, this device regulated target expression in a ‘two-way’ manner: the default state of target expression was ON; the expression was switched off by adding theophylline and restored to the ON state by adding IPTG without changing growth medium. We showcased the portability and two-way regulation of this device by applying it to the small RNA CsrB and the RpoS protein. Finally, the use of the hybrid device uncovered an inhibitory role of RpoS in acetate assimilation, a function which is otherwise neglected using conventional genetic approaches. Overall, this work establishes a portable riboswitch-LacP device that achieves sequential OFF-and-ON gene regulation. The two-way control of gene expression has various potential scientific and biotechnological applications and helps reveal novel gene functions.
INTRODUCTION
Inducible gene regulation is required for diverse applications such as gene function investigations, gene therapies and industrial uses. So far, inducible promoters are the major tools for specifically controlling target gene expression. However, there are only limited numbers of inducible promoters which, on some occasions, are insufficient for independent control of multiple genes by the addition of distinct inducers. It is, therefore, highly desirable to develop alternative approaches to enrich the repertoire of inducible systems. A promising candidate is riboswitch that is a RNA-based, ligand-dependent genetic control element located in non-coding regions of messenger RNAs. Riboswitches control biological processes at various regulatory levels. Lysine riboswitches prematurely halt RNA transcription by modulating the formation of a Rho-independent terminator (1–3). Translationally controlling riboswitches in the 5′-untranslated region modulate the ribosome access to the ribosome-binding site (RBS) (4–6). In addition, some riboswitches undergo self-cleavage upon ligand binding, resulting in rapid RNA degradation (7,8). Unlike inducible promoters, these RNA-based control elements are potentially numerous as a result of high-throughput screen for ligand-specific aptamers (the sensor domain of a riboswitch) (9–13).
In recent years, advancements have been made in the ability to convert ligand-specific aptamers to functionally active riboswitches by various random screening approaches (14–16) or rational design (4,5,8). However, library screening is labor intensive and time consuming, and successful rational design is not guaranteed even though a long list of design principles are followed, due to incomplete understanding of RNA-based gene regulation mechanisms. In addition, a common disadvantage of riboswitches as well as inducible promoters is that they only allow for one-way gene regulation. Once activated, a target gene cannot be turned off unless promoter inducers or riboswitch ligands are removed. The one-way regulation mode makes the existing regulation systems unsuitable for quick gene control in live animals or in vitro studies where intact cell cultures are required. Thus, there is a great need for a two-way regulation system, in which target gene can be switched off and on in response to distinct effector molecules.
Here, we set out to develop a portable and two-way regulatory device by integrating an ON theophylline-responsive riboswitch with the lac-inducible promoter system (abbreviated as LacP), in which the LacI repressor binds as a homotetramer to two LacI-binding sites positioned immediately downstream of the lac promoter. First, we used a library screening strategy to create ON and OFF theophylline-responsive riboswitches of the lacI gene that encodes the LacI transcriptional regulator. Then, combining an ON-riboswitch of LacI synthesized in the first part of this study with two LacI-binding sites (LacIbs), we established a portable hybrid device that acted as a versatile key for controlling gene expression without the need for further rational design or library screening. We demonstrated the portability of this device by applying it to rpoS (encoding RpoS, a master regulator of acid resistance) and csrB (encoding a small non-coding RNA CsrB). We showed that this portable device regulates target genes in a two-way manner, switching off the targets in response to theophylline and restoring the target expression in response to isopropyl β-d-1-thiogalactopyranoside (IPTG). The sequential two-way control of RpoS and CsrB by the riboswitch-LacP hybrid device reversibly fine-tuned virulence-associated cellular behaviors including acid resistance, intercellular autoaggregation and biofilm formation. Finally, we used this device to explore unidentified functions of RpoS and revealed for the first time that removing RpoS promotes acetate assimilation after the acetate switch is flipped. This finding is opposite to the previous ones made by using conventional rpoS mutants, which might have accumulated secondary mutations. Since the portable two-way device does not silence target genes until theophylline is added, its host cells are less likely to generate secondary mutations and therefore able to provide more reliable information.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
Escherichia coli strain MG1655 was used in this study. All bacterial mutants were grown at 37°C, with shaking at 220 rpm, in Luria–Bertani (LB) medium supplemented with 2 mM magnesium sulfate. The antibiotics ampicillin (50 µg/ml), kanamycin (50 µg/ml) and chloramphenicol (12.5 µg/ml) were used for selection when appropriate.
Mutagenesis
Gene deletion was performed using the recombineering system (17). Escherichia coli-K12 MG1655 was transformed with plasmid pSim6 (a gift from Dr Donald Court) from which the expression of the λ recombination proteins is induced at 42°C. PCR fragments encompassing a loxP-cat-loxP with homology (45 nt) to the regions immediately flanking each deletion locus were transformed via electroporation into MG1655 cells harboring pSim6. After induction of λred functions, recombinants were selected for chloramphenicol resistance (encoded by the cat gene) and were further verified by colony PCR.
Construction of randomized riboswitch libraries
For construction of theophylline-responsive riboswitch libraries on the chromosome, the loxP-cat-loxP fragment was linked to a theophylline-responsive aptamer followed by a 5-nt-random sequence by ligation PCR. The resulting PCR products with homology (45 nt) to the regions surrounding the ribosome-binding site of each target gene were then integrated into the chromosome using the above-described recombineering technique. In theory, each thus established library contains 45 riboswitch candidates.
Construction of chromosomal lacZ translational fusions and β-galactosidase assays
The loxP-cm-loxP selectable cassette was inserted immediately after the stop codon of the lacZ gene on the MG1655 chromosome using recombineering (as described above). Next, the lacZ-loxP-cm-loxP cassette was PCR amplified and inserted (in frame) immediately prior to the stop codon of the target gene on the chromosome. The inserted lacZ fragment started from the eighth codon of lacZ gene and was co-transcribed and translated with the fused genes. The expression of gene-lacZ fusions was quantified using a β-galactosidase assay as described previously (4,18). Levels of β-galactosidase were calculated using the following formula:
Acid resistance assay
Overnight cultures were treated with acid (pH 2) for 2 h and then serially diluted in neutral medium. After overnight culture, colony forming units (CFU) of acid-treated and untreated cells were determined. Acid survival (%) was calculated with the following formula:
Autoaggregation
Overnight cultures were diluted 1:500 and incubated at 37°C, with shaking at 220 rpm, in liquid LB medium. When cells aggregated (visualized macroscopically by the clumping or ‘fluffing’ of cells in liquid cultures), 100 µl of each cell suspension was transferred to flat-bottom 96-well plates (Iwaki, Tokyo, Japan) and the images of the cell aggregates were captured by scanning. To better visualize the cell aggregates, 1 µl of crystal violet (0.1%) was added to each cell suspension immediately prior to image capture. Cellular autoaggregation was also examined microscopically. Cell suspensions were spread on a microscope slide, heat-fixed, stained with DAPI (4′6-diamidino-2-phenylindol) and imaged using a fluorescence stereomicroscope SZX12 (Olympus, Tokyo, Japan) with DAPI filter sets.
Biofilm formation
Biofilms were formed on polystyrene, flat-bottom 96-well microtiter plates (Iwaki, Tokyo, Japan). Two hundred microliters of each cell suspension (105 cells/ml) was transferred into each well of a microtiter plate and incubated for 24 h at 37°C in a shaker at 75 rpm. Resulting biofilms were washed thrice with PBS and air dried. Then, biofilms were stained with 100 µl of 0.4% aqueous crystal violet solution for 15 min. Afterward, biofilms were washed thrice with sterile distilled water and immediately destained with 200 µl of 95% ethanol. After 30 min of destaining, 100 µl of destaining solution was transferred to a new well and measured with a microtiter plate reader (SpectraMAX 340 Tunable Microplate Reader; Molecular Devices Ltd) at 595 nm.
Semi-quantitative RT–PCR
Total RNA was isolated from overnight cultures in LB medium. Subsequently, 2 µg of RNA was reverse transcribed in a total reaction volume of 20 µl using the ThermoScript RT-PCR system (Invitrogen). Each reaction was incubated at 55°C for 50 min followed by 15 min at 70°C. Two microliters of the resulting reverse transcript products (cDNA) were then used for 18, 20, 22 and 24 rounds of PCR (30 s each at 94°C, 55°C, and 72°C) with Ex Taq DNA polymerase (Takara Bio, Inc.) and primers complementary to csrB (RT-CsrB-F: 5′-GTCAGACAACGAAGTGAACATCAGG-3′ and RT-CsrB-R: 5′-GGAGCACTGTATTCACAGCGCT-3′) and the 16S rRNA gene (RT-16S-F: 5′- CTCCTACGGGAGGCAGCAG-3′ and RT-16S-R: 5′-CTCCGTATTACCGCGGCTG-3′) to co-amplify the gene of interest and the internal control. PCR products were separated in 1.5% agarose gel.
Acetate determination
To quantify extracellular acetate, cell suspensions were centrifuged at 10 000 rpm for 5 min at 4°C. The resulting supernatants were then subjected to acetate determination using an acetate detection kit (Megazme, Bray, Ireland) according to the manufacturer's instructions.
Statistical analysis
Paired t-tests were used to compare the two means obtained from β-galactosidase assays; one-way ANOVA was used for the comparison of multiple means from the biofilm formation assay. P < 0.05 was considered statistically significant.
RESULTS
Establishment of a random library for engineering functionally active riboswitches on the chromosome
To construct a riboswitch-LacP hybrid device, our first task was engineering ligand-responsive riboswitches for the lacI gene, which encodes the LacI transcriptional regulator in the Lac system. Previously, we have successfully engineered a theophylline-responsive ON riboswitch residing upstream of the csrA gene in E. coli and managed to modulate the accessibility to ribosome binding and to flexibly control the csrA expression (4). However, the riboswitch did not work when grafted to lacI (data not shown), indicating that riboswitches besides the RBS are sensitive to changes in flanking regions and have to be specifically designed and constructed for each target gene. However, rational design is laborious and does not guarantee successful engineering of functionally active riboswitches. We, therefore, sought out to explore the possibility of establishing a chromosomal riboswitch library, with the aid of the recombineering technique, to screen for active riboswitches.
We used the theophylline-responsive aptamer as the sensor domain of our riboswitches. The riboswitches were proposed to adopt multiple conformations at equilibrium, some of which permitted downstream target expression whereas some of which did not. If theophylline binding to the aptamer favored the formation of conformations that permitted target expression, then the riboswitch was an ON switch. Conversely, if theophylline binding shifted the equilibrium distribution to conformations that repressed the target expression, the riboswitch was an OFF switch.
To engineer a theophylline-responsive riboswitch for lacI, we first constructed a riboswitch library and then screened for clones that could effectively control the target expression in response to theophylline (Figure 1A and B). We used the genomic DNA isolated from the previously reported switch-csrA strain (4) to PCR amplify the theophylline-specific aptamer fragment. Then, the aptamer was linked to a chloromphenical resistance gene (cat) by ligation PCR, generating a riboswitch cassette library containing a cat-aptamer-linker-random fragment on the chromosome. The cat gene facilitates recombineering of the cassette into the genome, and the 5-nt-random sequence allows construction of a riboswitch library. The cassette was integrated, using the recombineering technique, into the genome of the E. coli MG1655 strain right upstream of the RBS of target genes, resulting in numerous mutant strains each of which carried a riboswitch candidate (functional or nonfunctional) on the chromosome. In theory, the mutant library contained 45 riboswitch candidates.
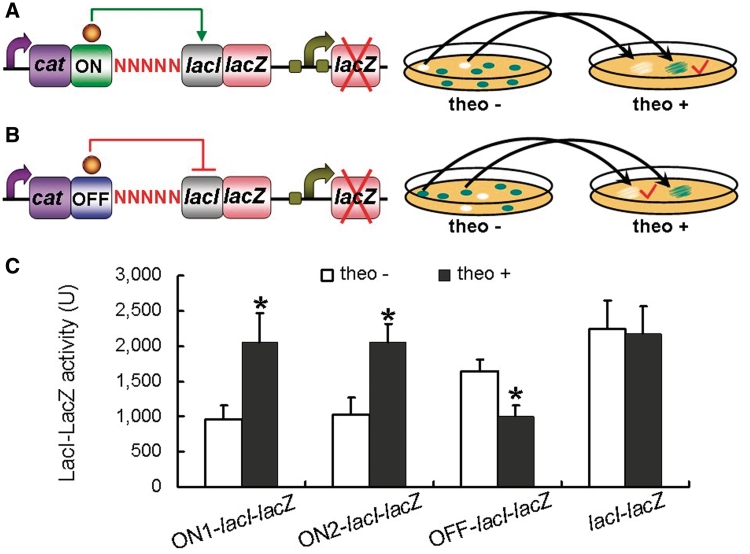
Figure 1.
Creation of ON and OFF riboswitches for the lacI gene using a library screening-based method. (A) A random cassette mixture containing a theophylline-responsive aptamer and five random bases was ‘recombineered’ into the chromosome of a MG1655 mutant that was defective in native lacZ but carried the lacI–lacZ translational fusion, generating a riboswitch library for lacI. Cells were placed on X-gal plates without theophylline but with 2 mM caffeine. Relatively whiter colonies were searched for and transferred to X-gal plates with 2 mM theophylline. Those that turned into blue were considered to carry ON riboswitches of lacI. (B) Cells were placed on X-gal plates without theophylline but with 2 mM caffeine. Blue colonies were searched for and transferred to X-gal plates with 2 mM theophylline. Those that turned into relatively white were considered to carry OFF riboswitches of lacI. (C) Evaluation of the ON and OFF riboswitches of lacI using β-galactosidase assay. The mutant strain (named lacI–lacZ) that carries the lacI–lacZ translational fusion without the ON riboswitch was used as a control.
Effective ON and OFF riboswitches of the lacI gene are constructed using the efficient library screening approach
LacI is a well-known repressor of lacZ and therefore the expression of lacI can be detected on X-gal plates. However, we did not use this strategy but instead constructed a lacI–lacZ translational fusion on the chromosome of a MG1655 lacZ null mutant, to illustrate that the library screening on X-gal plates allows for construction of riboswitches for any target genes fused with lacZ. To engineer an ON riboswitch of lacI, the riboswitch cassette cat-aptamer-linker-random sequence was integrated upstream of the RBS of the lacI–lacZ fusion on the chromosome by recombineering (Figure 1A). Then, the resulting mutant collection was placed on agar with 0.06 mg/ml X-gal and 2 mM caffeine (used as a negative control whose chemical structure differs from theophylline only by the additional presence of one methyl group) but without theophylline. Approximately 2000 colonies were checked for color, and two clones were whiter than others. Both clones turned into blue when re-streak on X-gal plates plus 2 mM theophylline. The riboswitches in the two clones were sequenced and the random sequence was TGTAT and CGTAT, respectively. The riboswitch carrying TGTAT was named ON1-lacI, and the one with CGTAT was named ON2-lacI. The effectiveness of the two ON riboswitches of lacI was confirmed by β-galactosidase assay comparing lacI–lacZ fusion levels in the presence of either caffeine or theophylline (Figure 1C).
To screen for effective OFF riboswitches of lacI, approximately 3000 blue colonies on the caffeine-supplemented X-gal plates were re-streak on plates plus 2 mM theophylline (Figure 1B). After 6 h culture, one clone gave whiter color relative to others. Its responsiveness to theophylline was verified by β-galactosidase assay (Figure 1C), indicating that the clone carries an OFF riboswitch of lacI. We named it OFF-lacI. Sequencing analysis revealed that the 5-nt random sequence in OFF-lacI is CTGGT.
The artificial riboswitches control the lacI expression via a long stem structure adjacent to the ribosome-binding site
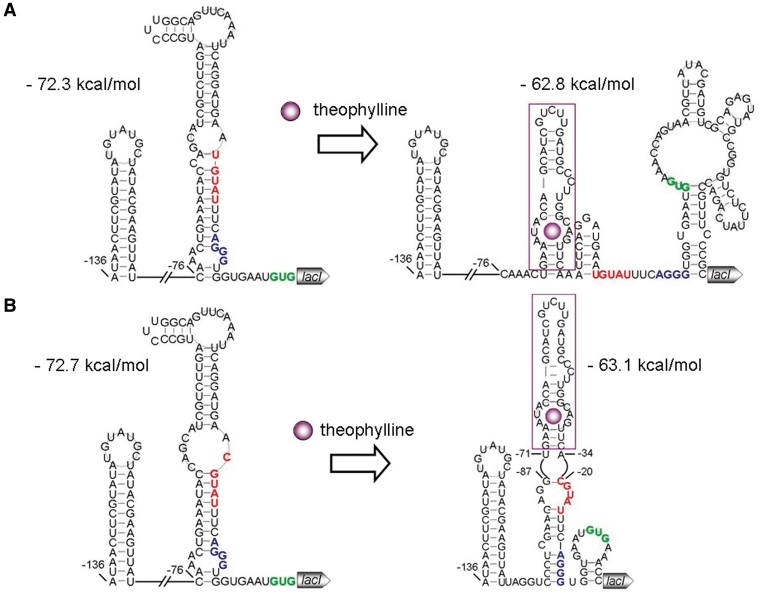
Secondary structures of the ON and OFF riboswitches of lacI were then predicted using RNAstructure 5.1 (19,20). The secondary structure prediction of all the riboswitches was performed for a 237-nt long fragment starting with a 33-nt stem–loop structure (−136 nt relative to the translational start site) and ending at the 100th nucleotide of lacI. For each riboswitch, the optimal structure with the minimum free energy was chosen for the structure analysis. To predict riboswitch structures in the ligand-bound form, we utilized the ‘force pair’ function of RNAstructure 5.1 to force the theophylline binding pocket to form.
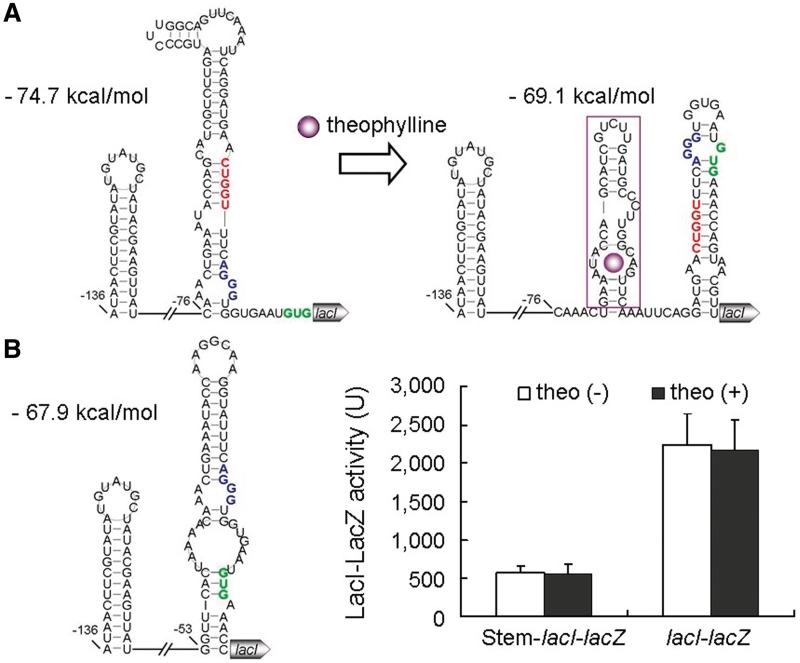
The structure analysis revealed that ON1-lacI in the non-ligand-bound state forms a 9-bp stem immediately adjacent to the RBS (Figure 2A). This long stem exists in not only the optimal structure but also alternative structures with higher free energy (data not shown). Except for the first nucleotide, the random sequence contributes to the formation of the stem. When theophylline binds to the riboswitch, the conformation changes and the long stem adjacent to the RBS does not form (Figure 2A). In ON2-lacI, the same was found (Figure 2B). Interestingly, the 5-nt random sequences of the two ON riboswitches share the latter 4 nt, which is unlikely to be coincident. We proposed that the latter four nucleotides GUAU is critical for the action of ON1-lacI and ON2-lacI by being part of the long stem immediately upstream of the RBS. If this is true, then the riboswitch OFF-lacI in the absence of theophylline would not contain a long stem next to the RBS, allowing for the gene translation; but would form the stem structure when bound by the ligand, repressing the expression. As shown in Figure 3A, the predicted structures of OFF-lacI in the presence and in the absence of theophylline are totally in agreement with this hypothesis. According to the prediction of free energy using RNA structure 5.1, all the structures in the non-bound state have lower free energy than the structures in the ligand-bound state (Figures 2 and 3), and therefore are proposed to predominate in the absence of theophylline. Only when theophylline binds to the riboswitches would the equilibrium distribution be shifted to the formation of structures with a theophylline-binding pocket, switching ON or OFF the target. To confirm the role of a long stem adjacent to RBS in the riboswitch-mediated gene control, we integrated a long stem (10 nt)–loop structure immediately upstream of the RBS of lacI fused with lacZ (Figure 3B). In the resulting mutant named Stem–lacI–lacZ, the expression levels of the lacI–lacZ fusion were reduced by 74% as a result of the insertion of the long stem (Figure 3B). Taken together, the synthetic riboswitches regulate lacI expression by forming or disrupting a long stem (9–10 nt) immediately upstream of the RBS in response to theophylline.
Figure 2.
Predicted structures of the riboswitches ON1-lacI (A) and ON2-lacI (B). In the non-ligand-bound state, there is a 9-bp stem structure immediately upstream of the ribosome binding site and is likely to block the gene translation. In the theophylline-bound state, the long stem does not form and consequently permits the gene translation. The random sequence is shown in red; the ribosome binding site in blue; and the start codon in green.
Figure 3.
Predicted structures of the riboswitch OFF-lacI and the long stem besides the ribosome binding site in the mutant Stem–lacI. (A) In OFF-lacI, a 9-bp stem structure forms immediately upstream of the ribosome binding site in the absence of theophylline but does not exist in the presence of the ligand. The random sequence is shown in red; the ribosome binding site in blue; and the start codon in green. (B) In Stem–lacI, there is a 10-bp stem structure located immediately upstream of the ribosome binding site. LacZ activity of the strain Stem–lacI–lacZ was determined using the β-galactosidase assay. The ribosome binding site is shown in blue, and the start codon in green.
ON-lacI–LacIbs acts as a magnified, tunable and two-way riboswitch device
Although the above-described riboswitches managed to fine-tune their target genes in response to theophylline and modulate corresponding biological processes, they had following limitations. First, they did not restore wild-type levels in the ON state (i.e. low ligand-saturating target levels) and failed to completely shut off the target genes in the OFF-state (i.e. high target leakage) (Figure 1C). Second, these riboswitches performed either ON or OFF function and thus were ‘one-way’ switches. It would be desirable to develop a ‘two-way’ device that not only switches off but also switches on gene expression without changing growth medium. Here, we utilized a ligand-responsive riboswitch in combination with the LacI protein and LacIbs to solve these problems. Given that the two ON ribowitches are equally effective in controlling lacI and both have a wider dynamic range than OFF-lacI (Figure 1C), we chose ON1-lacI for the construction of the riboswitch-LacP hybrid device.
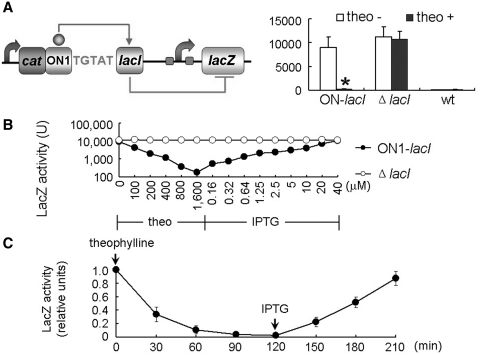
LacI is a transcriptional repressor and inhibits lacZ transcription by binding to two LacIbs upstream of lacZ. Although the cells carrying ON1-lacI displayed high basal levels of LacI in the ligand-free form and had only 1-fold increase in LacI levels in the ligand-bound form (Figure 1C), the narrow dynamic range of the ON1-lacI riboswitch would be magnified when the LacI repressor in turns controlled its downstream target. To test this possibility, we integrated the ON riboswitch upstream of the intact lacI gene (not fused with lacZ). The LacI protein represses expression of lacZ so that the ON riboswitch indirectly switches off lacZ in response to theophylline. In support of the above magnification hypothesis, ON1-lacI when equipped with two LacIbs (upstream of lacZ) displayed little leaky repression on lacZ in the presence of caffeine but in the absence of theophylline, and almost completely switched off lacZ when theophylline was added to the cells (Figure 4A). Thus, LacI as an intermediary modulator can greatly increase the dynamic range and improve the performance of the ON riboswitch. ON1-lacI equipped with LacI and two LacIbs was hereinafter referred to as ON-lacI–LacIbs.
Figure 4.
Magnified, tunable, two-way gene control mediated by the hybrid device ON-lacI–LacIbs. (A) Using the LacI repressor as a modulator, the ON-lacI riboswitch indirectly controlled lacZ expression and its regulatory effects on lacZ were magnified compared to its effects on lacI. (B) Tunable and two-way control of target gene mediated by the combinatorial use of the ON-lacI riboswitch and LacI-binding sites. Using LacI as a modulator, the ON-lacI riboswitches finely tuned lacZ expression in response to theophylline (0–1600 µM) and reversed lacZ expression by adding increasing concentrations of IPTG (0.16–40 µM) without removal of medium. Error bars represent standard deviation (*P < 0.05). (C) Kinetics of the LacZ activity in the strain carrying ON-lacI–lacZ. 1600 µM theophylline was added to exponentially growing culture at 0 min, and 40 µM IPTG was added to the culture at 120 min. Samples were collected for measurement of LacZ activity at the indicated time points. All data were normalized to the LacZ activity of the strain grown in LB medium plus 1600 µM caffeine.
Using LacI in the riboswitch device also makes possible sequential two-way gene regulation as the LacI protein can be inactivated by isopropyl β-d-1-thiogalactopyranoside (IPTG). Thus, adding theophylline should switch on lacI and repress target genes of LacI, whereas adding IPTG should inactivate LacI and derepress the targets. The sequential control was verified by determining β-galactosidase levels of the cells carrying the ON1-lacI switch and intact lacZ. Theophylline and IPTG regulated lacZ in opposite directions in a concentration-dependent manner (Figure 4B). The effective concentration range of theophylline was 100–1600 µM. Theophylline concentrations <100 µM gave little regulation and those higher than this range failed to cause additional repression. When IPTG was added to the medium containing 1600 µM theophylline, lacZ was derepressed. The effective range of IPTG was 0.16–40 µM. In addition, we performed time-course experiments to further characterize the response of ON-lacI–LacIbs to theophylline and IPTG. All the data were normalized to those of the same strain grown in LB plus 1600 µM caffeine. As shown in Figure 4C, the hybrid device switched off ~66% of the LacZ activity within 30 min after the addition of 1600 µM theophylline. Maximum silencing (~98% of the LacZ activity was switched off) was observed at 120 min. Forty micrometer IPTG was then added to the culture and switched on the lacZ expression rapidly. Within 90 min after the IPTG addition, ~85% of the LacZ activity was restored.
Application of ON-lacI–LacIbs to portable, tunable and two-way regulation of sigmaS (RpoS) subunit of RNA polymerase
Inducible gene control built on binding of LacI to LacIbs has been widely used for various non-lacZ genes in prokaryotic and eukaryotic cells (21–25), showing that the inducibility of the LacI-responsive promoters are less likely to be affected by downstream targets than that of riboswitches. Thus, the ON-lacI–LacIbs hybrid device should be capable of regulating any target gene when integrated upstream of the RBS. We next showcased the portability of this device by applying it to the rpoS gene.
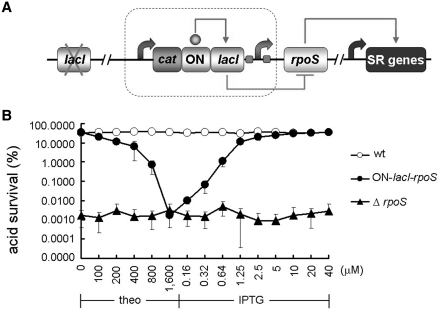
RpoS is an alternative sigma factor of RNA polymerase, governing expression of over 200 genes (26,27) and playing a critical role in survival of a diverse number of stresses such as acid shock (28,29), osmotic stress (30), heat shock (31), oxidative damage (29,32) and starvation (29,33). RpoS negatively regulates Salmonella virulence (29) and has, therefore, been mutated to engineer live attenuated vaccine strains (34,35). Given the significance of RpoS as a global response regulator and its potential medical use, it is desired that the rpoS gene could be placed under the control of the ON-lacI–LacIbs hybrid device for tunable and two-way expression control. To do this, we PCR amplified the ON-lacI–LacIbs cassette (2500 bp) from the genome of the strain carrying the ON riboswitch of lacI. The cassette is composed of the cat gene, the ON-lacI riboswitch, the lacI gene and the lacI–lacZ intergenic region containing the lac promoter (LacP) with two LacIbs. The cat gene facilitates integration of this cassette immediately upstream of any target gene via recombineering. Next, we simply integrated the riboswitch cassette upstream of the RBS of the rpoS gene, without adjusting the sequence of the device. Then, the native lacI gene was deleted from this strain, generating a new strain named ON-lacI–rpoS (Figure 5A). This strain was used to test if rpoS could be controlled by the theophylline-responsive and LacI-repressible regulation cassette. Since rpoS is required for acid resistance, we assayed survival of the strain under acidic conditions. We observed that the rpoS expression was on in the presence of caffeine and the absence of theophylline as revealed by normal survival percentage compared to the wild-type strain after 2-h acid treatment (pH 2) (Figure 5B). This was because the switch turned off lacI and therefore rpoS was in the ON state. When theophylline was added, the cells became sensitive to acid (Figure 5B), indicating that the addition of theophylline turned on the expression of lacI and the latter repressed the expression of rpoS. Acid survival in the absence of theophylline was close to the wild-type levels and decreased with increasing concentrations of theophylline (Figure 5B). Acid survival reached the minimum that was comparable to that of an rpoS null mutant when theophylline was increased to 1600 µM. When increasing concentrations of IPTG were added to the medium containing 1600 µM theophylline, the acid survival increased and was restored to the wild-type levels in the presence of 40 µM IPTG, consistent with the response curve of the ON1-lacI strain. These rpoS data not only confirmed the portability, tunability and two-way control features of the ON-lacI–LacIbs device, but also suggested the potential utility of this device in flexibly manipulating virulence and immunogenicity of live vaccines and bacteria for other medical purposes.
Figure 5.
Gene control mediated by the ON-lacI–LacIbs hybrid device. (A) The ON-lacI–LacIbs hybrid device. The boxed is a portable riboswitch cassette ON-lacI–LacIbs (LacI-binding sites) integrated immediately upstream of ribosome binding site of the rpoS gene that is a master regulator of stress resistance (SR). (B) Tunable and two-way control of rpoS via the ON-lacI–LacIbs device, as revealed by acid survival. The strain ON-lacI–rpoS that carried the ON-lacI–LacIbs device upstream of the ribosome binding site became increasingly sensitive to acid with theophylline (0–1600 µM). When increasing concentrations of IPTG were added to cells grown in media with 1600 µM theophylline, acid survival was increased and restored to wild-type levels in the presence of 40 µM IPTG. Error bars represent standard deviation.
ON-lacI–LabIs coupled with a small non-coding RNA CsrB for trans-acting regulation of autoaggregation and biofilm formation
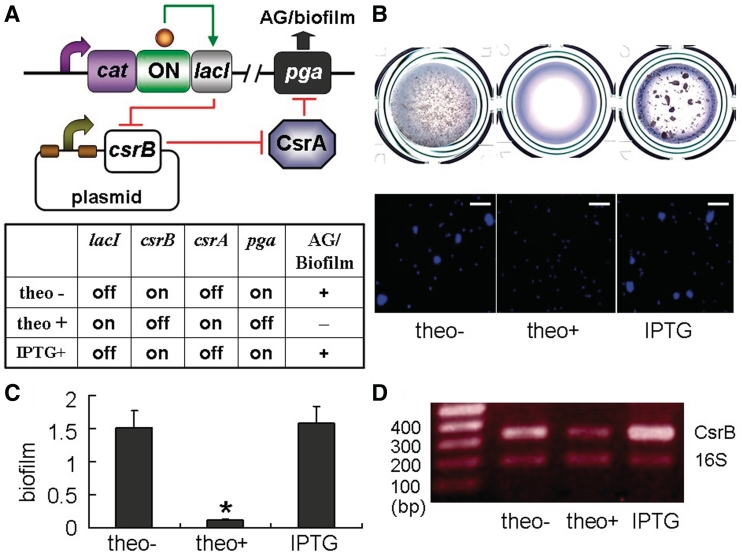
Next, we asked if we could accomplish trans-acting regulation of target expression using the ON-lacI–LacIbs device so that there is no need for chromosome engineering. We illustrated this using a naturally existing small non-coding RNA CsrB which is well known as an antagonist of the CsrA protein (36). It has previously been demonstrated that CsrB promotes cellular autoaggregation (4) and biofilm formation by indirectly activating the expression of the pgaABCD operon via CsrA (37). Considering these obvious phenotypes of CsrB-overproducing cells, we decided to couple CsrB with the ON-lacI–LacIbs device to control the pga operon (Figure 6A). We cloned csrB in a multi-copy vector downstream of the lac promoter to place the CsrB expression under the control of the ON1-lacI switch. Since one LacIb is after the transcriptional start site, the resulting CsrB slightly differs from the native one by the extra 21-nt sequence (i.e. LacIb). We induced the CsrB expression by IPTG and observed that autoaggregation (which serves as an indicator of CsrB expression) occurred (data not shown), indicating that this CsrB is functional. We introduced this CsrB-carrying vector into the ON1-lacI strain and examined autoaggregation and biofilm formation. In the presence of caffeine and the absence of theophylline, LacI levels were low and CsrB expression should be on (Figure 6A). As predicted, the cells autoaggregated and formed high levels of biofilms, showing that the pga expression was induced due to the overproduction of CsrB (Figure 6B and C). In the presence of theophylline, lacI expression is turned on and CsrB production should be repressed by LacI. The repression of CsrB is expected to result in upregulation of CsrA, which inhibits autoaggregation and biofilm formation (Figure 6A). In agreement with this, the cells were negative with autoaggregation and formed low levels of biofilms (Figure 6B and C). When IPTG was added together with theophylline, the activity of LacI was inhibited, and CsrB should be at high levels (Figure 6A). In support of this, autoaggregation and biofilm formation were restored in the presence of IPTG and theophylline (Figure 6B and C). The changes in CsrB expression in response to theophylline and IPTG were also confirmed by semi-quantitative RT–PCR (Figure 6D). These results demonstrate that the hybrid device can be used in combination with a small non-coding RNA to exert trans-acting control of target expression, alleviating the need for modification of the chromosome and expanding the applications of this riboswitch system.
Figure 6.
Trans-acting gene control through combinatorial use of the ON-lacI–LacIbs hybrid device and a small non-coding RNA CsrB. (A) Trans-acting system and predicted autoaggregation (AG) and biofilm phenotypic changes of the ON-lacI strain carrying pLacIb-CsrB in response to theophylline and IPTG. A csrB-carrying plasmid (pLacIb-CsrB) was transformed into the ON-lacI strain. The csrB gene was preceded by two LacI-binding sites and thus under the control of the ON1-lacI riboswitch. (B) Scanned images of cell suspensions (stained with crystal violet) of the ON1-lacI strain harboring pLacIb-CsrB in microtiter plates and fluorescence microscopic images of the corresponding cell suspensions (stained with DAPI) in the absence of theophylline, in the presence of 2 mM theophylline or in the presence of both 2 mM theophylline and 1 mM IPTG. (C) Biofilm formation of the ON1-lacI strain harboring pLacIb-CsrB in the absence of theophylline, in the presence of 2 mM theophylline or in the presence of both 2 mM theophylline and 1 mM IPTG. (D) Detection of intracellular CsrB small non-coding RNA levels by semi-quantitative reverse transcription-PCR. Total RNA was isolated from overnight cultures of the ON1-lacI strain carrying pLacIb-CsrB grown in the absence of 2 mM theophylline, in the presence of 2 mM theophylline or in the presence of both 2 mM theophylline and 1 mM IPTG. cDNA was amplified for 18, 20, 22 and 24 cycles using gene-specific primers. 16S rRNA was used as an internal control. PCR products formed after 18 cycles are included in the agarose gel image shown in the figure, along with a DNA ladder (100-, 200-, 300-, and 400-bp bands indicated). Densitometry indicated that for both the CsrB and 16S rRNA products, cycle 18 was within the linear range for the PCR amplification (data not shown). Expected product size for CsrB and 16S rRNA is 347 and 200 bp, respectively. Error bars represent standard deviation (*P < 0.05).
Use of ON-lacI–LacIbs to uncover a role of RpoS in acetate assimilation
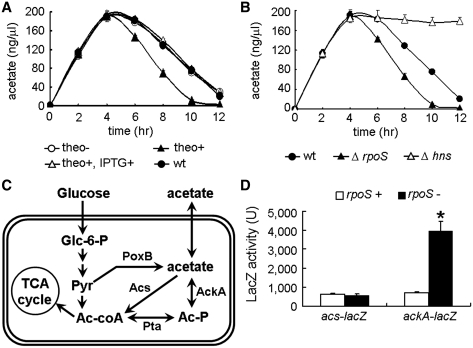
Next, we applied ON-lacI–LacIbs to identify new gene functions that may otherwise be neglected using traditional methods. It is well established that bacterial cells switch from rapid growth that produces and excretes acetate (dissimilation) in the presence of abundant nutrients to slower growth supported by the import and utilization of the excreted acetate (assimilation) when glucose is exhausted [for a review, see reference (38)]. This physiological event is defined as acetate switch, a survival response allowing cells to compete successfully during carbon starvation. It was previously reported that RpoS positively regulated consumption of acetate by activating the expression of acetyl-coenzyme A synthetase (Acs) (39,40). However, we obtained opposite results with ON-lacI–rpoS. ON-lacI–rpoS in the ON state in the absence of theophylline and IPTG exhibited acetate assimilation similar to that of the wild-type strain (Figure 7A). When grown in the presence of theophylline (2 mM) and absence of IPTG, the rpoS expression was shut off (detailed above) and accordingly, the acetate assimilation was accelerated. When IPTG (1 mM) was added to the medium to free up the rpoS expression, the acetate assimilation was restored to the wild-type levels (Figure 7A). Neither theophylline nor IPTG had effect on acetate assimilation of the wild-type strain (data not shown), excluding the possible non-specific effects of the two chemicals. The enhanced acetate assimilation was also observed with the newly constructed rpoS null mutant (Figure 7B). All the cell suspensions were adjusted to an OD of 0.08 prior to the acetate assays, and the cells with varying RpoS activities showed comparable cell growth rate (data not shown), ruling out the possible effects of growth rate on acetate assimilation. Taken together, RpoS represses acetate assimilation.
Figure 7.
A role of RpoS in acetate assimilation. (A) Extracellular acetate concentration plotted as a function of time for the wild-type and the strain ON-lacI–rpoS. (B) Extracellular acetate concentration plotted as a function of time for the wild-type strain and those deficient for rpoS or hns. (C) Pathways of acetate assimilation in E. coli. Glc-6-P, glucose 6-phosphate; Pyr, pyruvate; Ac-CoA, acetyl coenzyme A; Acetyl-P, acetyl phosphate. (D) LacZ activities of the strains carrying the acs–lacZ or ackA–lacZ translational fusion on the chromosome, with or without rpoS. Error bars represent standard deviation (*P < 0.05).
Next, we explored the underlying mechanism by which RpoS regulates the ability to scavenge for extracellular acetate. Acetate assimilation depends on the Acs and the phosphotransacetylase (Pta)–acetate kinase (AckA) pathway (38,41) (Figure 7C). We therefore proposed that RpoS inhibited the Acs and/or the Pta–AckA pathway and that deleting rpoS should increase their expression levels. To test this hypothesis, we incubated the cells for 6 h when acetate switch had been flipped and then examined the possible effects of RpoS on the levels of Acs and AckA–Pta. Our β-galactosidase data showed that the removal of rpoS slightly reduced the expression of an acs-lacZ translational fusion on the chromosome, excluding the first possibility. However, deleting rpoS resulted in a 5-fold increase in the expression levels of an ackA-lacZ translational fusion (Figure 7D). Pta and ackA are in the same operon. The ackA-lacZ data revealed that the Pta–AckA pathway was enhanced in the rpoS null mutant and the theophylline-grown ON-lacI–rpoS cells, relative to that of the wild-type. It is followed that the increased acetate assimilation as a result of the rpoS repression (ON-lacI–rpoS in the presence of theophylline) or deletion is at least partially due to the elevated activity of the Pta–AckA pathway.
It is notable that our results on the role of RpoS in acetate assimilation are opposite to those of previous studies using rpoS mutants (39,40). rpoS mutants frequently generate secondary mutations within the hns gene (encoding H-NS, an abundant nucleoid-associated protein) to compensate for the loss of RpoS function (42). It is, therefore, likely that rpoS mutants used in those studies might have accumulated analogous compensatory mutations and consequently exhibited the opposite phenotype. If this is true, then cells deleted for hns, compared to the wild type, should have a reduced ability to scavenge for extracellular acetate. As predicted, removing hns inhibited acetate assimilation as revealed by sustained high levels of extracellular acetate (Figure 7B), supporting the notion that the previous observations with the rpoS mutants may result from compensatory mutations in hns.
DISCUSSION
Here, we have used a library screening strategy for creating riboswitches residing upstream of the lacI RBS. Based on this strategy, we have further engineered a portable riboswitch-LacP hybrid device that efficiently regulates specific target genes without the need for rational design. Using the LacI protein as a signal converter, the hybrid device has accomplished portable and sequential two-way control of target expression in response to theophylline and IPTG. This two-way portable device can be used to uncover protein functions that would otherwise be neglected by using traditional genetic mutagenesis.
Two regulatory mechanisms have mainly been utilized for constructing synthetic riboswitches: translational repression (4,5,43) and mRNA destabilization based on hammerhead-mediated RNA self-cleavage (8,44). Here, we adopt the first strategy to create riboswitches for lacI. Riboswitches, like other RNA molecules, are sensitive to their sequence context and a subtle change in a single nucleotide often makes a big difference to their secondary structures and functionality. We take advantage of this feature and build up riboswitch libraries by grafting a theophylline-responsive aptamer followed by a 5-nt random sequence to target genes. The aptamer is used as the sensor domain that selectively binds theophylline, causes conformational changes and mediates downstream gene expression. This sensor is integrated upstream of RBS of target genes on the chromosome by recombineering. The five random bases in between the sensor and the RBS allow for generation of a library which in principle harbors 45 riboswitch candidates. Subsequent screening on X-gal plates can efficiently identify functionally active riboswitches. Unlike previously reported riboswitch libraries that are all carried on plasmids (14–16), our libraries are established on the chromosome with the aid of recombineering. Therefore, our riboswitches are more genetically stable than the previous plasmid-harbored ones. We built up a riboswitch random library for the lacI gene that was fused in frame with lacZ whose expression could be easily examined on X-gal plates. By checking the color of colonies grown on X-gal plates with or without theophylline, both ON and OFF riboswitches of lacI were created. Structure predictions revealed that the above riboswitches regulate the lacI expression by adopting or disrupting a long stem structure immediately upstream of the RBS in response to theophylline. The long stem forms in the OFF state and fails to exist in the ON state. Thus, the long stem besides the RBS is responsible for the gene silencing mediated by the riboswitches, consistent with previous reports (5).
Building on the synthetic riboswitches created in the first part of the work, we constructed a portable and two-way regulation device that act as a versatile key to any target without rational design or library screening. We achieved this by adopting the LacI repressor, which inhibits transcription by binding to LacIbs, as a modulator that links the ligand-responsive riboswitch to ultimate targets. As described above, effective ON riboswitches had been created for lacI. Upstream of the lacI gene in the strain ON-lacI is a riboswitch cassette that carries all necessary regulation elements including the CmR gene (cat), a constitutive promoter, the theophylline-responsive ON riboswitch of lacI, the lacI gene and two LacIbs. Without any sequence adjustment, we simply inserted this 2500-bp cassette immediately upstream of the RBS of the rpoS gene and showed that the ON-lacI riboswitch fine-tuned rpoS expression (as revealed by acid survival). When applied to the csrB gene, this device controlled the expression of the small non-coding RNA CsrB and consequently modulated biofilm formation and cellular autoaggregation. The success of this device in regulating the expression of lacZ, rpoS and csrB demonstrates its portability.
The combinatorial use of the ON riboswitch of lacI and LacIbs has other advantages in addition to portability. LacI can magnify the regulatory effects of the ON-lacI–LacIbs device and maximize its dynamic range that is defined as the ratio of the highest and lowest target levels. Previous regulation systems mediate control of gene expression in a one-way manner: target expression is turned on in response to a specific inducer and turned off when the inducer is removed by replacement of medium. However, this maneuver is infeasible in animal experiments and may cause problems in some demanding bacterial experiments where cultures have to be kept intact. The ON-lacI–LacIbs system overcomes this problem as its target gene is in the ON state in the absence of theophylline, switched off in response to the ligand, and switched back to the ON state when IPTG is added without removal of theophylline.
Finally, strains armed with this device are more genetically stable and therefore provide more reliable information compared to those constructed using traditional approaches. It has been demonstrated that deleterious mutations and those that cause reduced fitness can bring about secondary mutations that compensate for undesired changes (45–50). The compensatory mutations can give misleading results in gene function studies and destabilize geno- and phenotypes of genetically engineered bacteria for medical and industrial purposes. One solution to minimize the compensatory mutations is to engineer a strain in which the gene to be controlled is normally expressed and not shut off unless an inducer is applied. Moreover, it would be ideal if the wild-type levels of gene expression could be restored in response to a second effector molecule, to ensure that the observed mutation phenotype is not caused by pleiotropic effects of the inducer. The ON-lacI–LacIbs riboswitch device meets all these requirements. For example, the strain ON-lacI–rpoS exhibits normal rpoS expression in the absence of theophylline and IPTG and therefore has no fitness burden. Therefore, compared to an rpoS null mutant, ON-lacI–rpoS is less likely to accumulate compensatory mutations and thus more reliable. Only in the presence of theophylline, can ON-lacI–rpoS behavior as an rpoS mutant, allowing for rpoS function investigation. Phenotypes in the two ON states (in the absence of any inducer and in the presence of both theophylline and IPTG) should be identical, excluding the non-specific effects of theophylline. Here, the use of the ON-lacI–LacIbs device helped uncover a novel function of RpoS, which may otherwise be neglected or misunderstood by using traditional approaches. That is, RpoS inhibits acetate assimilation. Acetate assimilation is a critical process allowing the cell to efficiently utilize nutrient resources when favorable carbon sources are exhausted, and turning the waste product acetate into a carbon and energy source. Acetate assimilation is also responsible for managing levels of acetyl-coA and acetyl phosphate, both of which are important central metabolites [for a review, see reference (38)]. Interestingly, previous studies reported the opposite results that RpoS appeared to positively regulate acetate assimilation (39,40). One explanation of the discrepancy is the above-mentioned occurrence of compensatory mutations. Indeed, it has been demonstrated that secondary mutations within the hns gene frequently occur in strains carrying rpoS mutations to compensate for the loss of RpoS function (42). The action of H-NS is generally opposite to that of RpoS (51), and H-NS downregulates both rpoS mRNA translation and RpoS stability (52). This, together with our observation that deleting hns represses acetate assimilation makes it reasonable to speculate that compensatory mutations within hns may have occurred in the rpoS mutant strains used in the previous studies so that they observed opposite phenotypes with their rpoS mutants.
In summary, we have developed a portable device that performs sequential OFF-and-ON gene regulation in response to theophylline and IPTG, respectively. This study also indicates that the number of gene control elements available to us can be greatly increased by combining ligand-dependent riboswitches and inducer-responsive promoters. Suppose the number of riboswitch ligands is n and the number of inducers of inducible promoters is m, then n × m combinatorial control elements can be created, greatly enriching the repertory of gene regulatory elements. Although this work focuses on developing a riboswitch-LacP hybrid device in E. coli, similar strategy (i.e. a ligand-responsive riboswitch integrated with an inducible promoter system) may be extended to eukaryotic cells to achieve portable, two-way expression regulation.
FUNDING
Funding for open access charge: An Innovation and Technology Fund (ITS/148/09) from Hong Kong Innovation and Technology Commission, in part by a Research Fund for the Control of Infectious Diseases Commissioned Study of Food and Health Bureau of Hong Kong Government; a Research Grants Council CRF grant (HKU1/CRF/10 to J.-D.H.); a HKU small grant funding (201007176077 to Y.J.).
Conflict of interest statement. J.-D.H. and Y.J. declare competing financial interests in the form of a patent application whose value may be affected by the publication of this manuscript.
REFERENCES
- 1.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, Watt RM, Danchin A, Huang JD. Use of a riboswitch-controlled conditional hypomorphic mutation to uncover a role for the essential csrA gene in bacterial autoaggregation. J. Biol. Chem. 2009;284:28738–28745. doi: 10.1074/jbc.M109.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhounig A, Karcher D, Bock R. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc. Natl Acad. Sci. USA. 2010;107:6204–6209. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 10.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 11.Chushak Y, Stone MO. In silico selection of RNA aptamers. Nucleic Acids Res. 2009;37:e87. doi: 10.1093/nar/gkp408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collett JR, Cho EJ, Lee JF, Levy M, Hood AJ, Wan C, Ellington AD. Functional RNA microarrays for high-throughput screening of antiprotein aptamers. Anal. Biochem. 2005;338:113–123. doi: 10.1016/j.ab.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Lee HJ, Corn RM. Fabrication and characterization of RNA aptamer microarrays for the study of protein–aptamer interactions with SPR imaging. Nucleic Acids Res. 2006;34:6416–6424. doi: 10.1093/nar/gkl738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muranaka N, Abe K, Yokobayashi Y. Mechanism-guided library design and dual genetic selection of synthetic OFF riboswitches. Chembiochem. 2009;10:2375–2381. doi: 10.1002/cbic.200900313. [DOI] [PubMed] [Google Scholar]
- 17.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Y, Watt RM, Danchin A, Huang JD. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics. 2009;10:165. doi: 10.1186/1471-2164-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 22.Luukkonen BG, Seraphin B. Construction of an in vivo-regulated U6 snRNA transcription unit as a tool to study U6 function. RNA. 1998;4:231–238. [PMC free article] [PubMed] [Google Scholar]
- 23.Grilly C, Stricker J, Pang WL, Bennett MR, Hasty J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol. Syst. Biol. 2007;3:127. doi: 10.1038/msb4100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cronin CA, Gluba W, Scrable H. The lac operator–repressor system is functional in the mouse. Genes Dev. 2001;15:1506–1517. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong T, Kirchhof MG, Schellhorn HE. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol. Genet. Genomics. 2008;279:267–277. doi: 10.1007/s00438-007-0311-4. [DOI] [PubMed] [Google Scholar]
- 27.Schellhorn HE, Audia JP, Wei LI, Chang L. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J. Bacteriol. 1998;180:6283–6291. doi: 10.1128/jb.180.23.6283-6291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl Acad. Sci. USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokes NR, Murray HD, Subramaniam C, Gourse RL, Louis P, Bartlett W, Miller S, Booth IR. A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc. Natl Acad. Sci. USA. 2003;100:15959–15964. doi: 10.1073/pnas.2536607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sammartano LJ, Tuveson RW, Davenport R. Control of sensitivity to inactivation by H2O2 and broad-spectrum near-UV radiation by the Escherichia coli katF locus. J. Bacteriol. 1986;168:13–21. doi: 10.1128/jb.168.1.13-21.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 34.Coynault C, Robbe-Saule V, Norel F. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma S) regulon. Mol. Microbiol. 1996;22:149–160. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 35.Shi H, Santander J, Brenneman KE, Wanda SY, Wang S, Senechal P, Sun W, Roland KL, Curtiss R. Live recombinant Salmonella Typhi vaccines constructed to investigate the role of rpoS in eliciting immunity to a heterologous antigen. PLoS One. 5:e11142. doi: 10.1371/journal.pone.0011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 38.Wolfe AJ. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin S, Song SG, Lee DS, Pan JG, Park C. Involvement of iclR and rpoS in the induction of acs, the gene for acetyl coenzyme A synthetase of Escherichia coli K-12. FEMS Microbiol. Lett. 1997;146:103–108. doi: 10.1111/j.1574-6968.1997.tb10178.x. [DOI] [PubMed] [Google Scholar]
- 40.Rahman M, Hasan MR, Oba T, Shimizu K. Effect of rpoS gene knockout on the metabolism of Escherichia coli during exponential growth phase and early stationary phase based on gene expressions, enzyme activities and intracellular metabolite concentrations. Biotechnol. Bioeng. 2006;94:585–595. doi: 10.1002/bit.20858. [DOI] [PubMed] [Google Scholar]
- 41.Kumari S, Tishel R, Eisenbach M, Wolfe AJ. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desai SK, Mahadevan S. Accumulation of hns mutations specifically in stationary phase in an E. coli strain carrying an impaired rpoS locus. J. Genet. 2006;85:221–224. doi: 10.1007/BF02935336. [DOI] [PubMed] [Google Scholar]
- 43.Verhounig A, Karcher D, Bock R. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc. Natl Acad. Sci. USA. 107:6204–6209. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handel A, Regoes RR, Antia R. The role of compensatory mutations in the emergence of drug resistance. PLoS Comput. Biol. 2006;2:e137. doi: 10.1371/journal.pcbi.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bjorkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 48.DiNardo S, Voelkel KA, Sternglanz R, Reynolds AE, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 49.Robinson AC, Begg KJ, MacArthur E. Isolation and characterization of intragenic suppressors of an Escherichia coli ftsA mutation. Res. Microbiol. 1991;142:623–631. doi: 10.1016/0923-2508(91)90075-l. [DOI] [PubMed] [Google Scholar]
- 50.Ramos-Montanez S, Kazmierczak KM, Hentchel KL, Winkler ME. Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 2010;192:6390–6400. doi: 10.1128/JB.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of sigma S and many sigma S-dependent genes in Escherichia coli. J. Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y, Gottesman S. Modes of regulation of RpoS by H-NS. J. Bacteriol. 2006;188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]