Abstract
A cDNA clone encoding a thiol-protease (TPE4A) was isolated from senescent ovaries of pea (Pisum sativum) by reverse transcriptase-polymerase chain reaction. The deduced amino acid sequence of TPE4A has the conserved catalytic amino acids of papain. It is very similar to VSCYSPROA, a thiol-protease induced during seed germination in common vetch. TPE4A mRNA levels increase during the senescence of unpollinated pea ovaries and are totally suppressed by treatment with gibberellic acid. In situ hybridization indicated that TPE4A mRNA distribution in senescent pea ovaries is different from that of previously reported thiol-proteases induced during senescence, suggesting the involvement of different proteases in the mobilization of proteins from senescent pea ovaries. TPE4A is also induced during the germination of pea seeds, indicating that a single protease gene can be induced during two different physiological processes, senescence and germination, both of which require protein mobilization.
Senescence is the last step of plant development, leading to the death of a plant tissue or organ or the whole plant. There is evidence to indicate that senescence is a genetically controlled process (Gan and Amasino, 1997; Noodén et al., 1997). One of the major events taking place during the senescence process is the ordered degradation of cell constituents, and the degradation of proteins is a characteristic of senescence. Protease genes have been cloned from several senescent plants (Jones et al., 1995; Smart et al., 1995; Drake et al., 1996), and thiol-proteases are the most common proteolytic enzymes induced in senescent plant cells (Granell et al., 1998).
The unpollinated pea (Pisum sativum) ovary is a convenient system in which to study the natural senescence of reproductive organs in plants. When pollination is prevented, the ovary stops growing about 3 d after anthesis; natural senescence then starts, leading to ovary death and abscission within 2 or 3 d (Carbonell and García-Martínez, 1980). This senescence process is accompanied by the loss of sensitivity to GA3 treatments (García-Martínez and Carbonell, 1980) and by an increase in proteolytic activity at neutral pH (Carbonell and García-Martínez, 1985; Carrasco and Carbonell, 1988; Cercós et al., 1992). New proteases have been found associated with senescence in unpollinated pea ovaries (Granell et al., 1992; Cercós and Carbonell, 1993; Cercós et al., 1993). The senescence process is prevented when parthenocarpic fruit set is induced by treatment of the unpollinated ovaries with GA3 and other plant-growth regulators (García-Martínez and Carbonell, 1980).
Like senescence, seed germination involves the degradation and remobilization of stored nutrients. Thiol-proteases also play a key role in this remobilization step (Granell et al., 1998). It is not known whether the same thiol-proteases are involved in senescence and germination or whether there are different thiol-proteases for each process. There are few reports addressing this question (Griffiths et al., 1997), and none of them reports a single-copy gene being expressed in both senescence and germination.
In this study, we isolated and characterized a cDNA clone encoding TPE4A, a thiol-protease induced during senescence of unpollinated pea ovaries. The induction of TPE4A transcription during senescence and its repression after GA3 treatment were observed by northern-blot analysis and in situ hybridization. We also detected TPE4A transcription in other organs, including germinating seeds. We report evidence for a single-copy thiol-protease gene that is induced in both senescence and seed germination.
MATERIALS AND METHODS
Plant Material
Pea (Pisum sativum L. cv Alaska) plants were grown as described by Carbonell and García-Martínez (1985), and ovaries and fruits were collected as described by Cercós et al. (1992). After stamens and petals were removed 2 d before anthesis to avoid pollination, two types of samples were prepared: (a) presenescent and senescent ovaries, which were untreated and unpollinated ovaries collected between the day of anthesis and 4 d later; and (b) young fruits, in which fruit set was induced by treatment with GA3 on the day of anthesis (d 0) and fruits were collected between d 1 and 4 after anthesis.
Pea seeds were allowed to imbibe by placing them on top of sterile cotton swabs previously saturated with either sterile water or 50 μm STS. Seeds were kept in the dark at room temperature and collected after 0, 1, 2, 3, 4, and 6 d. Embryonic axes were removed in samples collected after 3 d of imbibition. Collected samples were stored at −80°C until use.
Cloning Strategy
Two degenerate sets of primers were designed according to conserved amino acid regions in the sequence of the papain family of plant thiol proteases. TP4 (5′-TGYGGNAGYTGYTGG-3′) was a sense degenerate set of oligonucleotides specific for the conserved CGSCW motif, which includes the catalytic Cys residue. TP7 (5′-NCCCCANGARTT-3′) was an antisense degenerate set of oligonucleotides specific for the conserved NSWG motif, which includes the catalytic Asn residue and a conserved Trp residue. For first-strand cDNA synthesis, an XSC adaptor (5′-GACTCGAGTCGACATCGAT-3′; Frohman et al., 1988) was added at the 5′ end of the TP7 oligonucleotide, generating the TP7-XSC oligonucleotide.
For first-strand synthesis, 0.1 μg of total RNA from senescent ovaries collected on d 4 after anthesis was reverse transcribed with 10 units of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) and 40 pmol of the TP7-XSC oligonucleotide in the presence of 10 units of RNase inhibitor (RNA-Guard, Pharmacia) for 1 h at 37°C. After alkaline hydrolysis of RNA and purification of the single-stranded DNA with Qiaex II (Qiagen, Chatsworth, CA), one-tenth of the purified single-stranded DNA was used for second-strand synthesis and PCR amplification.
First-strand cDNA was mixed with 10 pmol of TP4 oligonucleotide, 10 nmol of each nucleotide triphosphate, and 1 unit of Taq DNA-polymerase (Pharmacia). The second-strand synthesis reaction was carried out in a thermocycler (Perkin-Elmer) by incubating the mixture for 5 min at 94°C, 1 min at 40°C, and 15 min at 72°C. Next, 75 pmol of XSC oligonucleotide, 65 pmol of TP4 oligonucleotide, and 1 unit of Taq were added and the double-stranded cDNA was PCR amplified for 40 cycles at 94°C for 1 min, 40°C for 1 min, and 72°C for 1 min. The PCR product was electrophoresed onto a 1% (w/v) agarose gel, and a single band of about 400 bp was obtained (data not shown); this band was eluted with Qiaex II and cloned into the pT7-Blue vector (Novagen, Madison, WI).
To compare the positive clones and group the identical ones into families, the positions of the T residues in the nucleotide sequences of the clones were determined. DNA was denatured, annealed with a vector-specific primer (M13-20), and extended with Klenow fragment at 42°C for 5 min in the presence of 125 μm dCTP, 125 μm dGTP, 6.25 μm dTTP, 250 μm ddTTP, and 2 μCi of [α-35S]dATP (10 μCi/μL, 1000 Ci/mmol). The mixture was heat denatured and loaded onto a sequencing gel. Clones with the same T-band pattern were considered identical.
To generate the complete TPE4A cDNA, the flanking sequences for the initial PCR product were obtained by PCR. The template was a cDNA library generated with poly(A+) RNA from senescent pea ovaries on d 4 after anthesis in UNIZAP-XR vector (Stratagene) according to the manufacturer's instructions. The 5′ side was obtained by PCR with the T3 primer (specific to the vector) and the 13BR primer (specific for TPE4A; Fig. 1). The 3′ end was obtained by PCR with the T7 primer (specific to the vector) and the 13B primer (specific for TPE4A; Fig. 1). A single band was observed after each of the PCR products was loaded onto an agarose gel. The PCR bands were gel purified, cloned into pT7-Blue, and sequenced.
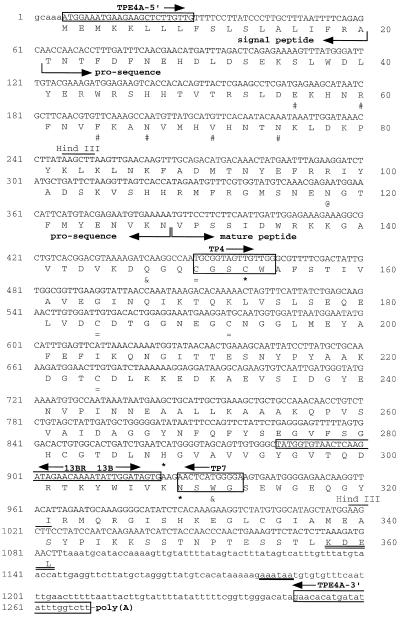
Figure 1.
Nucleotide and deduced amino acid sequences of TPE4A cDNA. Numbers on the left correspond to the nucleotide sequence, and numbers on the right correspond to the deduced amino acid sequence. The protein-coding region is shown in uppercase letters, and the 5′- and 3′-untranslated regions are shown in lowercase letters. Boxed sequences correspond to the target sequences for the oligonucleotides used in the PCR cloning experiments; the target amino acid sequences for the two degenerated oligonucleotides, TP4 and TP7, are also included in the boxes. Arrows next to the oligonucleotide names indicate the oligonucleotide direction (5′ to 3′). The proposed boundaries between the signal peptide, prosequence, and mature peptide are indicated by arrows. The boundary between the signal peptide and the prosequence was established based on the hydrophobicity plot of the protein (data not shown) and comparison with other protease sequences. Several motifs are highlighted: #, amino acids forming the ERFNIN motif; @, putative N-glycosylation site; *, essential catalytic amino acids; &, other conserved amino acid residues in the active site; and =, Cys residues involved in disulfide bridges. The ER retention signal (KDEL) is double underlined, the putative polyadenylation signal is single underlined, and the two HindIII restriction sites used for the genomic Southern-blot analysis are highlighted.
The entire TPE4A cDNA was generated by PCR using the same cDNA library as the template and the two specific primers for TPE4A, TPE4A-5′ and TPE4A-3′ (Fig. 1). The product was cloned in pT7-Blue as described above. To minimize the possibility of PCR artifacts, three independent PCR reactions were performed, and three positive clones from each of the reactions were sequenced. No differences were found between the nine sequenced clones with the exception that one of the three products was shorter (data not shown).
DNA Sequencing
Automatic DNA sequencing was carried out (PRISM 377, Applied Biosystems). Primers specific to the vector were used in the initial sequencing runs, and new primers specific to the insert sequences were synthesized after each run. Both DNA strands were completely sequenced. To minimize the possibility of PCR errors, nine TPE4A clones corresponding to three independent PCR reactions were completely sequenced. Sequences were analyzed with the University of Wisconsin Genetics Computer Group software package (Devereux et al., 1984).
Genomic Southern-Blot Analysis
Genomic DNA was isolated from pea leaves as described by Dellaporta et al. (1983). Twenty-microgram aliquots of genomic DNA were digested with the appropriate restriction enzymes and electrophoresed on a 0.6% (w/v) agarose gel in 1× TAE buffer (40 mm Tris-acetate and 1 mm EDTA, pH 8.0) as described by Sambrook et al. (1989). The gel was blotted onto Hybond N membranes (Amersham) according to the manufacturer's instructions and hybridized with the HindIII-HindIII fragment of the TPE4A cDNA (Fig. 1) according to the protocol described by Church and Gilbert (1984).
RNA Isolation and Northern-Blot Analysis
Total RNA was isolated from pea ovaries using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. Total RNA from seeds that had imbibed was extracted according to the procedure described by Bugos et al. (1995).
Ten-microgram aliquots of total RNA were electrophoresed in formaldehyde-agarose gels as described by Sambrook et al. (1989), blotted onto Hybond N membranes (Amersham) according to the manufacturer's instructions, and hybridized in the same conditions described above for Southern-blot analysis.
In Situ Hybridization
Localization of the TPE4A mRNA was determined by in situ hybridization as described by Jackson (1992). To generate the riboprobes, the TPE4A cDNA insert was subcloned in pBluescript SK− (Stratagene). Antisense probes and sense control probes were generated by in vitro transcription with T7 and T3 RNA polymerases, respectively. Probes were labeled with digoxigenin and immunodetected with an alkaline-phosphatase-conjugated anti-digoxigenin antibody. Alkaline phosphatase was detected by the 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium procedure. A microscope (Diaphot-TMD, Nikon) was used for sample visualization under phase contrast and photography.
RESULTS
Reverse Transcriptase-PCR Cloning of a New Senescence-Induced Thiol-Protease
A single reverse transcriptase-PCR band was obtained by reverse transcription with the TP7-XSC oligonucleotide and PCR with the TP4 and XSC oligonucleotides. Because several thiol-protease genes should be expressed in senescing pea ovaries, the PCR band should be a mixture of different cDNAs (actually, sequence alignments showed that the spacing between the two conserved sequences used for our oligonucleotide design is conserved in most of the plant thiol-proteases). To separate the different cDNA molecules present in the PCR band, 50 clones with insert were subjected to T-sequencing reactions. By comparing the T-track patterns, we were able to group the identical clones and found 11 different cDNA classes. One representative clone for each class was sequenced, and only two of them (classes 7 and 10) were similar to thiol-proteases. Clone 7 was identical to tpp, a previously reported thiol-protease cDNA from senescing pea ovaries (Granell et al., 1992). Clone 10 was a new thiol-protease cDNA expressed in senescing pea ovaries that we named TPE4A. It was different from tpp, with greatest similarity (90.8% at the protein level) to VSCYSPROA (Fig. 2), a thiol-protease cDNA isolated from germinating seeds of common vetch (Becker et al., 1997).
Figure 2.
Sequence similarities between TPE4A and other Cys proteases. The alignment was generated using the PileUp program of the Genetics Computer Group package. The compared proteases are: EP-C1 from bean (Ogushi et al., 1992); SEN102 (Valpuesta et al., 1995) from daylily; SEN11 (Guerrero et al., 1998) from daylily; SH-EP from Vigna mungo (Akasofu et al., 1990); vicilin peptide hydrolase from mung bean (VICILIN-PE; Lee et al., 1997); and VSCYSPROA from vetch (Becker et al., 1998).
Because both the TP4 and TP7 oligonucleotides were directed to internal conserved sequences of thiol-proteases, the cloned fragment of TPE4A was internal to the gene (Fig. 1). To obtain the complete cDNA clone, the remaining sequences were generated from a cDNA library by PCR using TPE4A-specific and vector-specific oligonucleotides. Because the TPE4A-specific oligonucleotides were directed to a nonconserved region of the TPE4A cDNA, these bands were expected to correspond to single cDNA species. After sequencing these PCR products, we found a perfect match of the overlapping sequences with the initial PCR fragment. Additionally, the new fragments aligned at the corresponding parts of the VSCYSPROA sequence. The entire TPE4A cDNA was obtained by PCR using oligonucleotides specific for the 5′ and 3′ ends.
Molecular Characterization of TPE4A
The isolated cDNA clone for TPE4A had an insert 1270 bp long, with an open reading frame encoding a 361-amino acid polypeptide (Fig. 1). Comparison of the deduced amino acid sequence with protein-sequence databases indicated homology with several plant thiol-protease genes (Fig. 2), especially with VSCYSPROA from common vetch (Becker et al., 1997), vicilin peptide hydrolase from mung bean (Lee et al., 1997), SH-EP from Vigna mungo (Akasofu et al., 1990), EP-C1 from bean (Ogushi et al., 1992), and SEN11 (Guerrero et al., 1998) and SEN102 (Valpuesta et al., 1995) from daylily. The putative thiol-protease activity of the TPE4A protein is supported by the conservation of the three catalytic amino acids of papain, Cys-154, His-298, and Asn-310 (Kamphuis et al., 1985), at positions 153, 288, and 309, respectively (Fig. 1). Other conserved amino acids involved in the catalytic activity of papain and some Cys residues forming disulfide bridges are also conserved in TPE4A (Fig. 1).
A putative signal sequence was found at the protein N terminus according to the rules proposed by Von Heijne (1983), with the peptide bond between Ala-20 and Thr-21 being the putative cleavage site with the highest probability. Downstream of the putative signal peptide, the consensus sequence of the ERFNIN motif (EX3RX3FX2NX3I/VX3N; Karrer et al., 1993) was found. The ERFNIN sequence has been described as a part of the proenzyme that is involved in the inhibition of the protease activity before processing (Karrer et al., 1993). The presence of this motif in TPE4A, as well as the comparison with the sequences of similar thiol-proteases, suggests that TPE4A is synthesized as a proenzyme and is processed before becoming an active enzyme. After alignment of the TPE4A amino acid sequence with the sequences of previously reported plant thiol-proteases, the putative position of the N-terminal residue in the mature protein was proposed to be Val-129 (Fig. 1). This agrees with the observation that in all of the ERFNIN-containing thiol-proteases, the seventh amino acid residue of the mature protein is Trp (Karrer et al., 1993).
Comparison of the TPE4A nucleotide sequence with the sequences of similar thiol-proteases suggested that the TPE4A cDNA was not full length, because only five nucleotides of the leader region were present in the clone (Fig. 1). Of the three independent PCR reactions carried out during TPE4A cloning, two of them started at the same point and the other was even shorter; this could suggest that a high degree of secondary structure and/or some kind of instability of the mRNA leader region would make the synthesis of a full-length cDNA difficult. However, the leader region should not be very long, because northern-blot analyses in ovaries and other tissues showed a single band that was smaller than the rRNA bands (1.8 kb) (data not shown).
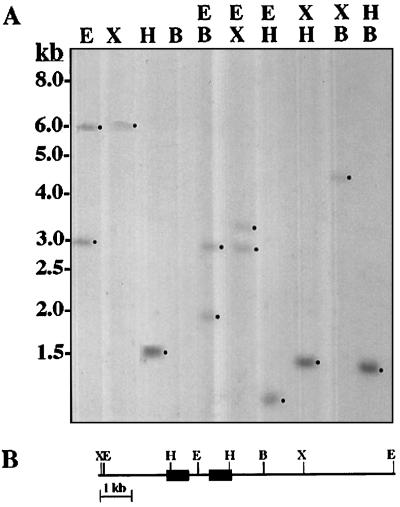
Genomic Southern-blot analysis (Fig. 3) indicated that TPE4A is encoded by a single-copy gene. Among the enzymes used for the restriction analysis, EcoRI, XbaI, and BamHI did not cut into the cDNA insert, whereas HindIII cut twice (Fig. 1). A single band of approximately 6 kb was found after XbaI digestion, suggesting that TPE4A is encoded by a single-copy gene. No band was detected after BamHI digestion, which could mean that the BamHI digestion product containing the TPE4A gene was longer than the sizes resolved in the gel. This was confirmed by the double-digestion results (Fig. 3A). The two bands formed in the EcoRI digestion indicated the presence of an EcoRI site in an intron. The difference in length of the HindIII fragment in the genomic DNA (about 1.5 kb; Fig. 3A) and in the cDNA clone (773 bp; Fig. 1) confirmed the presence of one or more introns between the HindIII sites, with a total intron length of about 700 bp. Figure 3B represents a proposed restriction map for the genomic DNA region containing the TPE4A gene.
Figure 3.
TPE4A is encoded by a single-copy gene. A, Genomic Southern-blot analysis showing that TPE4A is encoded by a single gene. Ten micrograms of pea genomic DNA was digested with EcoRI (E), XbaI (X), HindIII (H), and BamHI (B), and double digestions with combinations of these enzymes were also performed. The HindIII fragment of the cDNA clone was used as the probe. B, Deduced genomic organization of the TPE4A gene. Thick black lines represent the interrupted open reading frame, and thin lines represent the putative intron and the flanking regions of the gene. For simplicity, a single intron bearing the EcoRI site is shown.
Expression of the TPE4A Gene
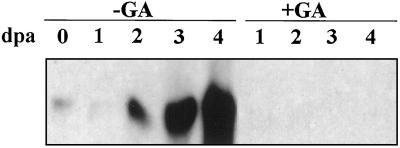
Northern-blot analysis of unpollinated pea ovaries indicated the induction of TPE4A transcription after the onset of senescence on d 2 after anthesis in nontreated ovaries (Fig. 4). No hybridization signal was detected in GA3-treated ovaries, whereas a slight signal was detected in presenescent ovaries, suggesting a repression of the basal expression levels after the hormonal treatment.
Figure 4.
TPE4A expression is induced during pea ovary senescence. Northern-blot analysis showing the progressive accumulation of the TPE4A transcript in presenescent (d 0, 1, and 2) and senescent (d 3 and 4) nontreated pea ovaries (−GA) and the repression of TPE4A expression in GA3-treated ovaries (+GA). dpa, Days postanthesis.
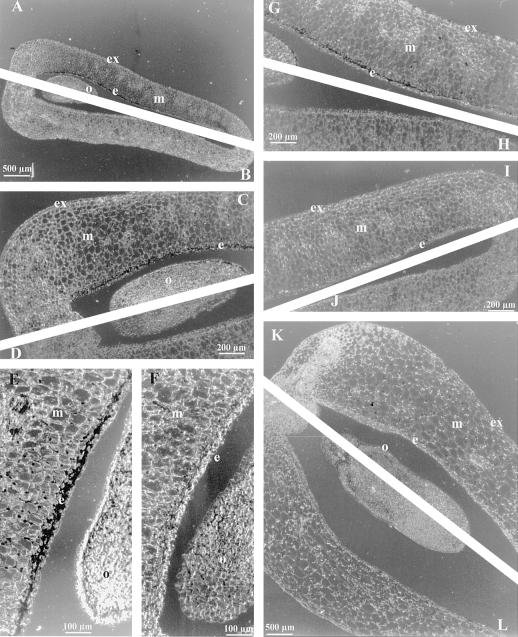
The distribution of the TPE4A transcripts in senescent pea ovaries was studied by in situ hybridization (Fig. 5). Using the antisense TPE4A RNA as the probe, we detected the presence of the TPE4A transcripts in the endocarp and, at lower amounts, in the outer cell layers of the ovule (Fig. 5, A, C, E, and G). The TPE4A transcript distribution was not uniform along the ovary endocarp. The concentration was highest in the endocarp area next to the ovule (Fig. 5, C and E) and almost undetectable at the opposite end of the transverse section (Fig. 5I), and an intermediate concentration was detected between these two areas (Fig. 5G). No hybridization occurred with the sense TPE4A RNA as a control probe (Fig. 5, B, D, F, and H). Some labeling was also detected associated with the cell walls of the mesocarp cells (Fig. 5E), but this could also be detected with the control probe (Fig. 5F), suggesting that it was not specific. This nonspecific labeling was also detected in the mesocarp cells of developing fruits on d 4 after anthesis (Fig. 5, K and L). No labeling was detected in the endocarp or ovules in developing fruits (Fig. 5, K and L). Northern-blot analysis indicated a complete suppression of TPE4A transcripts in developing fruits (Fig. 4); therefore, the detection of some labeling associated with the mesocarp cell walls in histological sections of developing fruits can be considered to be nonspecific.
Figure 5.
Detection of tissue-specific TPE4A expression by in situ hybridization. A through J, Cross-sections of senescent pea ovaries (d 4 after anthesis) hybridized with the TPE4A antisense riboprobe (A, C, E, G, and I) and the sense control probe (B, D, F, H, and J). C and D, Ovule and surrounding area of the sections shown in A and B at higher magnification; G, H, I, and J, other parts of the same sections. E and F, Detail of the ovule and surrounding endocarp area of different sections. K and L, Cross-section of the ovule and surrounding endocarp area of developing pea fruits (GA3-treated ovaries on d 4 after anthesis) hybridized with the TPE4A antisense riboprobe (K) and the sense control probe (L). Hybridization is indicated by the dark staining in the endocarp (e) area surrounding the ovule (o) and, at lower levels, in the outer cell layers of the ovule in senescent ovaries. m, Mesocarp; ex, exocarp.
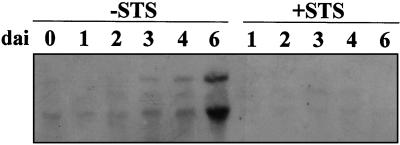
The TPE4A mRNA was also detected by northern-blot analysis in germinating pea seeds. When seeds were allowed to imbibe in water, the TPE4A band and another band of approximately 2.8 kb were induced after 3 d (Fig. 6); when seeds were allowed to imbibe in the presence of STS, these mRNA bands were not detected. Because Southern-blot analysis detected only one copy of the TPE4A gene in the pea genome (Fig. 3), and additional experiments conducted at lower-stringency conditions did not show any additional bands (data not shown), the 2.8-kb band could be either a cross-reacting mRNA expressed only in seeds or an artifact from the tissue- and RNA-extraction protocol.
Figure 6.
TPE4A expression is also induced during seed germination. Northern-blot analysis showing the accumulation of the TPE4A transcript (lower band) and another cross-reacting transcript (about 2.8 kb) during the imbibition of pea seeds in water (−STS) and STS (+STS). dai, Days after imbibition.
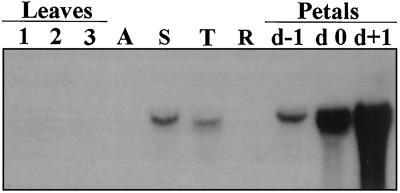
Northern-blot analysis of total RNA from different tissues of the pea plant showed the presence of the TPE4A transcript in the stem and the tendrils but not in the roots and the shoot apex. Expression of TPE4A was detected in petals, increasing in abundance during senescence (petals become senescent after anthesis). No signal was detected in young, mature, or senescing pea leaves. (Fig. 7).
Figure 7.
Expression of the TPE4A gene in different tissues of the pea plant. Northern-blot analysis showing the accumulation of the TPE4A transcript in leaves. Lane 1, Young leaves; lane 2, mature leaves; lane 3, senescent leaves; lane A, apex; lane S, stem; lane T, tendrils; lane R, roots. Petal lanes are as follows: d−1, 1 d before anthesis; d 0, day of anthesis; and d+1, 1 d after anthesis, when the petals were senescent.
DISCUSSION
We have cloned a thiol-protease cDNA (TPE4A) from senescing pea ovaries by reverse transcriptase-PCR using two degenerate primers derived from consensus sequences found in all plant thiol-proteases of the so-called papain family. With the two degenerate primers we used, only two protease genes were amplified: TPE4A and tpp, a previously reported thiol-protease gene induced in senescent pea ovaries (Granell et al., 1992). Because these two primers correspond to a subset of the possible primers encoding the target-protein sequences, the use of more primer combinations would probably allow the cloning of more senescence-related thiol-protease cDNAs.
TPE4A can be classified as a papain family member according to sequence similarity and the conserved positions of the amino acids forming the active site (Figs. 1 and 2). According to Karrer et al. (1993), TPE4A is a member of the ERFNIN-containing thiol-proteases (cathepsin L- and cathepsin H-like thiol-proteases), in that an ERFNIN motif can be found in its prosequence. All of the proteases of this family share several common structural properties that make them different from the cathepsin B-like thiol-proteases (Karrer et al., 1993). We have used all of these features to confirm the protein alignments in Figure 2 and the conserved features highlighted in Figure 1. The first 20 amino acids of the TPE4A deduced amino acid sequence can be considered a signal peptide according to the rules of Von Heijne (1983), suggesting that TPE4A enters the secretory pathway cotranslationally.
The deduced amino acid sequence of TPE4A contains a putative ER-retention signal in the C terminus (KDEL; Bednarek and Raikhel, 1992). Several plant thiol-proteases contain a similar signal at the C terminus (Fig. 2). The presence of a KDEL tetrapeptide at the C terminus of a fusion protein is in some cases sufficient for the retention of the protein in the ER (Bednarek and Raikhel, 1992). As proposed by Valpuesta et al. (1995) and Guerrero et al. (1998), a protease in the ER would be involved in homeostatic controls rather than protein mobilization, so further translocation to a different cell compartment is expected for a protease involved in the mobilization of storage proteins.
Studies carried out with VSCYSPROA, the vetch homolog of TPE4A (Becker et al., 1997), showed that the purified mature protein lacks the KDEL, indicating that it is removed after translation. It was proposed that VSCYSPROA is retained in the ER until it is needed for the germination process; the KDEL motif would then be removed and the protease translocated to the protein bodies. This would explain the time gap found between the transcription of the protease gene and proteolytic activity (Becker et al., 1997). Something similar could happen to TPE4A during the germination of pea seeds. The increase in the TPE4A mRNA amount 6 d after imbibition (Fig. 6) was not correlated with an increase in proteolytic activity in seed extracts (data not shown), suggesting that there is also a time gap between transcription of the TPE4A gene and the proteolytic activity in pea seeds. However, during ovary senescence no time gap has been found between transcription and proteolytic activities, suggesting that the ER-retention step would be unnecessary in the senescent ovaries. Further work concerning the changes in the subcellular localization of the TPE4A protein in senescent pea ovaries will be needed to clarify this point.
TPE4A is similar to several plant thiol-proteases (Fig. 2), with the closest similarity found with VSCYSPROA (Becker et al., 1997). The similarity between TPE4A and VSCYSPROA deduced amino acid sequences was 78% in the signal peptide, 91% in the prosequence, and 92% in the mature peptide. This close similarity is conserved even in sequences not shared with other similar thiol-proteases (Fig. 2). These data suggest that TPE4A and VSCYSPROA could be homolog genes. If this is true, they should have the same physiological functions, because pea and vetch belong to the same plant family. Furthermore, both proteases are expressed in the same tissues of the corresponding plants (compare Fig. 6 and Becker et al., 1997).
Because both senescence and germination processes are characterized by nutrient remobilization, thiol-proteases play a key role in both processes (Granell et al., 1998); however, there is no report demonstrating that a single protease gene can be induced in both processes. Griffiths et al. (1997) isolated a vacuolar thiol-protease (See1) from senescent maize leaves and found that it is very similar (99.3% in the cDNA sequence) to CCP2, a previously known thiol-protease associated with the germination of maize seeds. It was difficult to conclude whether See1 and CCP2 corresponded to different genes or to alleles of the same gene. These investigators also found two copies of the gene by Southern-blot and restriction fragment length polymorphism analyses (Griffiths et al., 1997), and they observed induction during both senescence and germination using northern-blot analysis with the See1 probe. However, because of this similarity, both mRNAs could be detected simultaneously in the northern-blot experiments, so it was not possible to conclude whether the two genes are expressed in both senescence and germination or if each gene is specific to one of the two processes. Because TPE4A is a single-copy gene (Fig. 3) and its expression was detected by northern-blot analysis in both ovary senescence (Figs. 4 and 5) and seed germination (Fig. 6), we conclude that this gene is expressed in both processes.
TPE4A accumulation in germinating seeds was suppressed when the seeds were allowed to imbibe in the presence of 50 μm STS, a known inhibitor of ethylene action (Davies et al., 1990). This suggests the possibility that, at least during seed germination, TPE4A transcription could be induced by ethylene. However, in the presence of STS, seed germination was slower during the first days of imbibition and seedling growth apparently stopped at approximately d 6. Therefore, it is not possible with our data to determine whether the observed repression of TPE4A expression was caused by the STS inhibition of ethylene action or by altered germination. In germinating cereal seeds, GA3 induces the transcription of thiol-proteases (Mikkonen et al., 1996). In legume seeds, there is a different control of protease expression; Cervantes et al. (1994) reported the first evidence of an ethylene-induced thiol-protease gene associated with the germination of chickpea seeds. We also found a possible induction of the TPE4A gene by ethylene; in addition, tpp, another thiol-protease gene induced in senescent pea ovaries, was also expressed during seed germination in pea (data not shown) and was repressed when seeds were allowed to imbibe in STS. It could be a general rule that in cereals germination-related thiol-protease genes are induced by GA3, whereas during the germination of legume seeds, they are induced by ethylene.
The data in Figure 4 indicate that TPE4A transcription is completely suppressed after treatment of the ovaries with GA3. Even the low background expression detected in presenescent ovaries is suppressed by GA3. These data suggest that TPE4A transcription is not controlled by GA3 in the same way as tpp transcription (greatly decreased but not completely suppressed by GA3; Granell et al., 1992). In addition, the TPE4A mRNA level increased from d 3 to 4, whereas tpp had a maximum level on d 3 and then decreased. Comparing the temporal patterns of gene expression and the evolution of proteases (Cercós and Carbonell, 1993; Cercós et al., 1993) with proteolytic activity (Cercós et al., 1992), we conclude that TPE4A evolution is more closely related to proteolytic activity changes than is tpp evolution.
The spatial distribution of TPE4A expression in senescent pea ovaries is also different from that of tpp (Granell et al., 1992) and a senescence-induced protease purified from senescent pea ovaries (Cercós et al., 1993). By combining the results found in the histological localization of the three known proteases induced in senescent pea ovaries, we hypothesize that the action of different proteases is needed for the proper mobilization of proteins from the senescent ovaries to the growing parts of the pea plant.
The accession number for the sequence reported in this article is AJ004958.
ACKNOWLEDGMENTS
We thank Dr. Antonio Granell for helpful suggestions. We also thank Dr. E. Grau for help with automatic DNA sequencing, Dr. M.D. Gómez for help with the in situ hybridization experiments, and Donnellan-Barraclough for help with the English language.
Abbreviation:
- STS
silver thiosulfate
Footnotes
This work was supported by grant no. GV-3208/95 from Generalitat Valenciana and by grant no. PB95-0029-C02-01 from Dirección General de Investigación Científica y Técnica, Spain.
LITERATURE CITED
- Akasofu H, Yamauchi D, Minamikawa T. Nucleotide sequence of the gene for the Vigna mungo sulfhydryl-endopeptidase (SH-EP) Nucleic Acids Res. 1990;18:1892. doi: 10.1093/nar/18.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Senyuk VI, Shutov AD, Nong VH, Fischer J, Horstmann C, Müntz K. Proteinase A, a storage-globulin-degrading endopeptidase of vetch (Vicia sativa L.) seeds, is not involved in early steps of storage-protein mobilization. Eur J Biochem. 1997;24:304–312. doi: 10.1111/j.1432-1033.1997.00304.x. [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Raikhel NV. Intracellular trafficking of secretory proteins. Plant Mol Biol. 1992;20:133–150. doi: 10.1007/BF00029156. [DOI] [PubMed] [Google Scholar]
- Bugos RC, Chiang VL, Zhang XH, Campbell ER, Podila GK, Campbell WH. RNA isolation from plant tissues recalcitrant to extraction in guanidine. Biotechniques. 1995;19:634–637. [PubMed] [Google Scholar]
- Carbonell J, García-Martínez JL. Fruit set of unpollinated ovaries of Pisum sativum L.: influence of vegetative parts. Planta. 1980;147:444–450. doi: 10.1007/BF00380186. [DOI] [PubMed] [Google Scholar]
- Carbonell J, García-Martínez JL. Ribulose 1,5-bisphosphate carboxylase and fruit set or degeneration of unpollinated ovaries of Pisum sativum L. Planta. 1985;164:534–539. doi: 10.1007/BF00395972. [DOI] [PubMed] [Google Scholar]
- Carrasco P, Carbonell J. Involvement of a neutral proteolytic activity in the senescence of unpollinated ovaries of Pisum sativum L. Physiol Plant. 1988;72:610–616. [Google Scholar]
- Cercós M, Carbonell J. Purification and characterization of a thiol-protease induced during senescence of unpollinated ovaries of Pisum sativum. Physiol Plant. 1993;88:267–274. [Google Scholar]
- Cercós M, Carrasco P, Granell A, Carbonell J. Biosynthesis and degradation of Rubisco during ovary senescence and fruit development induced by gibberellic acid in Pisum sativum. Physiol Plant. 1992;85:476–482. [Google Scholar]
- Cercós M, Harris N, Carbonell J. Immunolocalization of a thiol-protease induced during the senescence of unpollinated pea ovaries. Physiol Plant. 1993;88:275–280. [Google Scholar]
- Cervantes E, Rodríguez A, Nicolás G. Ethylene regulates the expression of a cysteine proteinase gene during germination of chickpea (Cicer arietinum L.) Plant Mol Biol. 1994;25:207–215. doi: 10.1007/BF00023238. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Hobson GE, Grierson D. Differential effect of silver ions on the accumulation of ripening related mRNAs in tomato fruit. J Plant Physiol. 1990;135:708–713. [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–22. [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake R, John I, Farrell A, Cooper W, Schuch W, Grierson D. Isolation and analysis of cDNAs encoding tomato cysteine proteases expressed during leaf senescence. Plant Mol Biol. 1996;30:755–767. doi: 10.1007/BF00019009. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Making sense of senescence. Molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez JL, Carbonell J. Fruit set of unpollinated ovaries of Pisum sativum L.: influence of plant growth regulators. Planta. 1980;147:451–456. doi: 10.1007/BF00380187. [DOI] [PubMed] [Google Scholar]
- Granell A, Cercós M, Carbonell J. Plant cysteine proteinases in germination and senescence. In: Barret AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. London: Academic Press; 1998. pp. 578–583. [Google Scholar]
- Granell A, Harris N, Pisabarro AG, Carbonell J. Temporal and spatial expression of a thiol protease gene during pea ovary senescence and its regulation by gibberellin. Plant J. 1992;2:907–915. doi: 10.1046/j.1365-313x.1992.t01-5-00999.x. [DOI] [PubMed] [Google Scholar]
- Griffiths CM, Hosken SE, Oliver D, Chojecki J, Thomas H. Sequencing, expression pattern and RFLP mapping of a senescence-enhanced cDNA from Zea mays with high homology to oryzain γ and aleurain. Plant Mol Biol. 1997;34:815–821. doi: 10.1023/a:1005896713830. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Calle M, Reid MS, Valpuesta V. Analysis of the expression of two thiolprotease genes from daylily (Hemerocallis spp.) during flower senescence. Plant Mol Biol. 1998;36:565–571. doi: 10.1023/a:1005952005739. [DOI] [PubMed] [Google Scholar]
- Jackson DP. In situ hybridization in plants. In: Bowles DJ, Gurr SJ, Pherenson M, editors. Molecular Plant Pathology: A Practical Approach. Oxford, UK: Oxford University Press; 1992. pp. 163–174. [Google Scholar]
- Jones ML, Larsen PB, Woodson WR. Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Mol Biol. 1995;28:505–512. doi: 10.1007/BF00020397. [DOI] [PubMed] [Google Scholar]
- Kamphuis IG, Drenth J, Baker EN. Thiol proteases: comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence information for cathepsins B and H, and stem bromelain. J Mol Biol. 1985;182:317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- Karrer KM, Peiffer SL, DiTomas ME. Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci USA. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Liu ZW, Tan-Wilson AL, Wilson KA. Transcript levels for a mung bean cysteine protease during early seedling growth. Seed Sci Res. 1997;7:359–372. [Google Scholar]
- Mikkonen A, Porali I, Cercós M, Ho TH-D. A major cysteine proteinase, EPB, in germinating barley seeds: structure of two intronless genes and regulation of expression. Plant Mol Biol. 1996;31:239–254. doi: 10.1007/BF00021787. [DOI] [PubMed] [Google Scholar]
- Noodén LD, Guiamét JJ, John I. Senescence mechanisms. Physiol Plant. 1997;101:746–753. [Google Scholar]
- Ogushi T, Tanaka T, Yamauchi D, Minamikawa T. Nucleotide sequence of a gene for an endopeptidase (EP-C1) from Phaseolus vulgaris. Plant Mol Biol. 1992;19:705–706. doi: 10.1007/BF00026797. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smart C, Hosken S, Thomas H, Greaves JA, Blais BG, Schuch W. The timing of maize leaf senescence and characterisation of senescence-related cDNAs. Physiol Plant. 1995;93:673–682. [Google Scholar]
- Valpuesta V, Lange NE, Guerrero C, Reid MS. Up-regulation of a cysteine protease accompanies the ethylene-insensitive senescence of daylily (Hemerocallis) flowers. Plant Mol Biol. 1995;28:575–582. doi: 10.1007/BF00020403. [DOI] [PubMed] [Google Scholar]
- Von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]