Abstract
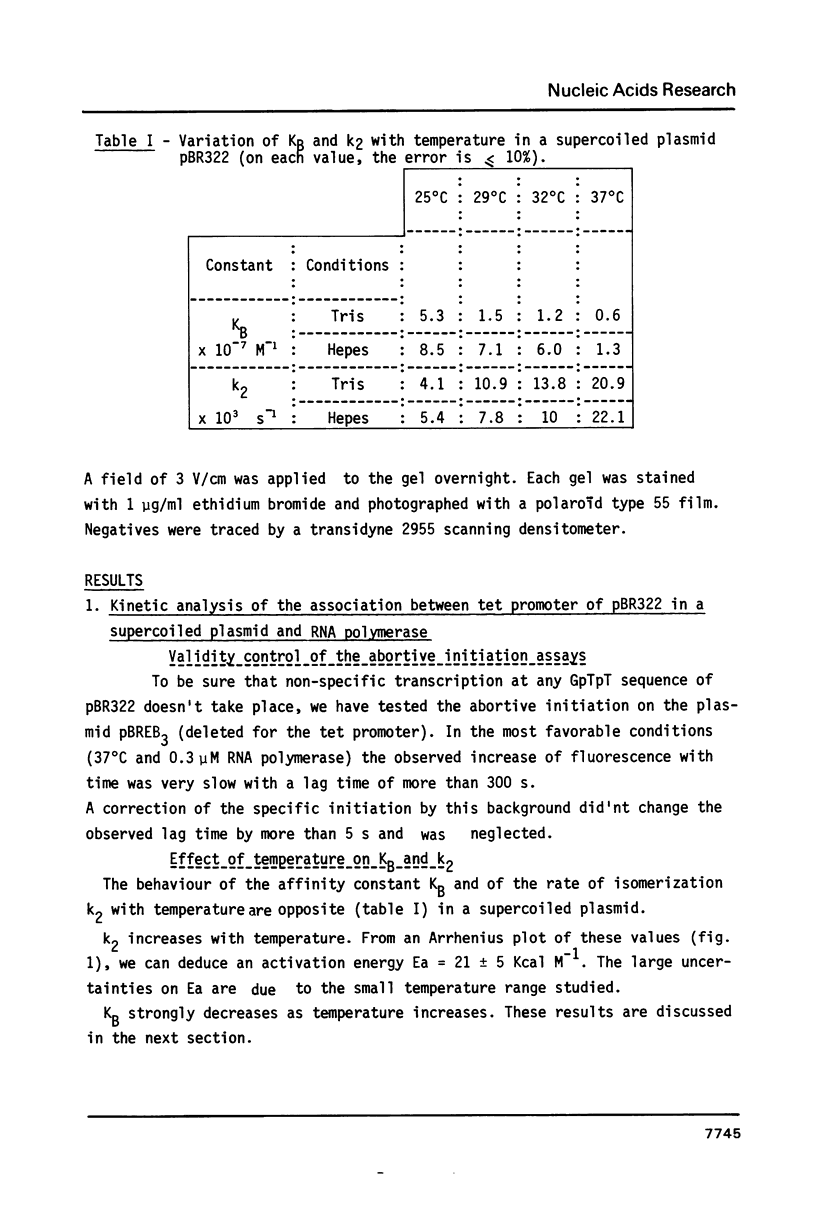
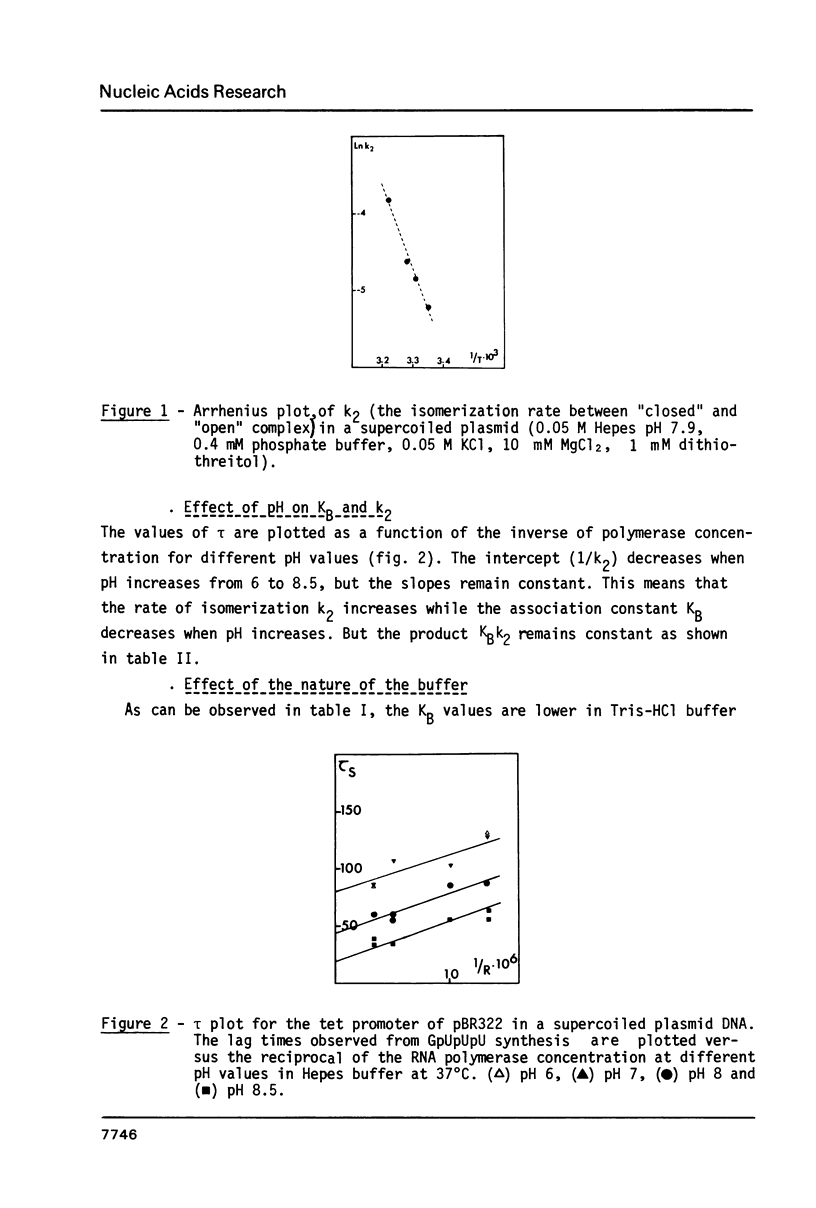
Supercoiling of DNA is now known to have considerable effects on transcription in bacteria. By abortive initiation reaction (6) we have determined the binding constant KB and the forward rate of isomerization k2 as a function of temperature, pH and buffer for the tet promoter in a supercoiled plasmid. If the activation energy of isomerization is very similar to that obtained previously under the same conditions on a linearized plasmid (6) (respectively 21 +/- 5 kcal/mole and 13 +/- 5 kcal/mole) the supercoiling introduces very important and not well understood changes in the thermodynamic parameters of the association polymerase - promoter. Using the technique of superhelical DNA relaxation by eukaryotic topoisomerase I, we have determined the specific unwinding by RNA polymerase of the tet promoter of pBR322 (430 degrees). This unwinding differs only slightly from the mean value (470 degrees) obtained for all the promoters of pBR322.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertrand-Burggraf E., Lefèvre J. F., Daune M. A new experimental approach for studying the association between RNA polymerase and the tet promoter of pBR322. Nucleic Acids Res. 1984 Feb 10;12(3):1697–1706. doi: 10.1093/nar/12.3.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan P., Wang J. C., Echols H. Effect of circularity and superhelicity on transcription from bacteriophagelambda DNA. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3077–3081. doi: 10.1073/pnas.70.11.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982 May;29(1):81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Gellert M., Menzel R., Mizuuchi K., O'Dea M. H., Friedman D. I. Regulation of DNA supercoiling in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):763–767. doi: 10.1101/sqb.1983.047.01.087. [DOI] [PubMed] [Google Scholar]
- Helene C., Maurizot J. C. Interactions of oligopeptides with nucleic acids. CRC Crit Rev Biochem. 1981;10(3):213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Wang J. C. Physiochemical studies on interactions between DNA and RNA polymerase. Ultraviolet absorption measurements. Nucleic Acids Res. 1978 Sep;5(9):3337–3345. doi: 10.1093/nar/5.9.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hélène C., Lancelot G. Interactions between functional groups in protein-nucleic acid associations. Prog Biophys Mol Biol. 1982;39(1):1–68. doi: 10.1016/0079-6107(83)90013-5. [DOI] [PubMed] [Google Scholar]
- Kolb A., Buc H. Is DNA unwound by the cyclic AMP receptor protein? Nucleic Acids Res. 1982 Jan 22;10(2):473–485. doi: 10.1093/nar/10.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Kano Y., Nakamura H., Nagata A., Imamoto F. In vivo enhancement of general and specific transcription in Escherichia coli by DNA gyrase activity. Gene. 1979 Oct;7(2):153–171. doi: 10.1016/0378-1119(79)90030-1. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., deHaseth P. L., Record M. T., Jr Pentalysine-deoxyribonucleic acid interactions: a model for the general effects of ion concentrations on the interactions of proteins with nucleic acids. Biochemistry. 1980 Jul 22;19(15):3522–3530. doi: 10.1021/bi00556a017. [DOI] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Stahl S. J., Chamberlin M. J. An expanded transcriptional map of T7 bacteriophage. Reading of minor T7 promoter sites in vitro by Escherichia coli RNA polymerase. J Mol Biol. 1977 Jun 5;112(4):577–601. doi: 10.1016/s0022-2836(77)80165-4. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Goodman H. M., Heyneker H. L., Shine J., Boyer H. W., Cozzarelli N. R. Interaction of bacteriophage T4 RNA and DNA ligases in joining of duplex DNA at base-paired ends. J Biol Chem. 1977 Jun 10;252(11):3987–3994. [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Jacobsen J. H., Saucier J. M. Physiochemical studies on interactions between DNA and RNA polymerase. Unwinding of the DNA helix by Escherichia coli RNA polymerase. Nucleic Acids Res. 1977;4(5):1225–1241. doi: 10.1093/nar/4.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough L. R., Schlageck J. G., Baughman M. Synthesis and properties of fluorescent nucleotide substrates for DNA-dependent RNA polymerases. J Biol Chem. 1979 Dec 10;254(23):12069–12073. [PubMed] [Google Scholar]