Abstract
West Nile virus (WNV) genome cyclization is mediated by two pairs of long-distance RNA/RNA interactions: the 5′CS/3′CSI (conserved sequence) and the 5′UAR/3′UAR (upstream AUG region) base pairings. Antisense peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs), designed to interfere with the 5′CS/3′CSI or 5′UAR/3′UAR base pairings, were previously shown to inhibit WNV. In this study, we selected and characterized WNVs resistant to a PPMO targeting the 3′UAR (3′UAR-PPMO). All resistant viruses accumulated one-nucleotide mutations within the 3′UAR, leading to a single-nucleotide mismatch or a weakened base-pairing interaction with the 3′UAR-PPMO. Remarkably, a one-nucleotide mutation within the 5′UAR was correspondingly co-selected; the 5′UAR mutation restored the base-pairing with the 3′UAR mutation. Mutagenesis of WNV demonstrated that the single-nucleotide change within the 3′UAR-PPMO-target site conferred the resistance. RNA binding analysis indicated that the single-nucleotide change reduced the ability of 3′UAR-PPMO to block the RNA/RNA interaction required for genome cyclization. The results suggest a mechanism by which WNV develops resistance to 3′UAR-PPMO, through co-selection of the 5′UAR and 3′UAR, to create a mismatch or a weakened base-pairing interaction with the PPMO, while maintaining the 5′UAR/3′UAR base pairings.
Keywords: West Nile virus, Flavivirus replication, Antiviral therapy, genome cyclization, RNA cis elements
INTRODUCTION
Many flaviviruses are emerging or reemerging pathogens, including the four serotypes of dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). DENV alone infects 50 to 100 million humans each year (Gubler, Kuno, and Markoff, 2007). Since the initial outbreak of WNV in New York in 1999, the virus has caused thousands of human cases each year, and has spread throughout North America (Kramer, Li, and Shi, 2007). There is no approved antiviral therapy for treatment of flavivirus infections. Human vaccines for flavivirus infections are currently available only for YFV, JEV, and TBEV. Vaccines for WNV are approved only for equine use (Davis et al., 2001; Minke et al., 2004; Ng et al., 2003). Development of a vaccine for DENV has been challenging, primarily because of the need to simultaneously immunize against all four DENV serotypes. The large disease burden caused by flaviviruses makes the development of effective therapies of acute importance to global public health.
The flavivirus genome is a single-strand, plus-sense RNA of approximately 11,000 nucleotides (nt). It consists of a 5′ untranslated region (UTR), a long open reading frame (ORF), and a 3′ UTR (Lindenbach, Thiel, and Rice, 2007). The single ORF encodes a polyprotein, which is processed by viral and cellular proteases into 10 mature proteins. Three structural proteins (capsid, premembrane or membrane, and envelope) are required for viral particle formation. Seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) are involved in viral RNA replication (Lindenbach, Thiel, and Rice, 2007); additionally, the nonstructural proteins play roles in virion assembly (Kummerer and Rice, 2002; Leung et al., 2008; Liu, Chen, and Khromykh, 2003; Patkar and Kuhn, 2008) and evasion of host innate immune responses (Best et al., 2005; Guo, Hayashi, and Seeger, 2005; Liu et al., 2005; Munoz-Jordan et al., 2005; Munoz-Jordan et al., 2003).
Flavivirus genome cyclization is essential for viral replication. As shown in Figure 1A using WNV genome as an example, two pairs of long-distance RNA/RNA interactions contribute to the genome cyclization for mosquito-borne flaviviruses. One RNA interaction involves base pairings between a 5′ conserved sequence (5′CS; located in the N-terminal coding region of the C gene) and a 3′CSI (located in the 3′ UTR) (Hahn et al., 1987). The 5′CS/3′CSI interaction is important for efficient RNA-dependent RNA polymerase (RdRp) activity (Filomatori et al., 2006; You and Padmanabhan, 1999), and for viral replication (Bredenbeek et al., 2003; Corver et al., 2003; Khromykh et al., 2001; Kofler et al., 2006; Lo et al., 2003; Men et al., 1996). A second long-range RNA/RNA interaction involves base pairings between a 5′UAR (upstream initiation AUG region) element and a 3′UAR (located in the 3′ terminal stem-loop of the genomic RNA). The 5′UAR/3′UAR interaction plays a critical role in DENV-2 (Alvarez, Filomatori, and Gamarnik, 2008; Alvarez et al., 2005b) and WNV (Zhang et al., 2008) replication.
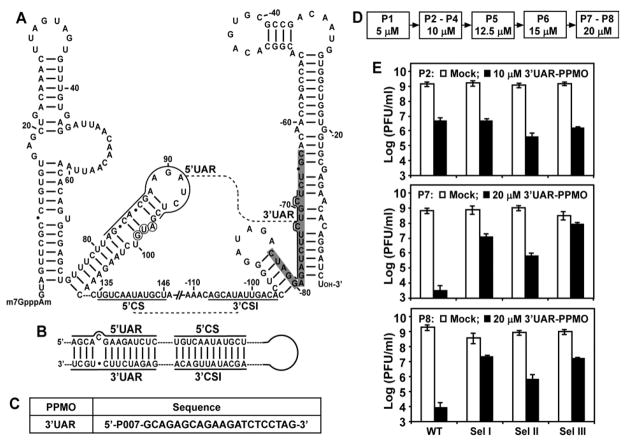
FIG. 1.
Selection of 3′UAR-PPMO resistant viruses. (A) Terminal secondary structures and potential long-distance RNA interactions of WNV genome. Stem-loop structures were calculated by the Mfold program (Mathews et al., 1999), and were verified by RNA structure probing (Dong, Zhang, and Shi, 2008). The two long-distance RNA interactions, 5′CS/3′CSI and 5′UAR/3′UAR, are indicated by dashed lines. The sequences for the 5′UAR, 5′CS, 3′CSI, and 3′UAR are labeled. The viral sequence is taken from GenBank accession no. AF404756. The AUG initiation codon of the open reading frame is circled. The nucleotides are numbered for both ends; “-” denotes numbering from the 3′ end of the genome. The nucleotides that are targeted by the 3′UAR-PPMO are shaded in grey. (B) The 5′UAR/3′UAR and 5′CS/3′CSI base-pairing interactions that contribute to the cyclization of WNV genomic RNA. (C) Antisense PPMO targeting 3′UAR of WNV RNA (3′UAR-PPMO). P007 represents an Arg-rich peptide that is conjugated to the 5′ end of the PMO to facilitate cellular uptake. The composition of P007 and structure of the PPMO backbone were described previously (Deas et al., 2005). (D) Selection scheme for 3′UAR-PPMO-resistant virus. Drug concentrations for each selection passage (P1 to P8) are indicated. (E) Resistance phenotype of viruses during selection. Vero cells were infected with the indicated viruses at an MOI of 0.1. After incubation of the infected cells with or without 3′UAR-PPMO for 42 h, viral titers in culture fluids were determined using plaque assays. Three independent selections were performed (Sel I, II, and III). Mean results from P2, P7, and P8 are presented for each selection. Error bars represent standard deviations from three experiments.
Phosphorodiamidate morpholino oligomers (PMOs) are uncharged, water-soluble, nuclease-resistant antisense agents that contain purine and pyrimidine bases attached to a backbone composed of morpholine rings joined by phosphorodiamidate intersubunit linkages (Summerton and Weller, 1997). An Arg-rich peptide was conjugated to the 5′ terminus of the PMO, creating peptide-conjugated PMO (PPMO), to facilitate cellular uptake of the oligomers (Deas et al., 2005). Using WNV, we previously showed that antisense PPMOs, with sequences designed to block the base pairings of 5′CSI/3′CSI or the 5′UAR/3′UAR, are potent inhibitors of WNV replication in cell culture (Deas et al., 2005; Zhang et al., 2008). Furthermore, a PPMO targeting the 3′CSI region was able to partially protect WNV-infected mice from viral disease (Deas et al., 2007). Similar antiviral activities of PPMOs targeting the 5′CSI/3′CSI interaction were reported for DENV in cell culture and mouse (Holden et al., 2006; Kinney et al., 2005; Stein et al., 2008). These results suggest that antisense PPMOs could be developed for antiviral therapy.
Here we select and characterize WNV that are resistant to a PPMO targeting the 3′UAR. The results show that WNV develops resistance to the 3′UAR-PPMO through accumulation of a single-nucleotide change within the 3′UAR sequence, resulting in a nucleotide mismatch or a weakened base-pairing interaction with the 3′UAR-PPMO. Interestingly, a single-nucleotide substitution within the 5′UAR was co-selected in the resistant viruses, leading to the restoration of perfect base pairing between the mutated 5′UAR and the mutated 3′UAR. Biochemical analysis showed that the single-nucleotide mismatch reduced the ability of the PPMO to interfere with the 5′UAR/3′UAR base pairing.
RESULTS
Selection of 3′UAR-PPMO-resistant WNV
Figure 1A shows the 5′ and 3′ stem-loop structures formed by the WNV genomic RNA, and the potential base pairings of 5′CS/3′CSI and 5′UAR/3′UAR, which cyclize the genomic RNA (Fig. 1B). We previously synthesized two PPMOs, to study the function of the 5′UAR/3′UAR interaction: one PPMO targeted the 5′UAR, and another PPMO targeted the 3′UAR. Viral titer reduction assay showed that the 3′UAR-PPMO was more potent than the 5′UAR-PPMO (Zhang et al., 2008). Therefore, we chose the 3′UAR-PPMO (Fig. 1C) for use in a resistance study. Resistant viruses were selected by culturing WNV in the presence of increasing concentrations of 3′UAR-PPMO (Fig. 1D). The highest concentration of selection was set at 20 μM, because higher concentrations of PPMO were previously shown to be cytotoxic (Zhang et al., 2008).
Three independent selections (Sel I, II, and III) were performed to examine the reproducibility of the resistance results (Fig. 1E). For each drug concentration, both the WT and selected viruses were tested for their sensitivities to PPMO inhibition. For example, at passage 2 (P2), the WT and three selected viruses were almost equally inhibited by 10-μM 3′UAR-PPMO (Fig. 1E, top panel). In contrast, at P7 (20-μM 3′UAR-PPMO), the titer of the WT virus was suppressed by >105-fold, whereas titers of the selected viruses were reduced by <104-fold (Middle panel). Further selection at 20-μM 3′UAR-PPMO (P8) did not enhance viral resistance (Bottom panel). Therefore, we terminated the selection at P8. Among the three selections, viruses from Sel I and III were more resistant than the virus from Sel II (Fig. 1E). The results suggest that WNV resistant to 3′UAR-PPMO can be developed in cell culture, although the selected viruses did not exhibit complete resistance to the inhibitor. In the absence of PPMO, the WT and three selected resistant viruses grew to similar titers during each passage (Fig. 1E). Plaques from the WT, Sel I, and Sel III viruses (P8) were similar, but were larger than those from the Sel II (P8) virus (Fig. 2A). These results indicate that the resistant viruses are not substantially compromised in their replication fitness.
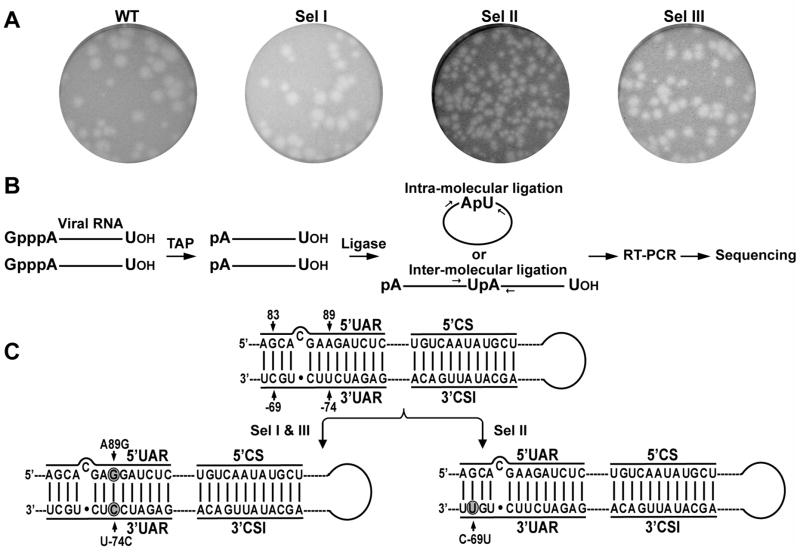
FIG. 2.
Plaque morphology and sequence analysis of the 3′UAR-PPMO-resistant WNVs. (A) Plaque morphology. Plaques are presented for WT and P8 viruses for Sel I, Sel II, and Sel III. (B) Scheme for RACE method. RNAs extracted from virions were treated with tobacco acid pyrophosphatase (TAP) to remove 5′ α- and β-phosphates, ligated intra- or inter-molecularly using T4 RNA ligase, and RT-PCR-amplified using primers annealing to the 5′ and 3′ terminal regions of the genomic RNA. Sequencing of the RT-PCR products allowed us to identify mutations in both ends of the viral RNA. (C) Mutations recovered from resistant viruses. Mutated nucleotides are shaded in circles. Two identical nucleotide changes (A89G in 5′UAR and U-74C in 3′UAR; symbol “-” denotes nucleotide numbering from the 3′ end of viral genome) were recovered for Sel I and Sel III resistance viruses. One nucleotide change (C-69U in 3′UAR) was recovered for Sel II resistant virus.
Mutations in 3′UAR-PPMO-resistant viruses
Using the RACE method depicted in Figure 2B, we sequenced the 5′ 684 nt and the 3′ 528 nt of the genomic RNA, derived from the P8 resistant viruses. Extracted virion RNAs were treated with tobacco acid pyrophosphatase (TAP) to remove 5′ α- and β-phosphates, ligated intra- or inter-molecularly using T4 RNA ligase, and RT-PCR-amplified using primers annealing to the 5′ and 3′ terminal regions of the genomic RNA. Sequencing of the RT-PCR products allowed identification of mutations accumulated in both ends of the viral RNA. For Sel I and Sel III viruses, identical nucleotide changes were recovered in both the 5′UAR and the 3′UAR. A nucleotide U at position -74 (“-” denotes nucleotide numbering from the 3′ end of viral genome), located in the 3′UAR, was mutated to C (U-74C); the U-74C mutation resulted in a single-nucleotide mismatch with the PPMO. Additionally, a nucleotide A at position 89, located in the 5′UAR, was changed to G (A89G); remarkably, the A89G in 5′UAR restored the base pairing with the U-74C in the 3′UAR (Fig. 2C). These results suggest that WNV develop escape mutants through creation of a single-nucleotide mismatch with the antisense PPMO, while maintaining a perfect base pairing of the 5′UAR/3′UAR interaction.
For Sel II virus, a nucleotide C at position -69, in the 3′UAR, was mutated to U (C-69U). The mutated C-69U could still base-pair with nucleotide G at position 83 in the 5′UAR, as well as to pair with the corresponding G residue within the PPMO. However, the bases, involved in the G/U base pairing between the PPMO and the 3′UAR, are derived from a morpholine chain and an RNA chain, respectively; whereas the bases, involved in the G/U base pairing between the 5′UAR and the 3′UAR, are both derived from RNA chains. It is likely that the G/U base pairing from the PPMO/RNA duplex is weaker than the G/U base pairing from the RNA/RNA duplex, leading to the decreased efficacy of the PPMO against the Sel II virus. However, experiments are needed to verify this possibility.
Chronological order of mutation emergence
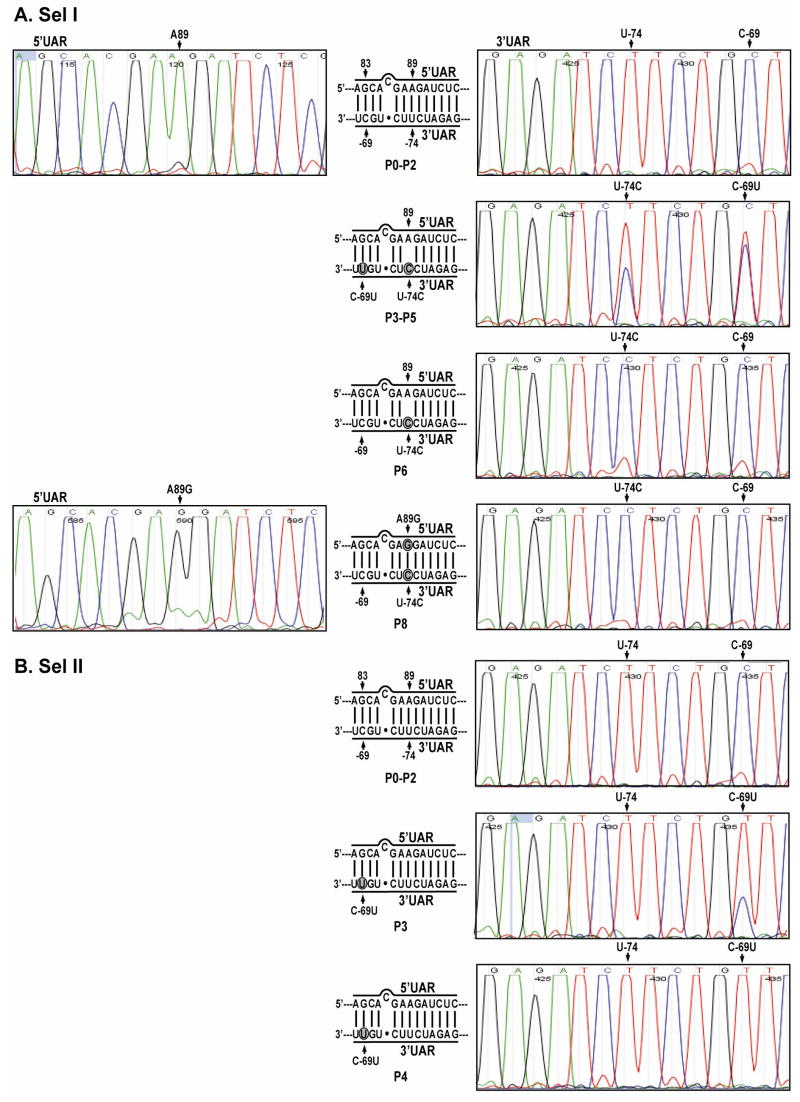
To examine the chronological order of mutation accumulation, we sequenced both ends of the viral RNAs, extracted from P0 to P6 viruses of Sel I and Sel II. Because the P8 viruses from Sel I and III exhibited identical mutations (Fig. 2C), Sel III viruses were not included in the analysis. For Sel I (Fig. 3A), C-69U and U-74C simultaneously appeared as mixed populations from P3 to P5. Since the sequences were derived by consensus sequencing, designed to define the most common sequence present at a specific location within a population, we are not able to conclude whether the C-69U and U-74C mutations were present in one or two different viral populations. Nevertheless, from P6 onward, the U-74C mutation dominated, whereas the C-69U mutation reverted to the WT sequence. Furthermore, at P8, the A89G mutation appeared in the 5′UAR.
FIG. 3.
Chronology of mutation emergence during development of resistance. Representative sequencing chromatograms are presented for P0 to P8 viruses for Sel I (A), and for P0 to P4 viruses for Sel II (B). The 5′UAR/3′UAR base-pairings are depicted; mutated nucleotides are indicated in shaded circles.
For Sel II (Fig. 3B), a C-69U mutation appeared at P3 as a mixed population. The C-69U mutation dominated from P4 onward. No other mutations were identified from any later passages of Sel II (data not shown). Overall, the results suggest that resistant WNVs arose through a single-nucleotide mismatch (Sel I and Sel III) or a weakened nucleotide base pairing (Sel II) between the 3′UAR and the PPMO; when the 3′UAR mutation resulted in a mismatched base pairing with the 5′UAR, a second-site adaptation occurred in the 5′UAR, leading to the restoration of perfect base pairing between the mutated 5′UAR and the mutated 3′UAR (Sel I and Sel III).
A single-nucleotide mutation within the PPMO-binding site confers resistance to 3′UAR-PPMO
We only sequenced the terminal regions of the viral genomes from the resistant viruses. Mutations outside the sequenced regions may contribute to the resistant phenotype. In addition, since we performed population sequencing (rather than sequencing of plaque-purified isolates), quasispecies within the viral population may act together to confer resistance. To exclude these two possibilities, we engineered the recovered mutations, both singly and in combination, into an infectious cDNA clone of WNV. For mutations recovered from Sel I and Sel III, three mutant WNVs were prepared: A89G, U-74C, and A89G + U-74C. For the mutation recovered from Sel II, one mutant C-69U virus was constructed. For all recombinant viruses, we first compared their plaque morphologies with that of the WT virus. As shown in Figure 4, plaques from mutant viruses U-74C and C-69U were smaller than those from the WT virus. The smaller plaques of the U-74C and C-69U viruses correlated with the weakened base pairing of the 5′UAR/3′UAR interaction: a single-nucleotide mismatch in the U-74C mutant virus (Fig. 4C), and a G/C→G/U change (mutated nucleotide underlined) in the C-69U mutant virus (Fig. 4E). In contrast, recombinant A89G and A89G + U-74C viruses developed plaque sizes equivalent to that of the WT virus (Fig. 4). Furthermore, mutant A89G + U-74C and C-69U viruses exhibited plaque morphology similar to those derived from the Sel I and III, and the Sel II viruses, respectively (Compare Fig. 4 with Fig. 2A).
FIG. 4.
Analysis of mutations responsible for 3′UAR-PPMO resistance. The plaque morphologies, 5′UAR/3′UAR base-pairings, and 3′UAR-PPMO resistance are shown for WT WNV (A), four recombinant viruses (B–E), and Sel I P8 virus (F). For resistance analysis, Vero cells were infected with the indicated virus at an MOI of 0.1 with or without 20 μM of 3′UAR-PPMO. Viral titers in culture fluids were measured by plaque assays at indicated time points post-infection. Mean values from three independent experiments are presented. Error bars represent standard deviations.
We next characterized the resistant phenotypes of the recombinant viruses by comparing the growth kinetics in the presence or absence of 20 μM 3′UAR-PPMO. WT (Fig. 4A) and Sel I viruses (Fig. 4F) were included as controls. In the absence of PPMO, the mutant viruses were not grossly incapacitated in replication, as indicated by similar viral titers derived from the mutant and WT viruses. However, mutant U-74C virus replicated to a slightly lower titer than the WT virus at 42 h post-infection (p.i.); this small growth disadvantage, due to the single-nucleotide A/C mismatch between the 5′UAR and the 3′UAR, could provide a selective pressure for the emergence of the compensatory A89G mutation in the 5′UAR after continuous passages, leading to the restoration of perfect base pairings between the 5′UAR and the 3′UAR sequences. In the presence of PPMO, viral titers of both WT and A89G viruses were significantly suppressed. In contrast, recombinant viruses U-74C, A89G + U-74C, and C-69U were not substantially inhibited by 3′UAR-PPMO. As expected, Sel I virus was not significantly suppressed by the inhibitor (Fig. 4F). These results demonstrate that a single-nucleotide mutation within the 3′UAR (i.e., U-74C or C-69U, leading to a mismatch or a weakened base-pairing interaction between the 3′UAR and the PPMO) is the determinant for resistance. In addition, the similarity in viral titer between cells that were mock-treated or 20 μM 3′UAR-PPMO-treated indicates that the 3′UAR-PPMO did not cause cytotoxicity.
Notably, the originally selected WNVs (P8 of Sel I, Sel II, and Sel III in Fig. 1E, bottom panel) were less resistant to 3′UAR-PPMO than were the recombinant viruses (A89G + U-74C and C-69U in Fig. 4). Since we did not sequence the complete genome of the selected viruses, it is not possible to conclude that the discrepancy was due to unidentified mutations in the Sel I, Sel II, and Sel III viruses. Alternatively, the discrepancy could arise from the quasispecies nature of the selected viruses.
A single-nucleotide mismatch reduces the ability of 3′UAR-PPMO to block the RNA/RNA interaction required for WNV genome cyclization
We hypothesized that the mechanism by which the 3′UAR-PPMO exerts antiviral activity is through interference with the 5′UAR/3′UAR base-pairing interaction, leading to reduced viral genome cyclization (Fig. 5A). In resistant viruses, a single-nucleotide change in the 3′UAR could be expected to result in weakened binding between the 3′UAR and the 3′UAR-PPMO. To directly test this hypothesis, we performed RNA binding assays to measure the effect of a single-nucleotide mismatch on the PPMO-mediated suppression of the RNA/RNA interaction required for genome cyclization. The assay involves two RNAs, one representing the 5′ terminal 190 nt (5′RNA; containing 5′UAR and 5′CS) and the other representing the 3′ terminal 111 nt (3′RNA; containing 3′CSI and 3′UAR). As shown in Figure 5B, 32P-labeled 3′RNA (lane 1) could be converted to an RNA duplex in the presence of unlabeled 5′RNA (lane 2) on a native polyacrylamide gel. Addition of 3′UAR-PPMO suppressed the formation of 5′/3′RNA duplex in a dose-responsive manner (lanes 3–6). Concurrently, a new band, probably representing 3′RNA/3′UAR-PPMO complex, appeared; however, the putative complex band was weak and smeared. A similar band pattern for the 3′RNA/3′UAR PPMO complex was observed in our previous study, and was attributed to the positive charge of the peptide component of the PPMO hindering entrance of the PPMO-RNA complex into the gel (Zhang et al., 2008). Nevertheless, the results from both studies confirm that the 3′UAR-PPMO disrupts the 5′/3′RNA duplex.
FIG. 5.
RNA binding assay. (A) Scheme of 3′UAR-PPMO-mediated competition with 5′UAR/3′UAR duplex formation. Two RNAs, one representing the 5′ terminal 190 nucleotides (5′RNA) and the other representing the 3′ terminal 111 nucleotides (3′RNA), could form 5′/3′RNA duplex when incubated together. Addition of 3′UAR-PPMO causes a reduction in the formation of 5′/3′RNA duplex, through binding to the 3′RNA. Sequences for both WT and mutant (MT) 5′RNA and 3′RNA are presented. Mutated nucleotides are indicated in shaded circles. (B) Competition assays. 32P-labeled 3′RNA probe (lane 1) was converted to a 5′/3′RNA duplex when incubated with cold 5′RNA (lane 2). Indicated amounts of 3′UAR-PPMO were added to the mixture of 5′RNA and 3′RNA (lanes 3–6 and 9–12). The reactions were then analyzed on a nondenaturing polyacrylamide gel. The effect of 3′UAR-PPMO on 5′/3′RNA duplex formation was analyzed using WT or MT RNA. The positions of free probe of 3′RNA, 5′/3′RNAs duplex, and 3′RNA/3′UAR PPMO complex are indicated on the left side of the gels; the concentrations of 3′UAR-PPMO are indicated on top of the gel. (C) Quantification of WT and MT 5′/3′RNA duplexes with increasing amounts of 3′UAR-PPMO. The amount of 5′/3′RNA duplex remaining after competition treatment with 3′UAR-PPMO was measured using a PhosphoImager. For each 3′UAR-PPMO concentration, the value = amount of 5′/3′RNA duplex with PPMO/amount of 5′/3′RNA duplex without PPMO × 100. Mean values from three experiments are shown.
Next, we prepared a mutant 5′RNA containing the A89G substitution in the 5′UAR, and a mutant 3′RNA harboring the U-74C change in the 3′UAR. Mixing of the two mutant RNAs, in which the 3′RNA was 32P-labeled, resulted in a mutant 5′/3′RNA duplex (Fig. 5B, lane 8). Competition assays showed that, although the 3′UAR-PPMO could disrupt the mutant 5′/3′RNA duplex (Fig. 5B, lanes 8–12), the efficiency was much lower than for its disruption of the WT RNA duplex (Fig. 5C). For instance, 10-μM 3′UAR-PPMO competed away 100% of the WT RNA duplex but less than 50% of the mutant RNA duplex. The results suggest that, due to a single-nucleotide mismatch, the 3′UAR-PPMO has a reduced ability to block the RNA/RNA interaction required for genome cyclization in the resistant viruses.
DISCUSSION
A number of approaches have been reported for development of flavivirus antiviral compounds (Shi, 2008). We have been exploring the possibility of inhibiting flavivirus infections through interference with cis elements of viral RNA. Results from several groups have shown that PPMOs designed to interfere with the 5′CS/3′CSI or 5′UAR/3′UAR base-pairing interactions can inhibit flaviviruses both in cell culture and in animal models (Deas et al., 2007; Deas et al., 2005; Holden et al., 2006; Kinney et al., 2005; Stein et al., 2008; Zhang et al., 2008). In this study, we have selected and characterized WNVs resistant to 3′UAR-PPMO. Our results suggest that WNV can develop resistance to 3′UAR-PPMO through co-selection of 5′UAR and 3′UAR elements that result in a 3′UAR/3′UAR-PPMO mismatch or a weakened 3′UAR/3′UAR-PPMO base pairing, while maintaining 5′UAR/3′UAR base pairings. These results represent the first demonstration that a secondary mutation outside of the PPMO target site can restore the ability of virus to establish an important RNA/RNA interaction with a region containing a primary mutation in the PPMO target site.
The successful selection of PPMO-3′UAR resistant viruses allowed us to identify nucleotide changes responsible for the drug resistance. Two types of mutations were recovered from the 3′UAR-PPMO-resistant WNVs (Fig. 2C). Type I mutation was derived from Sel II, and contained a nucleotide change (C-69U) that allows maintenance of base pairing with the 5′UAR. No mutation occurred in the 5′UAR region. Type II mutations were derived from both Sel I and Sel III, and contained a change (U-74C) that weakened the base pairing with the WT 5′UAR. Mutagenesis analysis demonstrated that both types of mutations contribute to the resistance phenotype (Fig. 4C–E). In agreement with the genetic results, biochemical binding analysis showed that, due to the single-nucleotide changes, the 3′UAR-PPMO had a reduced ability to block the RNA/RNA interaction required for genome cyclization (Fig. 5). Furthermore, in type II resistant viruses, one nucleotide in 5′UAR (A89G) was co-selected, leading to a restoration of perfect base-pairings between the 5′UAR and 3′UAR sequences. The co-evolution of changes in the 5′UAR and 3′UAR sequences during resistance development provides novel genetic evidence for the importance of the 5′UAR/3′UAR interaction in flavivirus replication
The 3′UAR-PPMO-resistant viruses (Sel I and Sel III) contained mismatch mutations within the PPMO-targeted region. These results are in agreement with our previous study on a PPMO targeting the first 20 nt of the WNV genomic RNA (5′-end–PPMO) (Deas et al., 2007). The 5′-end–PPMO-resistant virus accumulated mismatch changes within the PPMO-targeted region. In contrast, we found that WNV resistant to a PPMO targeting the 3′CSI (3′CSI-PPMO) accumulated mutations outside the PPMO-binding site; a single-nucleotide deletion or insertion, located in the 3′ terminal stem-loop of the genomic RNA, was responsible for the 3′CSI-PPMO resistance (Deas et al., 2007). Although both 5′CS/3′CSI and 5′UAR/3′UAR interactions contribute to flavivirus genome cyclization, it is puzzling why PPMOs targeting the 5′CS/3′CSI (3′CSI-PPMO) and the 5′UAR/3′UAR (3′UAR-PPMO) interactions produce resistance-determining nucleotide(s) outside and inside the targeted RNA sequences, respectively. Both the 5′CS/3′CSI and 5′UAR/3′UAR interactions have been shown to be critical for viral RNA synthesis in WNV (Khromykh et al., 2001; Lo et al., 2003; Zhang et al., 2008) and DENV (Alvarez, Filomatori, and Gamarnik, 2008; Alvarez et al., 2005a; Alvarez et al., 2005b; You et al., 2001; You and Padmanabhan, 1999; Yu et al., 2008). However, the two RNA interactions may function through distinct mechanisms during viral replication, leading to different outcomes in our resistance studies.
In summary, WNV develops resistance to 3′UAR-PPMO through co-selection of 5′UAR and 3′UAR sequences that each contains a single-nucleotide mismatch or a weakened base pairing with the PPMO, while maintaining the 5′UAR/3′UAR base-pairings. The single-nucleotide change within the 3′UAR reduces the efficacy of 3′UAR-PPMO to block the RNA/RNA interaction required for genome cyclization, while the 5′UAR mutation allows the 5′UAR/3′UAR base pairings to be maintained. More biochemical and genetic studies are required to illuminate the molecular mechanism of the 5′UAR/3′UAR interaction in the viral life cycle.
MATERIALS AND METHODS
Viruses and cells
Baby hamster kidney (BHK) cells and African green monkey kidney cells (Vero cells; American Type Culture Collection) were grown in 5% CO2 at 37ºC with DMEM plus 10% FCS. Confluent cell monolayers and virus-infected cells were maintained in DMEM plus 2% FCS. WNV was produced from an infectious cDNA clone pFLWNV by electroporation of BHK cells with in vitro-transcribed RNA (Shi et al., 2002).
RNA transcription and transfection
Replicon and genome-length RNAs were transcribed from the Xba I-linearized cDNA plasmids using T7 mMESSAGE mMACHINE kits (Ambion). The transcription reactions were performed according to the manufacturer’s protocols. For transfection, approximately 5 μg of RNA was electroporated into 8 × 106 BHK-21 cells in 0.8-ml ice cold phosphate-buffered saline (PBS; pH 7.5) in a 0.4-cm cuvette, with the GenePulser apparatus (Bio-Rad) using settings of 0.85 kV and 25 μF, pulsing three times at 3-s intervals. After a 10-min recovery at room temperature, the transfected cells were diluted in 25 ml of DMEM containing 10% FCS, and were transferred into a T-150 flask. Viruses in culture fluids were collected after incubation of the flask for 4 days at 37°C with 5% CO2. Aliquots of the viruses were stored at −80°C.
Plaque assay
A series of 1:10 dilutions were made by mixing 15 μl of virus sample with 135 μl of BA-1 diluent (Hanks M199 salts, 1% bovine serum albumin, 350 mg/l sodium bicarbonate, 100 U/ml penicillin, 100 mg/l streptomycin, 1 mg/l amphotericin B in 0.05 mol/l Tris, pH 7.6). One hundred microliters of each dilution were seeded to individual wells of 6-well plates containing nearly confluent Vero cells (6 × 105 cells per well, plated 3 days in advance). The plates were incubated at 37ºC with 5% CO2 for 1 h before the first layer of agar was added. A second layer of agar containing neutral red was added after 2 days of incubation at 37ºC with 5% CO2. Plaque morphology and numbers were recorded after incubation of the plates for a further 12 to 24 h.
Selection of 3′UAR-PPMO-resistant virus
3′UAR-PPMO-resistant WNV was selected by passaging of the wild-type (WT) virus in Vero cells with increasing concentrations of the 3′UAR-PPMO (Fig. 1D). Viral titers from each passage were determined by plaque assay. Three selections for 3′UAR-PPMO-resistant WNV were independently performed (selections I, II, and III). For each selection, Vero cell monolayers in a six-well plate were inoculated at a multiplicity of infection (MOI) of 0.1 with WT or previously passaged virus, in a total volume of 1 ml BA-1. After incubation at 37°C for 1 h, the cell monolayers were washed three times with BA-1 diluent. Three milliliters of DMEM with 2% FCS plus 3′UAR-PPMO were added to each well. The supernatants were harvested at 42 h p.i., and aliquots were stored at −80ºC. Supernatants from the 20 μM PPMO-selected viruses were subjected to sequencing analyses (see below).
5′ and 3′ rapid amplification of cDNA ends (RACE) sequencing of viral RNA
Viral RNA was extracted from virions in culture medium, using RNeasy kits (Qiagen). The extraction was performed following the manufacturer’s protocol. Viral RNA sample (5 μl) was treated with 10 units of tobacco acid pyrophosphatase (TAP) (EPICENTRE Biotechnologies) in a 10-μl reaction at 37°C for 1 h. The TAP-treated RNA (2 μl) was used for ligation with 5 units of T4 RNA ligase (Ambion) in a total volume of 10 μl at 37°C for 1 h. Ligated RNA sample (5 μl) was subjected to amplification using Onestep RT-PCR kit (Invitrogen) with primer pairs 10501v (5′-AGAAAGTCAGGCCGGGAAGTTC-3′ ′; sense sequence; representing nucleotides 10501 to 10522; GenBank no. AF404756) and 684c (5′-TGCTGACTTTGTGCACCAACA-3′; antisense sequence; complementary to nucleotides 664 to 684). The 1.2-Kb RT-PCR product, spanning the 5′ terminal 684 nucleotides and the 3′ terminal 529 nucleotides of the WNV genome, was gel-purified by QIAquick Gel Extraction kit (Qiagen), and was directly subjected to DNA sequencing.
Plasmid construction
WNV genome-length cDNA clones with 5′UAR and 3′UAR mutations were constructed by use of a modified pFLWNV (Shi et al., 2002) and two shuttle vectors. Shuttle vector A, used for mutagenesis of the 5′UAR element, was constructed by engineering the BamH I-Sph I fragment from the pFLWNV (representing the upstream end of the T7 promoter [used for transcription of the genome-length RNA] to nucleotide position 3627 of the WNV genome) into vector pACYC177 containing a modified cloning cassette (Zhou et al., 2007). A QuickChange II XL site-directed mutagenesis Kit (Stratagene) was used to engineer the 5′UAR mutations into shuttle vector A. The mutated DNA fragment was then pasted back into the pFLWNV clone at the BamH I and Cla I (nucleotide position 1089) sites.
Shuttle vector B, used for mutagenesis of the 3′UAR element, was constructed by engineering the Kpn I-Xba I fragment from the pFLWNV (representing nucleotides 7762 through the 3′ end of the WNV genome) into pcDNA3.1. A QuickChange II XL site-directed mutagenesis Kit (Stratagene) was used to engineer the 3′UAR mutations into the shuttle vector B. The mutated DNA fragment was pasted back into the pFLWNV clone at the Spe I (nucleotide position 8023) and Xba I sites. All constructs were verified by DNA sequencing.
Growth kinetics of WT and PPMO-resistant viruses in the presence and absence of PPMO
Vero cells (9 × 105 in 1 ml DMEM medium plus 10% FCS) were seeded in each well of a 12-well plate. After 12 h incubation, the cells were infected with WNV at an MOI of 0.1 in 100 μl of BA-1 solution. After 1 h infection, the cell monolayers were washed three times with BA-1 diluent. The infected cells were cultured with 1ml of DMEM plus 2% FCS with or without a final concentration of 20 μM 3′UAR-PPMO. Culture fluids were collected at various time points p.i. and stored at −80°C. Viral titers were determined using a plaque assay as described above.
Native gel analysis of 5′ and 3′RNA duplex formation
Cold and 32P-labeled RNAs representing the 5′ 190 nt or the 3′ 111 nt of the WNV genomic RNA were prepared for analysis of RNA/RNA interactions. The RNAs were transcribed from PCR products containing a T7-promoter sequence followed by the template sequence. For preparation of RNA probes, α-32P-GTP was internally incorporated into RNAs during in vitro transcription, using a MEGAscript T7 Kit (Ambion). After transcription, the template DNA was digested using DNase I, according to the manufacturer’s protocol. The reactions were passed through two G-25 columns (GE Healthcare) to remove unincorporated NTPs. The resulting RNA was measured using a scintillation counter.
For analysis of RNA duplex formation of the 5′ 190-nt and 3′ 111-nt RNAs, approximately 5 pmol of 3′RNA probe (2.5 × 104 to 4.5 × 104 CPM) was mixed with cold 5′RNA in a total volume of 15 μl buffer containing 50 mM Tris–HCl, pH 8.0, 200 mM NaCl, and 5 mM MgCl2. For analysis of the effects of antisense PPMOs on formation of the 5′ and 3′ RNA duplex, various concentrations of PPMOs were added during the reaction assembly. The reactions were then incubated at room temperature for 30 min and resolved on a native 8% polyacrylamide gel (15 × 15 cm2). The amounts of radioactivity were quantified by PhosphoImager.
Acknowledgments
We are grateful to Dr. Patrick L. Iversen and the Chemistry Group at AVI BioPharma for synthesis of the PPMO used in this study. We thank the Wadsworth Center’s Molecular Genetics Core for DNA sequencing and Cell Culture Facility for maintenance of BHK-21 and Vero cells. This work was supported partially by federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health (NIH), under contract NOI-AI-25490, and by NIH grants U01 AI061193 and U54-AI057158 (Northeast Biodefense Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez D, Filomatori C, Gamarnik A. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375(1):223–35. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005a;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005b;79(11):6631–43. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79(20):12828–39. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J Gen Virol. 2003;84(Pt 5):1261–8. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison R, Sando T, Strauss E, Strauss J. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol. 2003;77(3):2265–2270. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B, Chang G, Cropp B, Roehrig J, Martin D, Mitchell C, Bowen R, Bunning M. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J Virol. 2001;75(9):4040–4047. doi: 10.1128/JVI.75.9.4040-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas TS, Bennett CJ, Jones SA, Tilgner M, Ren P, Behr MJ, Stein DA, Iversen PL, Kramer LD, Bernard KA, Shi P-Y. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob Agents Chemother. 2007;51:2470–82. doi: 10.1128/AAC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas TS, Binduga-Gajewska I, Tilgner M, Ren P, Stein DA, Moulton HM, Iversen PL, Kauffman EB, Kramer LD, Shi P-Y. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J Virol. 2005;79(8):4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi P–Y. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology. 2008 doi: 10.1016/j.virol.2008.07.040. (in press) [DOI] [PubMed] [Google Scholar]
- Filomatori C, Lodeiro M, Alvarez D, Samsa M, Pietrasanta L, Gamarnik A. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20(16):2238–49. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler D, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5. Vol. 1. Lippincott William & Wilkins; Philadelphia, Pa: 2007. pp. 1153–1253. [Google Scholar]
- Guo J, Hayashi J, Seeger C. West nile virus inhibits the signal transduction pathway of alpha interferon. J Virol. 2005;79(3):1343–50. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198(1):33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Holden K, Stein D, Pierson T, Ahmed A, Clyde K, Iversen P, Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem-loop structure. Virology. 2006;344(2):439–52. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J Virol. 2001;75(14):6719–28. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R, Huang C, Rose B, Kroeker A, Dreher T, Iversen P, Stein D. Inhibition of dengue virus serotypes 1 to 4 in vero cell cultures with morpholino oligomers. J Virol. 2005;79(8):5116–5128. doi: 10.1128/JVI.79.8.5116-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler RM, Hoenninger VM, Thurner C, Mandl CW. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J Virol. 2006;80(8):4099–113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer L, Li J, Shi P. West Nile virus. The Lancet Neurology. 2007;6:171–182. doi: 10.1016/S1474-4422(07)70030-3. [DOI] [PubMed] [Google Scholar]
- Kummerer BM, Rice CM. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol. 2002;76(10):4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. Role of nonstructural protein NS2A in flavivirus assembly. J Virol. 2008;82(10):4731–41. doi: 10.1128/JVI.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel H–J, Rice CM. Flaviviridae: The Virus and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4. Lippincott William & Wilkins; 2007. [Google Scholar]
- Liu W, Wang X, Mokhonov V, Shi P, Randall R, Khromykh A. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol. 2005;79(3):1934–42. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Chen HB, Khromykh AA. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J Virol. 2003;77(14):7804–13. doi: 10.1128/JVI.77.14.7804-7813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Tilgner M, Bernard K, Shi P-Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus using a reporting replicon that differentiates between viral translation and RNA replication. J Virol. 2003;77(18):10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded Sequence Dependence of Thermodynamic Parameters Improves Prediction of RNA Secondary Structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Men R, Bray M, Clark D, Chanock RM, Lai CJ. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol. 1996;70(6):3930–7. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke JM, Siger L, Karaca K, Austgen L, Gordy P, Bowen R, Renshaw RW, Loosmore S, Audonnet JC, Nordgren B. Recombinant canarypoxvirus vaccine carrying the prM/E genes of West Nile virus protects horses against a West Nile virus-mosquito challenge. Arch Virol Suppl. 2004;(18):221–30. doi: 10.1007/978-3-7091-0572-6_20. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of Alpha/Beta Interferon Signaling by the NS4B Protein of Flaviviruses. J Virol. 2005;79(13):8004–13. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci USA. 2003;100(24):14333–8. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T, Hathaway D, Jennings N, Champ D, Chiang YW, Chu HJ. Equine vaccine for West Nile virus. Dev Biologicals. 2003;114:221–7. [PubMed] [Google Scholar]
- Patkar CG, Kuhn RJ. Yellow Fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol. 2008;82(7):3342–52. doi: 10.1128/JVI.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY. Novel therapeutics against West Nile virus. In: Diamond MS, editor. West Nile Encephalitis Virus Infection: Viral pathogenesis and the Host Immune Response. Springer Publisher; 2008. (In press) [Google Scholar]
- Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol. 2002;76(12):5847–56. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D, Huang C, Silengo S, Amantana A, Crumley S, Blouch R, Iversen P, Kinney R. Treatment of AG129 mice with antisense morpholino oligomers increases survival time following challenge with dengue 2 virus. J Antimicrob Chemother. 2008 doi: 10.1093/jac/dkn221. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense & Nucleic Acid Drug Dev. 1997;7(3):187–95. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. Journal of Biological Chemistry. 2001;276(19):15581–91. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem. 1999;274(47):33714–22. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Yu L, Nomaguchi M, Padmanabhan R, Markoff L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008;374(1):170–85. doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008;(273):1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard K, Shi P, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]