Abstract
Background and Purpose
DNA repair assays to identify radiosensitive patients have had limited clinical implementation due to long turn-around times or limited specificity. This study evaluates γ-H2AX Irradiation Induced Foci (IRIF) kinetics as a more rapid surrogate for the ‘gold standard’ colony survival assay (CSA) using several known DNA repair disorders as reference models.
Materials and Methods
Radiosensitive cells of known and unknown etiology were studied. γ-H2AX-IRIFs were quantified over 24 hours, and the curves were fitted by combining logarithmic growth and exponential decay functions. Fitted values that differed from radionormal controls were considered aberrant and compared to CSA results.
Results
We observed 87% agreement of IRIF data with the CSA for the 14 samples tested. Analysis of γ-H2AX-IRIF kinetics for known repair disorders indicated similarities between an RNF168−/− cell line and an RS cell of unknown etiology. These cell lines were further characterized by a reduction in BRCA1-IRIF formation and G2/M checkpoint activation.
Conclusion
γ-H2AX-IRIF kinetics showed high concordance with the CSA in RS populations demonstrating its potential as a more rapid surrogate assay. This method provides a means to globally identify defective DNA repair pathways in RS cells of unknown etiology through comparison with known DNA repair defects.
Keywords: Radiosensitivity, BRCA1, H2AX, Foci, Kinetics
Introduction
Defects in DNA double strand break repair proteins result in varying clinical phenotypes which, thus far, can be included under the umbrella of the XCIND syndrome (X-ray irradiation sensitivity, Cancer susceptibility, Immunodeficiency, Neurological impairment, and Double strand breakage repair) [1]. Many deficiencies such as Ataxia-Telangiectasia (A-T), Nijmegen Breakage Syndrome (NBS), DNA LIGASE IV deficiency (LigIV), RNF168 deficiency (RNF168), and polynucleotide kinase 3’-phosphotase (PNKP) deficiency are now diagnosed by routine immunoblotting or sequencing. The large number of potentially defective DNA repair proteins, however, makes these techniques impractical for diagnosis of new double strand break (DSB) DNA damage response (DDR) deficiencies. To address this challenge our lab has sought to utilize functional DNA repair assays that can rapidly identify pathways of underlying repair deficiency.
The DNA repair disorders mentioned above have been extensively studied to elucidate the role the responsible gene contributes to DSB processing. In some cases, these associations are overly simplistic and only provide working models for diagnostic evaluations. Ataxia-Telangiectasia Mutated (ATM) is activated within minutes after irradiation (IR) and is responsible for many downstream events in the DSB-DDR [2]. Nibrin plays an early role in recruiting ATM to the damage site and initiating homologous recombination [3]. DNA LIGASE IV is responsible for ligation of the broken DNA in non-homologous end joining [4]. RNF168 and PNKP have been characterized more recently and represent defects in the chromatin ubiquitin ligase cascade (CULC) [5–7] and 3’–5’ end processing after DSBs, respectively [8, 9]. We have evaluated the utility of γ-H2AX-Irradiation Induced Foci (IRIF) kinetics as a general screen for radiosensitivity and characterized the unique kinetics in known and unknown DNA repair disorder models.
Materials and Methods
Cell Panel
The lymphoblastoid cell lines (LCLs) used in this study were derived mainly from patients with an XCIND-like disorder or a reduced survival fraction (SF%) by CSA. They were coded for anonymity in accordance with an approved protocol for the study of human subjects (Exemption #4). No human subjects were recruited for this study. Peripheral blood lymphocytes (PBLs) were isolated and transformed with EBV as previously described [10]. LCLs were screened by immunoblotting to confirm the deficiencies reported and compared to radionormal or wild type (WT) daily controls. Transformed LCLs were maintained in RPMI1640 media, 10% fetal bovine serum (FBS) (Hyclone, Logan, Utah) and 1% penicillin/streptomycin (Gibco BRL, Grand Island, NY) at 37°C and 5% CO2.
Colony Survival Assay (CSA)
The average SF% for LCLs was assessed by CSA, using normal and RS ranges, previously defined with 29 WT and 104 A-T LCLs [10]. Briefly, cells were seeded in a 96-well plate and treated with 1 Gy γ-IR at a dose rate of 4.5 Gy/min (Mark 1 Cs137 Irradiator) or mock irradiated. Plates were returned to 37°C for 10 to 13 days at which time they were stained with MTT dye (tetrazolium-based colorimetric assay, Sigma, St. Louis, MO). SF% of <21% was interpreted as RS and SF% >36% was considered radionormal [10].
Immunofluorescence detection of γ-H2AX-IRIFs
Immunofluorescence detection of γ-H2AX has been previously described in many laboratories [11, 12]. Briefly, LCLs were irradiated, spread on coverslips, and fixed with a 4% formaldehyde solution at the indicated time points. The cells were permeabilized in 0.5% Triton-X-100, incubated with appropriate primary and secondary antibodies then mounted on slides using Vectashield with DAPI. Cells were quantified by fluorescent microscopy and nuclei containing 4 foci/cell or more were scored as positive.
G2/M Checkpoint Assay
Histone, H3, phosphorylated on serine 10 (H3pS10) has been described previously as a marker for the G2/M checkpoint [13]. Briefly, LCLs were irradiated or mock irradiated and returned to 37°C for 1.5 hours. Cells were fixed in 70% ethanol and incubated with appropriate primary and secondary antibodies followed by staining with propidum iodide (Invitrogen, Carlsbad, CA) before analysis by flow cytometry.
Antibodies
Immunofluorescence antibodies were: γ-H2AX 1:300 (Upstate, Charlottesville, VA), 53BP1 1:400 (Santa Cruz Biotechnology, Santa Cruz, CA), BRCA1 1:300 (Novus, Littleton, CO), RAD51 1:100 (Santa Cruz Biotechnology, Santa Cruz, CA), Alexa Fluor 488 anti-mouse 1:300, and Alexa Fluor 568 anti-rabbit 1:400 (Invitrogen, Carlsbad, CA). The H3pS10 antibody was used at 1:100 (Cell Signaling, Beverly, MA) and the FITC secondary antibody was used at 1:100 (Jackson Biologicals, West Grove, PA).
Kinetics Model and Statistics
The kinetics of γ-H2AX-IRIF formation has been observed to be logarithmic in nature while the resolution of IRIFs is exponential [14, 15]. These characteristics were expressed in a non-linear fit function combining logarithmic growth and exponential decay functions. An Interior-Trust-Region non-linear optimization algorithm was implemented to fit the data with this model [16]. A visual reference of this technique is shown in Supplemental Figure 1. P-values are based on the two samples of unequal variance two-tailed student’s t-test calculated in Microsoft Excel (Microsoft, Redmond, WA).
Results
γ-H2AX IRIF kinetics identifies ‘fingerprint’ curves for known DNA repair disorders
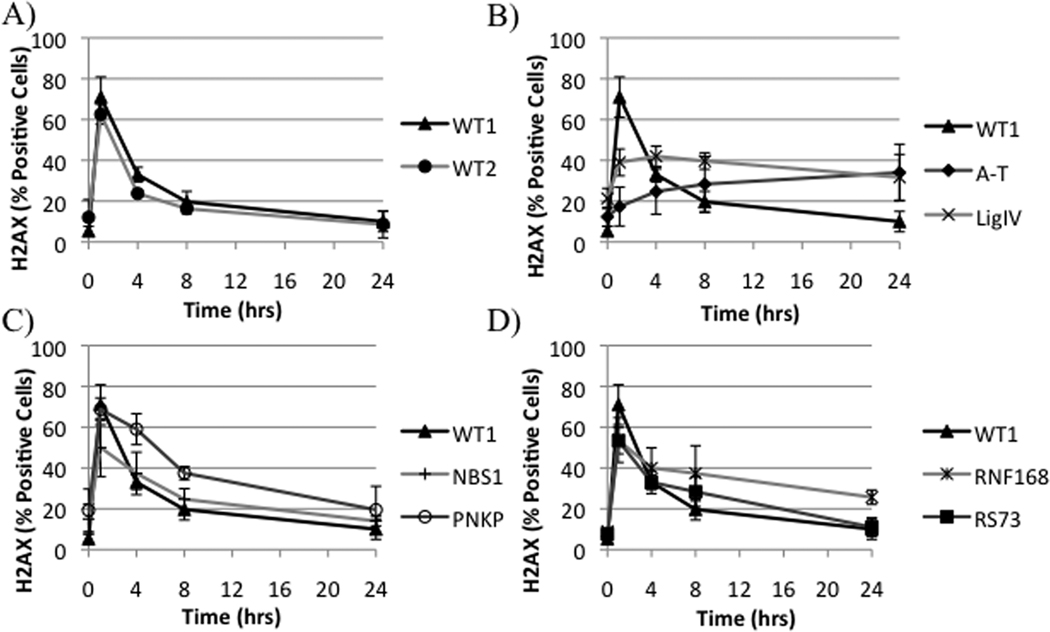
LCLs with formation and resolution of IRIF constants statistically different from the WT were considered RS. This resulted in a 60%, 80%, and 87% concordance with the CSA for the formation of IRIF, resolution of IRIF, and combination of constants, respectively (Table 1). A significant relation between SF% and the individual fit constants was found by linear regression analysis with a correlation coefficient of 0.54 for each constant (Table 1). The kinetics of A-T was characterized by a delay and reduction in the formation of γ-H2AX-IRIFs with poor resolution of IRIFs at 24 hours. Similarly, DNA LigIV exhibited poor resolution of IRIFs at 24 hours but formed IRIFs at a higher efficiency than A-T LCLs at 1 hour (Figure 1B). PNKP and NBS, conversely, displayed WT-like curves characterized, however, by a slower resolution kinetic. NBS was characterized by reduced positive cells at one hour (Figure 1C). Lastly, the RNF168-LCL displayed WT-like repair rates until four hours post-IR at which time a slower, linear repair rate was observed (Figure 1D).
Table 1.
γ-H2AX-IRIF comparison with the CSA
| Cell Line | 24 Hour1 | Formation2 | Resolution3 | Combined |
|---|---|---|---|---|
| WT2 | N | N | N | N |
| A-T | N | D | D | D |
| NBS | N | D | D | D |
| LigIV | N | D | D | D |
| PNKP | N | N | D | D |
| RNF168 | D | N | D | D |
| RS36 | N | N | D | D |
| RS73 | N | D | D | D |
| RS70 | N | D | D | D |
| RS39 | N | N | N | N |
| RS26 | N | D | N | D |
| RS100 | N | D | D | D |
| RS29 | D | N | D | D |
| RS37 | D | N | N | N |
| Concordance With CSA (%) | 27 | 60 | 80 | 87 |
| Regression Slope | −0.04 | 2.25 | 0.008 | NA |
| P-value | 0.79 | 0.001 | 0.001 | NA |
| Correlation Coefficient | 0.005 | 0.54 | 0.54 | NA |
N: Normal compared to WT1 daily control.
D: Defective. Values outside of WT range (WT1 daily control).
Previously used single time point IRIF technique (24 hour post-IR).
Formation of IRIF constant from fit function.
Resolution of IRIF constant from fit function.
Figure 1. γ-H2AX kinetic ‘fingerprint’ of repair disorders.
The unique groups of γ-H2AX-IRIF kinetics characterized in WT1, WT2, A-T, LigIV, NBS, PNKP, and RNF168 LCLs are shown. IRIFs were evaluated after 1 Gy IR.
‘Fingerprint’ γ-H2AX kinetic identifies defective repair pathway in unknown RS-LCL
We focused on RS73, which displayed γ-H2AX-IRIF kinetic changes at four hours similar to the RNF168-LCL (Figure 1D). This suggested that RS73 might have a defect in the same pathway as the RNF168-LCL prompting us to analyze 53BP1-IRIF kinetics, a biomarker of the entire CULC pathway [7]. A-T and RNF168 LCLs displayed large differences in the γ-H2AX and 53BP1 kinetics (Supplemental Figure 2D&E). In WT, NBS, and PNKP LCLs, the γ-H2AX and 53BP1 kinetic curves closely mimicked each other in that early (1hr) and late (24hrs) repair were normal (Supplemental Figure 2A,B,C). 53BP1-IRIF kinetic’s of RS73 mimicked those of γ-H2AX-IRIF kinetics suggesting a defect independent or downstream of 53BP1-IRIF formation (Supplemental Figure 2F).
RS73 confers a reduction in both BRCA1 foci formation and G2/M checkpoint activation after IR
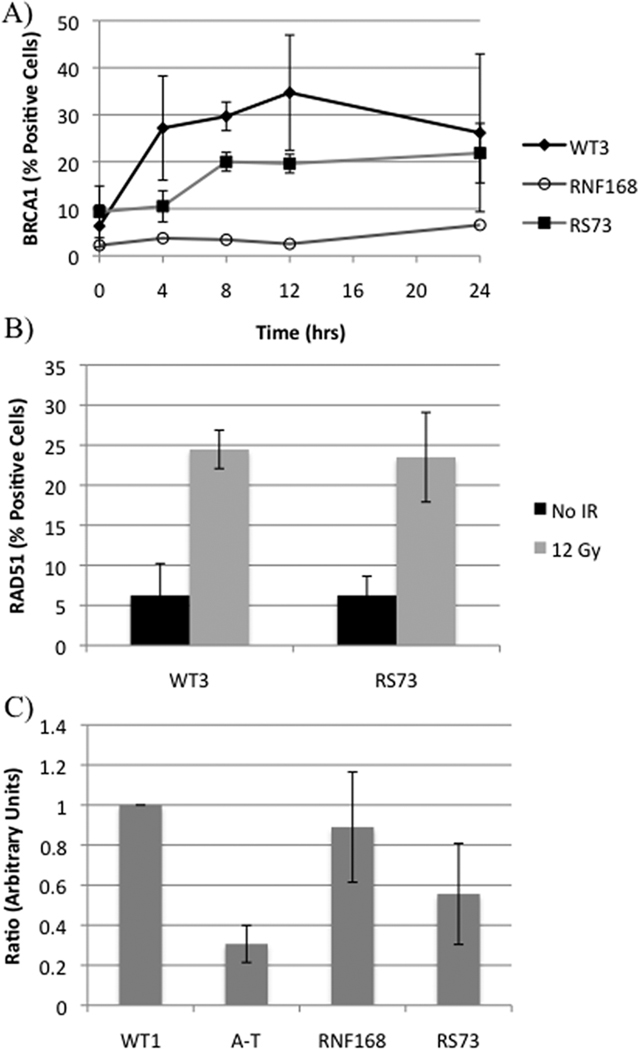
The above results prompted us to test BRCA1-IRIF kinetics. The BRCA1A complex has been shown to function downstream of 53BP1-IRIFs through ubiquitin binding [17]. RS73 displayed a delay in BRCA1-IRIF formation but did not display the severe reduction observed in the RNF168-LCL (Figure 2A). RAD51-IRIF formation in RS73 was similar to the WT-LCL, suggesting that the core of the homologous recombination pathway was intact [18] (Figure 2B). Previous experiments have demonstrated that adequate function of the BRCA1A complex is required for a functioning IR-induced G2/M checkpoint [17, 19]. RS73 exhibited reduced activation of the G2/M checkpoint post-IR (Figure 2C) strongly supporting this operational model.
Figure 2. Follow-up analysis of RS73.
A) BRCA1-IRIF kinetics evaluated after 12 Gy IR. B) RAD51-IRIF evaluated at 8 hours post-IR. C) Results of the G2/M checkpoint assay. Values are the ratio of H3pS10 positive cells before and 1.5 hrs after IR (−IR/+IR). The data were normalized to WT1, a WT-LCL included in the same experiment.
Discussion
Developing a predictive assay for radiosensitivity is a central challenge in radiation biology [20]. Previously, genetic association and molecular techniques have been applied to this problem [21, 22]. The CSA SF% remains the ‘gold standard’ and identifying a more rapid surrogate assay would make measuring radiosensitivity practical in oncology patients. It would also improve diagnostic studies of patients with an XCIND phenotype. Such efforts could also impact upon the analysis of large populations with exposure in nuclear accidents. Previous studies have found significant correlations between SF% and γ-H2AX-IRIF methods in cell lines with repair defects, however, in more heterogeneous populations significant correlations have been difficult to achieve [14, 15, 22, 23]. It is also unclear whether persistent DNA damage is predictive of side effect severity in radiotherapy [24].
The present study demonstrates strong qualitative agreement between γ-H2AX kinetics and the CSA. Low correlation coefficients, however, indicate this γ-H2AX kinetic method is unable to adequately account for the variability observed in RS-LCLs and quantitatively predict SF%. The utility of this method, instead, is its ability to characterize repair kinetics to identify global DSB repair defects while qualitatively identifying potentially RS populations.
The concept of synthetic lethality has gained recent popularity and current trials using PARP-1 inhibitors for breast cancer patients with BRCA1 mutations have provided a convincing argument for even broader implementation in oncology [25]. However, rapidly identifying DNA repair defects in cancers with unknown backgrounds remains a central challenge to identifying promising candidates and targets for synthetic lethality-based treatments [26, 27]. The efficacy and implementation of synthetic lethality could be vastly improved if an assay(s) could clearly evaluate all major DNA repair pathways and indicate targets for specific inhibitors.
This study begins to build a library of global repair kinetics from known disorders. Defects in repair function, as those found in A-T and LigIV LCLs, are characterized by persistent IRIFs at 24hrs, representing residual, unrepaired DNA damage, and minimal resolution of γ-H2AX-IRIFs over 24 hours. Reduced positive cells early after IR in NBS and A-T LCLs illustrates the impact of sensory protein defects. RNF168 and PNKP deficiencies exhibit delayed resolution kinetics and relatively flat curves from 8 to 24 hours, suggesting the presence of damage that requires these proteins for timely repair [28].
RS73 illustrates the use of γ-H2AX-IRIF kinetics to evaluate unknown repair defects. RS73 was submitted to our lab for RS testing because the patient had developed T-cell leukemia early in life and had an adverse reaction to radiotherapy, strongly suggesting the presence of a DNA repair defect. Analysis of the γ-H2AX and 53BP1-IRIF kinetic profiles in the RNF168-LCL and RS73 cells suggested a defect downstream of 53BP1 in RS73. The BRCA1A complex has been described previously as being recruited to polyubiquitin chains [19] at the sites of 53BP1-IRIFs and its retention at these sites is instrumental for homologous recombination repair [29]. A functioning, or partially functioning, homologous recombination pathway is suggested by the formation of a)BRCA1-IRIFs (although delayed) and b) RAD51-IRIFs, which may explain the WT-like γ-H2AX-IRIF levels at 24 hours.
The G2/M checkpoint assay further supports a defect in the G2/M checkpoint pathway in RS73 that is more severe than that of the RNF168-LCL. This G2/M checkpoint defect appears in other deficiencies in the CULC pathway [30, 31] and interactions of the BRCA1A complex at damage sites plays a central role in the activation of this checkpoint [13, 17, 19]. The data for RS73 and the RNF168-LCL suggest that adequate formation of BRCA1-IRIFs at the sites of polyubiquitin chains is necessary for a functional G2/M checkpoint following IR. These data provide an example of applying kinetic measurements to the diagnostic assessment of DNA repair disorders.
Supplementary Material
Shown is a visual representation of the kinetics model used in this study. Growth and decay functions are combined to fit the γ-H2AX-IRIF data. IRIFs were assessed at the indicated time points after 1 Gy IR.
γ-H2AX and 53BP1 IRIF kinetics are compared for WT2, A-T, NBS, RNF168, PNKP, and RS73 LCLs. Both IRIF experiments were evaluated after 1 Gy IR.
Acknowledgements
This work was supported by grants from the NIH (NS052528 and AI067769), A-T Medical Research Foundation, and the Joseph Drown Foundation. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health (CA-16042 and AI-28697) and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors report no conflict of interests.
References
- 1.Gatti R, Boder E, Good R. Immunodeficiency, radiosensitivity, and the XCIND syndrome. Immunologic Research. 2007;38:87–101. doi: 10.1007/s12026-007-0018-y. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair. 3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Mahaney BL, Meek K, Lees-miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devgan SS, Sanal O, Doil C, et al. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. doi: 10.1038/cdd.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart GS, Panier S, Townsend K, et al. The RIDDLE Syndrome Protein Mediates a Ubiquitin-Dependent Signaling Cascade at Sites of DNA Damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 7.Doil C, Mailand N, Bekker-Jensen S, et al. RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JNM. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends in Biochemical Sciences. doi: 10.1016/j.tibs.2011.01.006. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Gilmore EC, Marshall CA, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Becker-Catania SG, Chun HH, et al. Early diagnosis of ataxia-telangiectasia using radiosensitivity testing. The Journal of Pediatrics. 2002;140:724–731. doi: 10.1067/mpd.2002.123879. [DOI] [PubMed] [Google Scholar]
- 11.Lobrich M, SA, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. GammaH2AX Foci Analysis for Monitoring DNA double-strand Break Repair: Strengths, Limitations and Optimization. Cell Cycle. 2010;9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 12.Nahas SA, Gatti RA. DNA double strand break repair defects, primary immunodeficiency disorders, and 'radiosensitivity'. Current Opinion in Allergy and Clinical Immunology. 2009;9(6):510–516. doi: 10.1097/ACI.0b013e328332be17. [DOI] [PubMed] [Google Scholar]
- 13.Xu B, Kim S-t, Kastan MB. Involvement of Brca1 in S-Phase and G2-Phase Checkpoints after Ionizing Irradiation. Mol. Cell. Biol. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid TE, Dollinger G, Beisker W, et al. Differences in the kinetics of <gamma>-H2AX fluorescence decay after exposure to low and high LET radiation. International Journal of Radiation Biology. 2010;86:682–691. doi: 10.3109/09553001003734543. [DOI] [PubMed] [Google Scholar]
- 15.Beels L, Werbrouck J, Thierens H. Dose response and repair kinetics of <gamma>-H2AX foci induced by in vitro irradiation of whole blood and T-lymphocytes with X- and <gamma>-radiation. International Journal of Radiation Biology. 86:760–768. doi: 10.3109/09553002.2010.484479. [DOI] [PubMed] [Google Scholar]
- 16.Coleman TF, Li Y. An Interior Trust Region Approach for Nonlinear Minimization Subject to Bounds. SIAM Journal on Optimization. 1996;6:418–445. [Google Scholar]
- 17.Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 Form a BRCA1 Protein Complex Required for the DNA Damage Response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch K, Wrona A, Dikomey E, Borgmann K. Impact of homologous recombination on individual cellular radiosensitivity. Radiotherapy and Oncology. 2009;90:265–272. doi: 10.1016/j.radonc.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 19.Kim H, Chen J, Yu X. Ubiquitin-Binding Protein RAP80 Mediates BRCA1-Dependent DNA Damage Response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 20.Andreassen CN. Searching for genetic determinants of normal tissue radiosensitivity - Are we on the right track? Radiotherapy and Oncology. 97:1–8. doi: 10.1016/j.radonc.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Zschenker O, Raabe A, Boeckelmann IK, et al. Association of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivity. Radiotherapy and Oncology. 97:26–32. doi: 10.1016/j.radonc.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Taneja N, Davis M, Choy JS, et al. Histone H2AX Phosphorylation as a Predictor of Radiosensitivity and Target for Radiotherapy. Journal of Biological Chemistry. 2004;279:2273–2280. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 23.Banath JP, MacPhail SH, Olive PL. Radiation Sensitivity, H2AX Phosphorylation, and Kinetics of Repair of DNA Strand Breaks in Irradiated Cervical Cancer Cell Lines. Cancer Research. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 24.Olive PL, Ban·th JP, Keyes M. Residual [gamma]H2AX after irradiation of human lymphocytes and monocytes in vitro and its relation to late effects after prostate brachytherapy. Radiotherapy and Oncology. 2008;86:336–346. doi: 10.1016/j.radonc.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 26.Moeller BJ, Arap W, Pasqualini R. Targeting Synthetic Lethality in DNA Damage Repair Pathways as an Anti-Cancer Strategy. Curr Drug Targets. doi: 10.2174/1389450111007011336. [DOI] [PubMed] [Google Scholar]
- 27.Rodemann HP. Molecular radiation biology: Perspectives for radiation oncology. Radiotherapy and Oncology. 2009;92:293–298. doi: 10.1016/j.radonc.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura AJ, Rao VA, Pommier Y, Bonner WM. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle. 9:389–397. doi: 10.4161/cc.9.2.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes & Development. 2011;25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huen MSY, Grant R, Manke I, et al. RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Capetillo O, Chen H-T, Celeste A, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown is a visual representation of the kinetics model used in this study. Growth and decay functions are combined to fit the γ-H2AX-IRIF data. IRIFs were assessed at the indicated time points after 1 Gy IR.
γ-H2AX and 53BP1 IRIF kinetics are compared for WT2, A-T, NBS, RNF168, PNKP, and RS73 LCLs. Both IRIF experiments were evaluated after 1 Gy IR.