Abstract
Amid researchers’ growing need for study data management, the CTSA-funded Institute for Translational Health Sciences developed an approach to combine technical and scientific resources with small-scale clinical trials researchers in order to make Electronic Data Capture more efficient. In a 2-year qualitative evaluation we found that the importance of ease of use and training materials outweighed number of features and functionality. EDC systems we evaluated were Catalyst Web Tools, OpenClinica and REDCap. We also found that two other systems, Caisis and LabKey, did not meet the specific user needs of the study group.
Keywords: Clinical trials, Electronic data capture, biomedical informatics
1.0 Introduction
The data collection process for clinical trials can be a tedious and error-prone process, and even a barrier to initiating small-scale studies. Electronic Data Capture (EDC) software can meet the need for faster and more reliable collection of case report forms (CRFs), but these informatics solutions can also be difficult for researchers to set up. One solution is a centrally supported Clinical Trials Management System (CTMS), which might feature not only EDC but also billing support and integration with a Master Patient Index. We investigated commercial CTMS options, but at the current time resource constraints at the University of Washington (UW) make establishing an institution-wide full-featured CTMS ecosystem an unrealistic short-term goal.
Additionally, we serve an audience wider than just UW. In 2007 a consortium of institutions including the University of Washington, Seattle Children’s, Fred Hutchinson Cancer Research Center and several other local and regional partners received funding under the NIH Clinical and Translational Science Awards (CTSA) to establish the Institute of Translational Health Sciences (ITHS). Our ITHS Biomedical Informatics Core (BMI) has a mandate to serve small-scale investigators at all institutions, including K scholars and internally mentored scientists, who lack funds to purchase commercial study management tools.
We have strong evidence that study data collection tools and informatics support are a valuable resource to these emerging scholars as well as other scientists. In an extensive qualitative study with over 100 local researchers, UW Information Technology and the UW eScience Institute found a strong need for data management expertise and infrastructure, including database design and management, data storage, backups, and security [1]. Researchers “do not want to spend much time finding data management solutions or solving technology support problems—they would rather spend their time doing research.” Due to lack of time or money to invest in alternatives, they often use general-purpose office applications, such as spreadsheets, that are poorly suited to data management [2].
Though we lack funding for an ITHS-wide commercial CTMS, we can connect researchers who have some data management funding with other local purchasers of commercial EDC software for discounts. However, even with discounts cost often limits the use of commercial EDC software to large studies, though several vendors have expressed interest in developing a support model for medium sized studies such as an NIH R01. In any case, at the present time commercial EDC software remains out of reach for small-scale studies at our institution.
To serve this group, BMI embarked on a “bottom up” approach by partnering with researchers to evaluate low-cost EDC systems. The evaluation periods overlapped through 2008-2009, but this paper is organized into distinct sections for each tool. At the end of the evaluation period we chose one system to offer as a resource, and will propose future work to extend and integrate this system.
2.0 Method
In 2008, we developed a concept map of EDC system features that facilitate translational research [3]. Based on these findings, we selected the subset of those concepts that were most applicable to small-scale clinical trials as criteria for our qualitative evaluation:
Availability of training materials and documentation
Ease of designing CRFs including automated error detection (edit checks)
Ability to create a patient visit schedule for data entry
Presence of site and user roles and permissions
Effort taken in exporting or importing data in a standards-compliant way
We began with an extensive list of EDC systems gathered from collaborators, referrals, and web searches. We narrowed the field by limiting our options to EDC software that is: 1) open source, free, or very low cost; 2) web-based on any platform to prevent the need for researchers to install software; and 3) accessible via an application programming interface (API) for integration with other data sources such as freezer inventory or instrumentation output. While not the focus of this paper, we are making use of these APIs to continue our lightweight approach to data integration [4]. We also expect to make use of these APIs for data exchange communication such as used in the caBIG Clinical Trials Suite [5].
Our narrowed list had seven systems. Of these, we discarded two that had no recent project activity, Visitrial [6] and TrialDB [7] and initially investigated two systems, Caisis [8] and LabKey [9], but the versions available at that time did not include a way for the study team to create their own CRFs. Therefore, we focused on the remaining three EDC systems, about which over the past two years we piloted and prepared a whitepaper [10]. These systems were Catalyst Web Tools from University of Washington Learning and Scholarly Technologies [11], OpenClinica from Akaza Research [12], and REDCap from Vanderbilt University and the REDCap Consortium [13].
We then established partnerships with local translational research teams to evaluate these systems in depth. These partnerships were ad hoc, based on a pragmatic need to evaluate multiple systems within our time and staffing constraints. While using all systems in parallel on a single study would have provided a more direct comparison, the duplicated effort would have been prohibitively difficult for both the study team and BMI. Also, piloting multiple heterogeneous studies provided more insight into the challenges that real-world clinical trial data pose in our CTSA research environment.
In the initial phase described in this paper, we provided the technical expertise to install an EDC system for each study team, configure backups and security, and set up the system for data collection. Study staff performed all data entry and provided us with valuable user insights. ITHS BMI performed a qualitative evaluation of each system for this paper.
3.0 Results
The following sections describe our experiences with these systems during our pilot studies.
3.1 Catalyst Web Tools
Catalyst Web Tools is an open source suite of web applications developed by the University of Washington primarily for internal support of teaching and research. It includes surveys, secure file management, content management, and project workspaces.
A big advantage in our environment is that Catalyst is well known to the local Institutional Review Board (IRB) and many researchers at both UW and local collaborating institutions. Catalyst is also financially and technically supported completely separately from BMI, and has extensive online tutorials and some in-person training courses.
BMI partnered with a local researcher conducting a prospective clinical trial with approximately 800 subjects at 3 sites. The initial skeleton workspace had basic project information and links to study protocol and documents, discussion area, forms, and an automatically generated list of current study team members. Our partner project study coordinator was already familiar with Catalyst and performed the setup of data collection forms.
However, we found that Catalyst lacked multiple features specific to clinical trials research. There is no support for automated scheduling of data entry events, so the study team had to manually keep track of followup visits. Additionally, each visit required collection of multiple forms but they could not be automatically linked together by visit. While approved for human subjects research, Catalyst lacks support for hiding or de-identifying sensitive data such as Protected Health Information (PHI). Catalyst is integrated with the UW campus single sign-on authentication system and allows approved outside users via Internet2’s Shibboleth [14], but does not have any support for managing multiple collection sites.
Exports are available at any time point in multiple formats, but form modifications created an unexpected level of complexity in the data exports. When the study team changed the wording of a question or available options, the Catalyst export would include an entire new export column for the updated wording. While this is understandable from the standpoint of data integrity, it required unnecessary manual edits for analysis and we would have preferred an option to allow single export columns for approved small updates to the data collection protocol. Importing data required developing a script to feed the data into Catalyst web services API.
In summary, we had high hopes for the simplicity and wide local knowledge of Catalyst, but found that its design makes it difficult to use for EDC. Additionally, although it is available as an open source download, to our knowledge the Catalyst Web Tools are not deployed at any other institution so there are limited opportunities for broader collaboration, a must in the CTSA environment.
3.2 OpenClinica
OpenClinica is an open source EDC system developed by Akaza Research. We installed OpenClinica on BMI servers located in a secure server room and performed backup and security updates ourselves. We evaluated the free community edition, but Akaza also offers paid commercial support and instructor-based training for an enterprise edition, including an optional regulatory compliance validation service.
Our initial pilot collaboration was with a mentored scholar who received an internal pilot grant to build a prospective disease registry, and later two research centers with more complex needs. OpenClinica is designed exclusively for EDC and has an impressive array of functionality allowing complex CRFs, but we also found that it has a very steep learning curve. Simply creating a new study requests completion of six sections of details. Some of the items such as a ClinicalTrials.gov unique identifier, while not required, indicated that the software was not intended for small internal projects like our initial pilot. Our other partners made more use of OpenClinica’s advanced features.
Learning to design CRFs was a significant hurdle. OpenClinica provides a blank template Excel file with four sheets of options including revision notes, sections and groups, and exact placement of questions and labels. It took a lot of trial and error to discern which columns were necessary. Previous revisions of CRFs continue to show up in the CRF administration interface but are marked as “removed.” During the learning process, creating a clean starting point required frequently rebuilding an empty database. Additionally, more complex edit checks required writing poorly documented XML rules files referring to internal OpenClinica Event OIDs such as “I_MALA_MALA11_2804”.
On the other hand, OpenClinica supports detailed linkage of forms with subject visits, including both scheduled followups and unscheduled occurrences such as adverse events. Each event has a table that clearly indicates which CRFs to complete. There is also a Subject Matrix giving a quick overview of which subjects and visits are complete, in progress, or not yet scheduled, though OpenClinica does not provide automated reminders to the study team.
OpenClinica has extensive site management, including contact details and allowing different versions of a CRF to be used at certain sites as might happen in a large study. It also has very granular permissions based on a user role, and can hide PHI from unauthorized users while still allowing access to other study data.
Data export in OpenClinica requires a cumbersome process of creating a dataset for specified events and CRFs, setting the longitudinal scope, and establishing filtering criteria before actually exporting the data. Again this would be potentially useful in a large study but was an annoyance for our small-scale pilot collaborations. Exports are formatted as Comma Separated Values (CSV), SPSS, or CDISC Operational Data Model (ODM) XML files. Imports can only be ODM XML files, and the study, subjects, and events must already exist in OpenClinica before data can be imported. However, OpenClinica version 3.0 and higher, which was released during our evaluation period, also supports a web services API, which can perform more flexible imports.
In all, OpenClinica was clearly the most powerful open source EDC system we piloted, though it is still missing features found in more expensive commercial EDC systems. However, the significant time investment required to create a study and CRFs made OpenClinica difficult to fit into our use case of supporting small-scale investigators with limited resources.
3.3 REDCap
REDCap (Research Electronic Data CAPture) is an EDC software package available to academic institutional partners of Vanderbilt University. It is not open source, and the license specifically prohibits using REDCap “as the basis for providing a contract service to any commercial (for profit) entity.” However, there is no charge for academic use of REDCap. Our Office of Technology Transfer was happy to execute the license agreement, especially after we mentioned the costs associated with licensing commercial EDC systems. Support for REDCap comes from the REDCap Consortium, which is led by Vanderbilt University but has various working group participants from a wide number of member institutions (including several CTSA sites) and an active email list. There is currently no paid enterprise support option; a study wanting to undergo regulatory compliance validation would need to perform the work themselves, though a working group has identified potential technical configurations of REDCap, which along with standard operating procedures could accomplish 21 CFR 11 or FISMA compliance.
As with OpenClinica, we installed and configured REDCap on BMI servers and performed backups and security updates. Our pilot partner study for REDCap was an NIH R21 Exploratory/Developmental Grant collecting data on approximately 200 subjects in Cairo, Egypt, with analysis taking place at UW Seattle and Stanford University. A colleague referred the principle investigator to REDCap because of its secure file repository feature, which allows easy uploading and sharing of any file.
The biggest strength of REDCap is the copious amount of training materials: prerecorded webinars, online tutorials, and help documentation, most of which is integrated into the software itself. For example, when using the web-based CRF designer, there is a link at the top of the page to a 5-minute video on Field Types and near the choice and skip logic sections there are help links titled “How do I manually code the choices?” and “How do I use this?” Additionally, a “REDCap Demo Database” includes example forms to highlight available functionality. New REDCap projects begin in “Development” mode, which allows changes to forms and events, but after switching to “Production” mode any changes must be approved by a REDCap administrator. REDCap supports somewhat complex skip logic with some limitations; for example, you cannot hide an entire CRF. In our initial pilot project, a graduate student statistician on the study team set up the forms so that he could code each response how he wanted.
REDCap has the ability to create patient visit schedules based on days offset for each event, which can then be shown on an online project calendar and edited for each subject as needed. However, REDCap does not alert the study team of upcoming scheduled visits; each staff member must visit the REDCap installation to see the calendar. The event module is somewhat confusing in that recurring or unscheduled events still have some arbitrary “days offset” defined. Weeks, months, and years also must be converted to days, and any studies with multiple events per day (such as with infants) have to work around the “days” nomenclature. The REDCap development team has identified this as an area for improvement.
REDCap has extensive support for user and group roles and permissions. While the instructional materials mention groups as a way to set up multi-site studies, there is no functionality specific to managing sites other than the group names. Each REDCap project has its own distinct user and group rights, and once a user is approved to use REDCap the study team can assign permissions on their own. Reading, editing, or viewing CRFs can be assigned on a form-by-form basis to individual users or groups, as can access to data points marked as PHI and each of sixteen different modules such as data exports, file repository, reports, and so on. Each REDCap project shows a matrix summary of permissions of current users. Like the online CRF editor, the user rights module has links to short tutorial videos and online help.
Data exports are very straightforward in REDCap. There is a button to select every field in the database for very quick data access, or a large number of options for selecting individual forms or data points and various export de-identification options such as removing fields tagged as PHI and shifting dates. Upon completing the export, the user is also given a configurable “citation notice” requesting that any publications cite the support of our CTSA grant as well as the REDCap project. Exported data automatically comes in SPSS, Excel/CSV, SAS, R, and STATA formats including both data and labels and option values. Imports are done using a CSV import template, though events for imported data must be assigned manually. REDCap version 3.3 marked the formal release of an API, which allows more flexibility in importing and exporting data.
The emphasis in REDCap on ease of use and quick turnaround worked very well in our environment. While the licensing creates additional complexity in potential use cases, we believe it is an appropriate risk in order to provide a useful informatics tool for translational researchers.
4.0 Discussion
When we began our pilot process, we assumed that the EDC functionality present in each system would be the most important aspect. Therefore our whitepaper mentioned above provides details about each system’s ability to organize visit and event based data, the range of data entry widgets available, support for complex edit checks and skip patterns, and so on.
However, as we worked with various study teams it became clear they were willing to work around limitations in the EDC software. On reflection, this makes sense as many small-scale investigators use paper forms with data points transcribed into spreadsheets as the best available data collection method that they can afford. Any web-based EDC software is a step up. Therefore for our qualitative evaluation we focused on the relative ease of use of the functionality that was present.
In early 2010, BMI’s data management decided on REDCap as the preferred EDC software to support small-scale studies, and added a description to the resource catalog on the ITHS website. Since then usage has steadily increased. As of May 2011 there were 164 active REDCap users and 30 production studies at the University of Washington, Seattle Children’s, Fred Hutchinson Cancer Research Center, and Bastyr University, with collaborators from many other institutions.
REDCap had a very clear advantage due to its extensive tutorials and online training materials. OpenClinica has more functionality, for example in complex CRF design and site management, but there is less documentation and what is available is written in technical language. We plan to continue supporting OpenClinica for our existing pilot users but for new studies will focus our available staff resources on REDCap. The limitations of Catalyst Web Tools are too great to manage a clinical trial, though could be used for some parts of a trial such as initial screening.
Since we began this project we have learned that serving a variety of sizes of studies is a common need. Several CTSA-funded institutions, including Vanderbilt University, support REDCap as an entry-level EDC solution but also recommend various custom or commercial EDC systems where appropriate.
Post-evaluation, in addition to maintaining the ITHS BMI installation of REDCap we are concentrating on future work in two areas: partnerships with investigators to enhance the local usage of REDCap, and informatics research to solve problems in data integration and interoperability.
Our top priorities in local partnerships are developing a local user group, where “power users” can assist others in form design and other issues, and creating a local library of form templates to supplement the REDCap Consortium’s Shared Library of validated instruments. We have also held in-person REDCap practice sessions.
ITHS BMI has a longstanding involvement with informatics research on ontologies and data integration. Most recently, we have contributed to the Ontology of Clinical Research (OCRe) project which seeks a “systematic description of, and interoperable queries on, human studies and study elements” [15]. We are developing tools to bring OCRe-defined study elements into REDCap as a foundation to improve interoperability and study design efficiency. Additionally through our i2b2 Cross-Institutional Clinical Translational Research (CICTR) project we have identified use cases for moving data between REDCap and i2b2, which in our installation contains only high-level anonymized descriptive characteristics of population-level data [16]. Lastly, in keeping with our “bottom up” philosophy we are applying lightweight data integration techniques to query across REDCap and other low cost systems, such as a freezer inventory management system we have developed.
Fig. 1.
Catalyst Web Tools from the University of Washington.
Fig. 2.
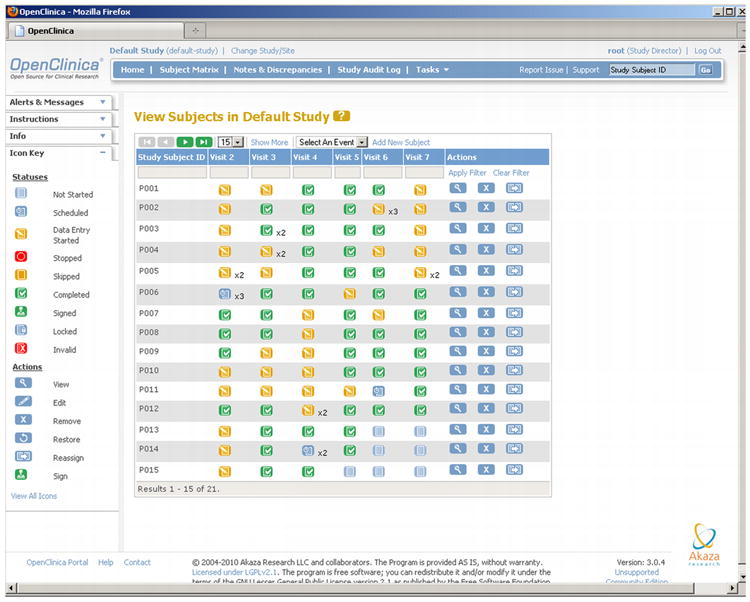
Openclinica from Akaza Research.
Fig. 3.
REDCap from Vanderbilt University and the REDCap consortium.
Table 1.
Qualitative evaluation of UW Catalyst Web Tools
| Criteria | Rating |
|---|---|
| Training materials and documentation | Good |
| Ease of designing CRFs including edit checks | Good |
| Create a patient visit schedule for data entry | Poor |
| Site and user roles and permissions | Fair |
| Effort taken in exporting or importing data | Poor |
Table 2.
Qualitative evaluation of OpenClinica from Akaza Research
| Criteria | Rating |
|---|---|
| Training materials and documentation | Fair |
| Ease of designing CRFs including edit checks | Poor |
| Create a patient visit schedule for data entry | Good |
| Site and user roles and permissions | Good |
| Effort taken in exporting or importing data | Poor |
Table 3.
Qualitative evaluation of REDCap from Vanderbilt University
| Criteria | Rating |
|---|---|
| Training materials and documentation | Good |
| Ease of designing CRFs including edit checks | Good |
| Create a patient visit schedule for data entry | Good |
| Site and user roles and permissions | Fair |
| Effort taken in exporting or importing data | Good |
Table 4.
Summary of Qualitative evaluation
| Criteria | Catalyst | OpenClinica | REDCap |
|---|---|---|---|
| Training materials and documentation | Good | Fair | Good |
| Ease of designing CRFs including edit checks | Good | Poor | Good |
| Create a patient visit schedule for data entry | Poor | Good | Good |
| Site and user roles and permissions | Fair | Good | Fair |
| Effort taken in exporting or importing data | Poor | Poor | Good |
Acknowledgments
Funded by ITHS grants UL1 RR025014, KL2 RR025015, and TL1 RR025016, and NLM training grant T15LM007442. Additionally we wish to acknowledge our pilot partner investigators and their grants: de Boer, Ian R01HL096875, Landau, Ruth NCT00799162. Ringold, Sarah, Seattle Children’s Research Institute CCTR mentored scholar. Stevens, Anne, R01 AR051545. Walson, Judd, FWA00006878. Wennberg, Richard R21HD060901.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lane C, Fournier J, Lewis T. Scientific advances and information technology: meeting researchers’ needs in a new era of discovery. EDUCAUSE Center for Applied Research, Case Study 3. 2010 available from http://www.educause.edu/ecar.
- 2.Anderson N, Lee ES, Brockenbrough JS, Minie ME, Fuller S, Brinkley JF, Tarczy-Hornoch P. Issues in Biomedical Research Data Management and Analysis: Needs and Barriers. J Am Med Inform Assoc. 2007;14:478–488. doi: 10.1197/jamia.M2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidry AF, Brinkley JF, Anderson NR, Tarczy-Hornoch P. Concept mapping to develop a framework for characterizing electronic data capture (EDC) systems. Proc AMIA Symp. 2008:960. [PubMed] [Google Scholar]
- 4.Detwiler LT, Sucui D, Franklin JD, Moore EB, Poliakov EV, Lee ES, Corina DP, Ojemann GA, Brinkley JF. Distributed XQuery-based integration and visualization of multimodality brain mapping data. Front Neuroinform. 2009;3(2) doi: 10.3389/neuro.11.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speakman J. The caBIG Clinical Trials Suite. In: Ochs MF, Casagrande JT, Davuluri RV, editors. Biomedical Informatics for Cancer Research. Springer; 2010. pp. 203–213. [Google Scholar]
- 6. [March 1, 2011];Archive of Visitrial website. 2007 Available from: http://web.archive.org/web/20070329221856/http://devctr.visitrial.com/
- 7.TrialDB. [March 1, 2011];website. 2007 Available from: http://ycmi.med.yale.edu/trialdb/
- 8.Caisis. [March 1, 2011];2011 Available from: http://www.caisis.org/
- 9.LabKey. [March 1, 2011];2011 Available from: http://www.labkey.com/
- 10.Oldenkamp P. Guide to low cost electronic data capture systems for clinical trials. Seattle Children’s Research Institute and Institute for Translational Health Sciences. 2010 Available from http://sigpubs.biostr.washington.edu/archive/00000249/
- 11. [March 1, 2011];University of Washington Catalyst Web Tools. 2011 Available from http://www.washington.edu/lst/web_tools.
- 12. [March 1, 2011];OpenClinica from Akaza Research. 2011 Available from: https://www.openclinica.com/
- 13.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) -- a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009 Apr;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. [March 1, 2011];Shibboleth System from Internet2. 2011 Available from: http://shibboleth.internet2.edu/
- 15. [March 1, 2011];The Ontology of Clinical Research (OCRe) 2010 Available from: http://rctbank.ucsf.edu/home/ocre.
- 16.CICTR, Cross-Institutional Clinical Translational Research. [March 1, 2011];2010 Available from: http://www.i2b2cictr.org/


