Abstract
The hypothesis that host prion protein (PrP) converts into an infectious prion form rests on the observation that infectivity progressively decreases in direct proportion to the decrease of PrP with proteinase K (PK) treatment. PrP that resists limited PK digestion (PrP-res, PrPsc) has been assumed to be the infectious form, with speculative types of misfolding encoding the many unique TSE agent strains. Recently, a PK sensitive form of PrP has been proposed as the prion. Thus we re-evaluated total PrP (sensitive and resistant) and used a cell-based assay for titration of infectious particles. A keratinase (NAP) known to effectively digest PrP was compared to PK. Total PrP in FU-CJD infected brain was reduced to ≤0.3% in a 2hr PK digest, yet there was no reduction in titer. Remaining non-PrP proteins were easily visualized with colloidal gold in this highly infectious homogenate. In contrast to PK, NAP digestion left 0.8% residual PrP after 2hr, yet decreased titer by >2.5logs; few residual protein bands remained. FU-CJD infected cells with 10x the infectivity of brain by both animal and cell culture assays were also evaluated. NAP again significantly reduced cell infectivity (>3.5 logs). Extreme PK digestions were needed to reduce cell PrP to <0.2%, yet a very high titer of ≥7.8 logs remained. Our FU-CJD brain results are in good accord with the only other report on maximal PrP digestion and titer. It is likely that one or more residual non-PrP proteins may protect agent nucleic acids in infectious particles.
Keywords: proteinase K, keratinase, Transmissible Spongiform Encephalopathies, scrapie, infectious particles, agent strains, cell culture
Transmissible Spongiform Encephalopathies (TSEs) include endemic sheep scrapie and epidemic BSE, human kuru, and rapidly spreading cervid TSEs. These endemic and epidemic infections continue to present major public health and economic problems. According to the protein-only prion hypothesis, a misfolded form of host prion protein (PrP) is the causal infectious agent [Prusiner, 1998]. Misfolded PrP was first identified on gels as a 27-29kd protein that survived limited proteolysis with proteinase K (PK) in infected brain fractions. This resistant molecular and presumably infectious form is called PrP-res or PrPsc. PrP-res was simultaneously identified ultrastructurally in scrapie and CJD preparations as infection-associated amyloid fibrils [Diringer et al., 1983; Merz et al., 1983]. Infectious fractions with no PrP fibrils, but only fluffy PrP aggregates, rapidly formed amyloid fibrils upon exposure to PK [Manuelidis et al., 1989]. Although it is often stated that PrP amyloid fibrils are a consequence of detergent treatment, masses of such fibrils have been identified ultrastructurally inside fixed infected cells not treated with detergents [Manuelidis et al., 2007]. PrP amyloid fibrils are also the major, but not sole constituent of end-stage extracellular PrP plaques in infected brains with vacuolar neurodegeneration and reactive glial changes [Manuelidis et al., 1997].
Substantial and often ignored evidence indicates that PrP-res is part of a pathological response to infection, rather than the causal agent that incites disease [Manuelidis, 2003; Manuelidis, 2007]. Many viruses require a specific host protein for entry and replication, and infectious TSE particles similarly require host PrP for effective spread from cell to cell [Manuelidis et al., 2009a]. Merely increasing PrP, even from a different species, enhances transmission of foreign TSE agents [Manuelidis et al., 2009a; Manuelidis et al., 2009b]. Diverse published data that are inconsistent with an infectious form of PrP include, but are not limited to: i) the variety of unique and mutable agent strains, a property of nucleic acid not protein [Kimberlin et al., 1989; Manuelidis et al., 1997]; ii) TSE agents breed true in various tissues, cell cultures, and cross-species transmissions whereas PrP patterns do not [Manuelidis et al., 2009b; Arjona et al., 2004; Nishida et al., 2005]; iii) a preventable environmental source of infection in which epidemic outbreaks disappear after infectious material is removed (as in BSE and kuru) strongly implicates a foreign source of infection rather than a “spontaneously generated” host PrP self-conversion into an infectious form [Colby and Prusiner, 2011; Prusiner, 1998]; iv) infection elicits early innate immune responses that indicate host recognition of an invading foreign entity and these responses, not educed by host PrP-res, occur well before PrP-res begins to accumulate [Lu et al., 2004] and can be protective [Sethi et al., 2002]; v) microglia with barely detectable PrP, and no PrP-res, contain high levels infectivity [Baker et al., 2002]; vi) all detectable forms of PrP are digested in the gastrointestinal tract yet the invasive infectious particle (as many conventional viruses) is not destroyed [Jeffrey et al., 2006; Scherbel et al., 2007]. Finally, vii) infectious particles of viral size (~25nm diameter) with protected nucleic acids can be separated from the majority of host PrP and other proteins, and GdnSCN disruption of these particles into protein and nucleic acid components reduces infectivity by >99.8%. Some of these released nucleic acids have been sequenced such as capsid-protected endogenous retroviral RNAs, mitochondrial DNA, and newly discovered circular Sphinx DNAs of ≥1.8kb that have large regions that are not in the database [Akowitz et al., 1994; Manuelidis, 2010].
Most investigators who work with TSE agents concentrate on PrP, and do not evaluate nucleic acids in their infectious fractions using modern molecular techniques such as PCR [Manuelidis, 2010]. Additionally, there are few reports on proteins other than PrP in more purified preparations with high levels (titers) of infectivity. This is surprising because a “protein X” has long been postulated to be required for conversion of non-infectious PrP to infectious PrP-res [Telling et al., 1995]. Therefore a fundamental re-examination of infectious titer with respect to all forms of PrP, as well as other proteins, was warranted. Previous experiments in our laboratory uncovered proteins that are more resistant to PK digestion than PrP, including endogenous retroviral capsids [Akowitz et al., 1994], and highly infectious agent particle preparations additionally revealed nucleic acid binding and other proteins when separated from most pathologic PrP in gradients of CJD infected hamster brain lysates [Manuelidis et al., 1995]. Essentially all infectivity was similarly recovered from 22L scrapie infected N2a cell culture lysates in a comparable gradient fraction with low PrP. These N2a cells, unlike brain, displayed no complicating degenerative changes [Sun et al., 2008].
The original report claiming PrP-res is directly proportional to infectious titer was based on PK digestion of 263K scrapie infected hamster brain samples pretreated with PK at 4°C, and then gradient purified [McKinley et al., 1983]. The percent of total starting brain infectivity recovered in the PrP-res enriched fraction was not reported in that paper. After 4hrs incubation with 100μg/ml PK at 37°C, the radioiodinated gradient protein, presumed to be 100% PrP-res, was no longer visible, and a <3 log protein loss yielded a 6 log loss of infectivity. In another plot (their Fig. 2) the reverse was seen; an ~4 log reduction of iodinated protein yielded only a ≤2 log loss of infectivity. With the development of antibodies specific for PrP, a more precise analysis of PrP is possible. Remarkably, we could find only one other titration of an apparently complete PrP digestion. This was done with a similar 263K scrapie PrP-res brain fraction, but showed only a very small 0.5 log reduction in infectivity that accompanied the disappearance of all PrP forms [Suzuki et al., 2008]. Because subcellular brain fractions typically contain ≤15% of starting infectivity, and precipitation and detergent treatments can affect enzyme accessibility to molecular aggregates, we reevaluated infectious titers with respect to total PrP, PrP-res, and residual minor proteins after digestion. To rule out preparative artifacts we tested PK digestion on total brain and cell homogenates from high titer FU-CJD mouse brain and FU-CJD infected GT1 cells by quantitative Western blotting. Infectivity was titrated by a rapid and verified incubation end-point cell culture assay [Liu et al., 2008; Miyazawa et al., 2011]. This study of total PrP has become most pertinent because the newly modified prion proposal now posits that only a possibly minor and still uncharacterized PK-sensitive form of PrP is the real infectious entity [Colby and Prusiner, 2011; Cronier et al., 2008].
Fig. 2.
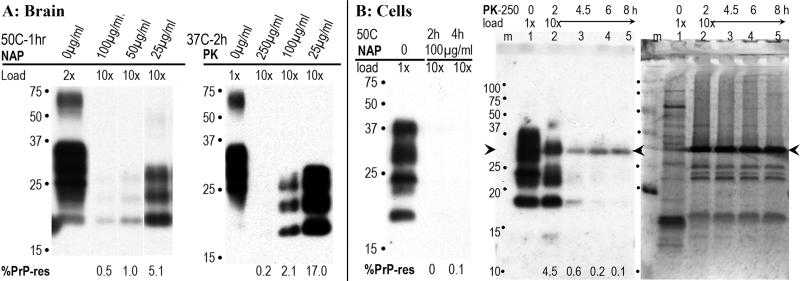
Representative digestion of brain (A) and cells (B) under different digestion conditions. In brain, different concentrations of NAP were tested for 1hr and compared with no enzyme controls (0μg/ml). At 100μg/ml 99.5% of the PrP was digested and can be seen as faint bands in the 10x gel loaded lane. A 2hr incubation with NAP did not increase digestion of PrP (data not shown). With PK, 99.8% digestion of brain PrP required 250μg/ml for 2hr. Panel B shows NAP digested almost all the PrP of FU-CJD cells at 2hr, i.e., was slightly more effective than in brain. Longer 4h digests did not further reduce the PrP (0-0.1% by photon counting) as noted. In contrast to brain, 250μg/ml PK left 4.5% PrP-res at 2hr, and longer incubations were tested as shown. It required 6-8hr to digest PrP to 0.1-0.2%. The PK enzyme in this blot bound PrP antibody (arrowhead) and corresponds to the PrP band seen with colloidal gold staining. Remarkably, exhaustive PK digestion left many obvious non-PrP protein bands between 15-29kd. The 6 and 8hr PK cell digests shown here were titered for infectivity.
In parallel experiments we also compared the effect of keratinase digestion on PrP and infectivity. Keratinases, produced by yeast and bacteria, are very effective in solubilizing highly ordered proteins and β-sheet structures including feathers. They have been shown to reduce PrP-res to undetectable levels, even when dried on plastic [Yoshioka et al., 2007]. We found no reports on TSE titers after keratinase digestion, and therefore tested a keratinase that was chromatographically pure and capable of PrP digestion under non-extreme pH and temperature conditions [Mitsuiki et al., 2006]. Because the GT1 indicator cell line used to titrate infectivity is susceptible to many different mouse-adapted TSE agents, including human derived vCJD, kuru, and FU-CJD, as well as sheep derived 22L, RML and 263K scrapie agents, they also provide a base for comparing resistance properties of these unique agent strains in a tightly controlled setting [Manuelidis et al., 2009a; Manuelidis et al., 2009b; Nishida et al., 2005],.
Materials and Methods
FU-CJD brain and cell homogenate preparation
Mouse passaged FU-CJD infected brains were collected from clinically ill CD-1 mice and rapidly frozen at -80°C. Frozen brains (1-2gm total) were subsequently pooled, homogenized to 10% (wt/vol) in cold phosphate-buffered saline (PBS) and aliquots of the 10% homogenate immediately frozen at -80°C until use. A gram of brain or of monotypic cells contains ~109 cells, and a 10% brain homogenate (10-1 dilution) contains 108 cells/ml [Arjona et al., 2004; Liu et al., 2008]. Thus 30μl of a 1% homogenate, the standard maximal intracerebral mouse dose, contains 3e5 cell equivalents (CE). CE are used here to facilitate direct comparisons of infectivity as well as PrP-res in brain and cells homogenates. Infected and control GT1 cells were counted and homogenized by syringing through progressively smaller bore needles and the total cell lysate was then sonicated for thorough dispersal. Standard protein assays were used to confirm cell counts.
Keratinase (NAP) and proteinase K (PK) digestions
NAP is an alkaline serine protease purified from a Bacillus strain and was a generous gift of S. Mitsuiki [Mitsuiki et al., 2006]. It has an activity of 1,000-1,500 units/mg. PK (EMD-Merck powder from Tritirachium album) was assessed on the parallel aliquots. A series of experiments were done to determine minimal enzyme and optimal buffer and detergent conditions necessary to digest total PrP by ~3 logs (99.8%) as assessed by quantitative Western blotting. In typical brain experiments, 3μl of a 10% FU-CJD brain homogenate (3e5 CE) was digested with each enzyme at different concentrations and times as representatively shown in the results. We used a pH of 9.6, less than the optimal pH of 10.5 for NAP. Previous studies showed a pH of 9 rapidly disaggregated all visible hamster brain PrP so that it became completely sensitive to PK digestion; this pH also simultaneously elevated CJD titers [Sklaviadis et al., 1989]; a pH of 10.5 was not evaluated because it could reduce infectivity. We also used 50°C incubations to preserve infectivity rather than the NAP temperature optimum of 60-70°C. Even though TSE agents are often claimed to be unaffected by temperatures of over 100°C, some agents can lose substantial titer at temperatures ≥70°C [Cronier et al., 2008; Somerville et al., 2011]. Detergents including Triton X-100, NP-40, and sarkosyl all enhanced NAP as well as PK activity to a similar extent (data not shown); all digestions for parallel infectivity titrations were done simply with 0.1% Triton X-100.
Parallel PrP Western blot and infectivity assays
For comparative protein and infectivity studies we digested samples with NAP at 100μg/ml in freshly prepared optimal NAP buffer (100mM TrisCl pH9.6, 0.1% Triton X-100) for 2 hours at 50°C. PK was most effective at 250μg/ml in 100mM TrisCl pH7.5, 0.1% Triton X-100 whereas maximal preservation of PrP-res was obtained using PK at 25μg/ml for 30min at 37°C. Untreated and digested aliquots were analyzed on standard Western blots as previously with a commercial goat anti-PrP antibody (M20, Santa Cruz Biotechnology, Santa Cruz, CA) for the detection of total PrP and PrP-res [Arjona et al., 2004; Manuelidis, 1998; Manuelidis and Fritch, 1996]; ChemiGlow (Alpha Innotech) was used for quantitative photon signal detection and spanned a linear range of >3 logs as previously with Fluorochem software. Loads of 10x the undigested samples as well as long film exposures further confirmed the reductions in total PrP and PrP-res. Colloidal gold staining of blots was done after antibody detection.
Infectivity assays were performed as detailed [Liu et al., 2008], using a standard reference end-point dilution curve previously determined in detail for dilutions of the FU-CJD agent in mouse brain and in GT1 cell homogenates over 13 sequential passages [Miyazawa et al., 2011]. Both brain and cells gave the same best-fit curves. Briefly, control and digested samples with known CE were serially diluted with feeding medium and then applied to target GT1 cells plated in replicate wells and passaged to determine the tissue culture infectious dose (TCID). De novo production of PrP-res by GT1 target cells indicates positive infection. For PrP-res readout, 5-10x the standard target cell load was applied for Western blot quantitation as indicated above lanes. When very high loads (20x) were required to show minute amounts of de novo PrP-res, low speed cytoplasmic supernatants rather than whole cell homogenates were used to minimize nuclear viscosity artifacts. FU-CJD titers were previously verified by standard long LD50 animal assays and compared with the rapid TCID end-point agent replication culture assay for infectious particles; TCID were only 2-3 fold less sensitive than the long LD50 animal assay of the FU-CJD agent [Liu et al., 2008; Miyazawa et al., 2011].
Results
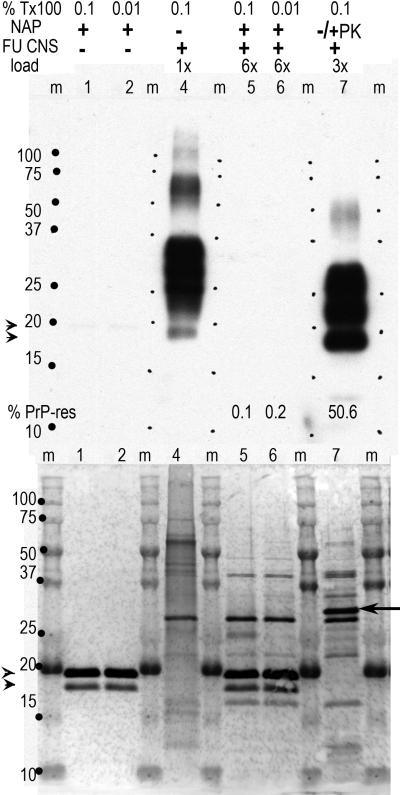
We tested the effectiveness of keratinase (NAP) under various conditions as specified in the methods. Fig. 1 shows a representative gel blot of PrP antibody binding (top) with the colloidal gold stain to detect other proteins beneath it. Because silver stains of gels show a marked preference for PrP, colloidal gold was used to detect minor proteins that may be missed. Two concentrations of Triton X-100 (Tx100) were applied to FU-CJD brain homogenates prior to enzyme treatment. Controls showed NAP does not bind the PrP antibody used here, as shown in lanes 1 and 2; the NAP however was clearly visualized as two close bands of slightly less than 20kd in the corresponding gold-stained lanes. No other proteins are visible in the gold stained NAP-only lanes. As in all standardized experiments, 3e5 CE were loaded for the 1x undigested controls so that both total PrP (including PrP that is PK sensitive) and PrP-res from the same sample are visualized. Digestion for 2hrs with NAP (lanes 5 & 6) yielded invisible total PrP even with a 6x load (1.8e6 CE); photon capture however, showed a slight residue of PrP-res with 0.01% Tx100 (0.2 vs 0.1% PrP-res). As in all previous TSE experiments, the % PrP-res is defined as the fraction of total PrP that remains, and PrP-res bands are of lower Mr than the starting PrP because the NH2 terminus is digested as previously demonstrated [Arjona et al., 2004]. Increasing Tx100 to 1% did not further increase NAP digestion of PrP and hence we chose 0.1% as the standard detergent treatment. FU-CJD brain homogenates treated with PK at only 25μg/ml for 30min at 37°C with 0.1% Tx100 showed 50.6% residual PrP-res (lane 7). This amount is the same as found with previous standard digestions of FU-CJD brain where NP40-DOC or sarkosyl rather than Tx100 was used [Arjona et al., 2004; Manuelidis, 1998]. Even with virtually complete PrP digestion by NAP, a few residual host proteins, in addition to the NAP bands can be seen, as shown in the colloidal gold stained lanes 6 and 7. The gold stained partially digested PK lane also shows other residual protein bands unrelated to PrP. These are different than those seen with NAP. The added PK is the sharp 29kd band at arrow.
Fig. 1.
Digestion of FU-CJD brain homogenate (CNS) with NAP (100μg/ml pH9.6, 50°C for 2hr) at different Tx100 concentrations as indicated. Top blot shows PrP antibody binding bands and bottom shows the same blot stained with colloidal gold for total proteins. Relative sample gel loads are indicated. Control lanes 1-2 are NAP only, lane 4 is undigested homogenate, lanes 5-6 are homogenate with NAP and lane 7 contains homogenate treated under standard low PK conditions (25μg/ml at 37°C for 30 min) to reveal the maximal PrP-res. The % PrP-res is indicated under each lane. PrP-res is invisible in the NAP digested lanes, whereas it is 50.6% in the PK lane. The gold stain reveals no other material in the NAP only lanes. In lane 4 (undigested brain) there are many overlapping proteins whereas after NAP digestion few bands are seen, as well as the two NAP bands (arrowheads). After limited PK digestion, even at lower sample loads, there are many residual bands in addition to the added PK (at arrow). Marker proteins and lanes (m) are indicated.
The effectiveness of NAP and PK digestions on brain and infected monotypic cells differed. Fig. 2 shows PrP-res after incubation with different concentrations of each of these enzymes on brain homogenates (panel A). Control incubations for 1h at 50°C without NAP (0μg/ml lane) gave no change in the profile of total PrP (compare with Fig. 1, lane 7). PrP-res was increasingly digested with concentrations from 25 to 100μg/ml NAP, and even at 100μg/ml, 0.5% residual PrP-res was still present. Higher concentrations of NAP did not abolish the PrP-res signal. This residue can be seen in the 10x loaded lane. Even with 2hr NAP incubations, similar residual amounts up to 0.8% PrP-res were reproducibly seen in independent brain homogenates. Thus NAP did not completely digest PrP. According to infectious prion protein predictions, such samples should have a reduction in titer of ~2 logs. The amounts of PK needed to digest >99% of brain PrP after a 2hr incubation at 37°C are shown in panel A. At 100μg/ml, PK still left a strong signal of 2.1% PrP-res whereas a 250μg/ml PK consistently reduced total PrP by >99.5%. This would be expected to reduce titer by close to 3 logs.
FU-CJD cell homogenates displayed no residual PrP with NAP at 2hr (Fig. 2B). In longer 4hr incubations with NAP, 0.1% PrP-res was detected by photon counting, and this low value may not represent a real or accurate PrP residue, i.e., the Western blot antibody assay consistently achieved a span of 3 logs but values of 0.1-0% could not be reproducibly distinguished in repeat experiments. More importantly, a dramatic difference in PrP susceptibility to PK was reproducibly identified in infected cells as compared to brain. In contrast to brain homogenates, very long incubations were required to reduce PrP >99% in infected cells. For reference, FU-CJD cells have 25% maximal PrP-res, yet produce 10x higher infectious titers than FU-CJD brain with ~55% PrP-res [Miyazawa et al., 2011]. As shown in Fig. 2B (lane 2), after a 2hr incubation with 250μg/ml PK, 4.5% PrP-res remained and was easily visualized with PrP antibody, in addition to the strong PK band at 29kd (arrowhead) that can non-specifically bind PrP antibodies [Sklaviadis et al., 1989] . This PrP residue was 22x greater than the PrP-res remaining in brain after digestion with 250μg/ml PK for 2hrs. With increasing incubation times, as shown in lanes 3-5, total GT1 cell PrP could be reduced to <0.2%. The corresponding colloidal gold stained blot of these cells treated with PK is also shown in Fig. 2B. Notably, there are many protein bands of 16-25kd after this stringent PK digestion, and these proteins are unrelated to either PrP or PK. Similar non-PrP bands between 16-25kd were also seen in PK digested brain (data not shown), but were not seen after the NAP digestions. PK digested cell samples shown in this blot were also assayed for infectivity.
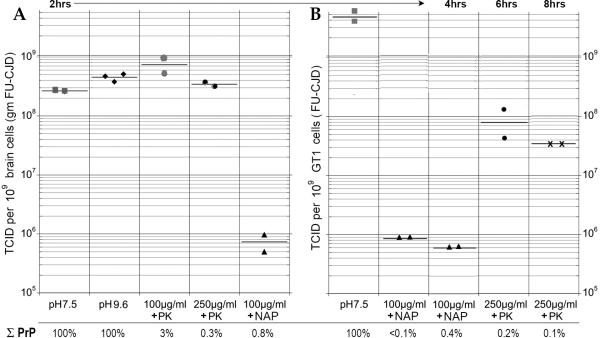
Fig. 3 shows the tissue culture infectious dose (TCID) of starting samples incubated in control buffers and parallel aliquots after PK and NAP digestion. The total remaining %PrP that was detectable, as determined from Western blots of these samples, is noted below the graph bars. Brain homogenate without incubation in Tx100 had 4e8 TCID/gm, reconfirming our rapid cell assay that showed only a 2-3 fold less sensitive readout than the FU-CJD animal assay of 1e9 LD50/gm, equivalent to 1 LD50 per brain cell [Arjona et al., 2004; Manuelidis, 1998]. Panel A shows that TCID of brain aliquots were unchanged after a 2hr 37°C incubation at pH of 7.5, (2.9e8 TCID). Total PrP in the undigested control was also unchanged (100%). Similarly, the control incubation at 50°C in pH 9.6 buffer, as used for NAP digestions, also failed to significantly alter infectivity (4.5e8 TCID) or to reduce PrP. The slight elevation of titer at the higher pH is consistent with previous observations of enhanced disaggregation of infectious particles at a pH of 8.9 [Sklaviadis et al., 1989]. Surprisingly, digestion of PrP with 100 μg/ml PK in the sample with a markedly reduced PrP residue of 3%, displayed an even higher titer (7.1e8 TCID), again suggesting a disaggregation effect with reduction of PrP amyloid aggregates. The prion prediction for this sample should have shown a significant titer reduction of ~2logs. Reduction of PrP to 0.3% in brain samples treated with 250μg/ml PK also failed to significantly reduce titer (3.5e8 TCID) as plotted in panel A. This data implicates residual PK resistant molecules other than PrP are essential components of infectious TSE agents. In contrast to the PK digests, the residual NAP non-PrP proteins are unlikely to be critical components of the infectious particle. Brain homogenates digested with NAP showed a marked and significant 2.5 log reduction of infectivity even though more PrP (0.8%) remained in the NAP digested sample than in the 250μg/ml PK treated sample. This data further challenges the claim that either a sensitive or a resistant form of host PrP is proportional to TSE agent titer.
Fig. 3.
Tissue culture infectious doses (TCID) determined from at least 2 serial dilutions at sequential passages in replicate or triplicate wells where each point represents the well average for two readouts. The % PrP remaining (of untreated homogenates) is shown beneath each condition of incubation and pH. FU-CJD brain homogenate titers in control and enzyme treatment are shown in panel A, and FU-CJD cell homogenates are shown in panel B. Note there is no significant reduction in TCID when PrP is reduced by 99.7% whereas the NAP sample with more residual PrP reduced titer by ≥2.5 logs. In cells (with a 10 fold greater titer than brain), NAP reduced TCID by >3.5 logs. However, PK, even at 6hr and 8hr incubations, yielded only a ≤2 log titer reduction with an ~3 log reduction of PrP. The TCID readout for the FU-CJD agent is based on previously detailed standard end-point curves that were verified by animal inoculation [Miyazawa et al., 2011]
Panel B shows parallel aliquots of whole FU-CJD cell homogenates subjected to NAP and PK digestion. Since these cells reproducibly contain 10-fold more infectious particles than FU-CJD brain [Arjona et al., 2004; Manuelidis, 1998], the starting culture material incubated for 2hr contains 4.5e9 TCID. NAP digestions at both 2hrs and 4hrs yielded a titer reduction of >3.6 logs. Notably, the residual PrP in the 4hr digest was clear (0.4% PrP-res) yet this sample had slightly less infectivity than the 2hr digest with undetectable PrP. This data further undermines the presumption that any PrP form is the real infectious entity. PK digestions of infected cells further undermined the assumption of a minor form of infectious PrP. Incubations with 250μg/ml PK at 6hr and 8hr, the conditions required to reduce PrP to ~ 0.1%, (3 logs) were much less effective at reducing titer than the NAP digestions of this sample that left a larger PrP residue. These long maximal PK digestions did reduce titer by 1.6-2.1 logs, unlike the no titer change found in 2hr brain incubations with PK. Most remarkable was the extremely high titer that remained in cells (>7.7 logs by TCID, equivalent to ~8.3 LD50 by mouse assay), even after a 3 log reduction in PrP.
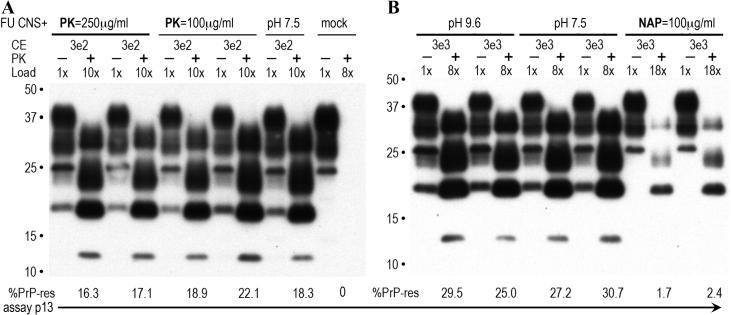
Relevant examples of the primary brain assay data that showed no loss of infectivity after virtually complete PK digestion of PrP are seen in Fig. 4, panel A. When a low cell number of PK digested FU-CJD brain homogenates were applied to replicate wells of GT1 target cells, they induced large amounts of de novo PrP-res by passage 13 (p13). These high levels of PrP-res are indicative of a high titer and are essentially the same as those seen in parallel undigested controls. The low inputs (3e2 CE) of FU-CJD agent shown here, as in previous studies, induced no detectable PrP-res production at p5 and only at later passages did indicator PrP-res appear [Miyazawa et al., 2011], i.e., de novo PrP-res accumulates linearly from 4-25% in sequential passages, and the TCID dilution endpoint is the fewest applied CE able to induce detectable PrP-res by p13. The loads and PrP-res values for each sample lane are indicated in Fig. 4, and values between 25-30% represent the maximal readout. Visual inspection of the PK digests in panel A show that duplicate wells at p13 give comparable PrP-res as the controls, even from the sample with a 99.7% reduction in starting total PrP (250μg/ml PK). Minor differences in the % PrP-res in these lanes were not significant, and 2-3 sequential passage readouts were combined for the determination of each of the TCID duplicates graphed in Fig. 3. In contrast to PK, NAP gave an obvious reduction in titer in this cell-based assay. As shown in Fig. 4B, a 10 fold higher CE sample input of 3e3 for the NAP digested brain required a load of 18x to clearly visualize the induced PrP-res (only ~2% in the duplicate well lanes) at p13 whereas the undigested control at a 10-fold lower CE input of 3e2 displayed 25-30%, past its maximal TCID readout. In summary, all forms of PrP are very sensitive to PK, but the infectious particles are remarkably resistant to digestion by this enzyme. Either no infectivity was lost when PrP was reduced by 99.7%, as in brain, or very high amounts of infectivity were still intact in the cultured cells (~1e8 TCID) even after 6-8hr digestions with 250μg/ml PK. The fact that NAP consistently yielded 2 log greater reductions in titer in the cell culture samples that contained more residual PrP, further emphasizes molecular components other than PrP are essential for infection.
Fig. 4.
Representative blot for GT1 indicator cell assay of TCID. For easy comparison the de novo PrP-res produced in indicator GT1 cells is shown at the same passage (p13), with initial CE applied to the indicator cells as well as the load in each lane indicated. An undigested aliquot of each duplicate well with a PK digest for maximal PrP-res detection (25μg/ml for 30 min) allowed quantitation of the indicated % PrP-res (under each lane). In FU-CJD infected cells the maximal PrP-res ranges from 25-30% and only values below 25% are in the linear range. Note the mock lane in panel A shows no detectable PrP-res, and the undigested controls and PK treated samples have high similar values of de novo PrP-res, indicating insignificant differences in titer. In panel B, control pH and NAP digestions are shown with a higher 3e3 input CE to bring out the low signal in the NAP digests. Control buffer and temperature conditions at this CE are already maximal and out of the linear range by p13 whereas the NAP samples required an 18x load to detect ~2% de novo PrP-res in the indicator cells.
Discussion
The above data are not in accord with the PK digestion results used to establish the prion hypothesis [McKinley et al., 1983]. While it has long been obvious from a variety of transmission and agent purification studies that host PrP-res does not correlate with infectious titers, only recently have prion adherents been forced to acknowledge this major discrepancy. In part, this has been brought about by in vitro protein conversion cyclic amplification studies (PMCA). These studies most often demonstrate the ability to produce enormous amounts of PrP-res, assayed by limited PK digestion, but no significant infectivity (reviewed in [Manuelidis, 2007]). In these cell free reactions, “spontaneous conversion” was also shown with recombinant PrP (recPrP) alone, and these “synthetic prions” were not infectious. Moreover, parallel “bona fide recPrPsc” (first spiked with infectious brain), showed no infectivity in animals even after extended incubation times [Atarashi et al., 2007]. A single new report of high infectivity generated from recPrP remains questionable until repeated [Wang et al., 2010], especially because the agent recovered was indistinguishable, in the neuropathology shown, from the scrapie strain used in the laboratory. In contrast to technically variable test-tube PrP-res PMCA studies, direct PK digestion of a standard 263K scrapie brain fraction previously demonstrated only a minimal reduction in titer (0.5logs) in samples without detectable PrP bands; this slight loss of titer was probably caused by the pretreatment of samples at 100°C to facilitate proteolysis [Suzuki et al., 2008]. In contrast, ≥2M GdnHCl, a treatment known to disrupt viral particles as well as amyloid fibril structures, yielded a ≥2 log reduction in titer. Our data repeats and extends this observation considerably. Careful quantitation showed that when PK digests all detectable forms of PrP in total brain homogenates by 3 logs, no infectivity is lost. Since visible non-PrP proteins remained, one or more of these is probably an essential component of the infectious particle. The previous independent gastrointestinal studies also point to an infectious agent constituted by something other than PrP. The obsession to explain every facet of TSEs with PrP [Manuelidis, 2003] has led to a failure to evaluate other proteins, including nucleic acid binding proteins, that are likely to be structural agent components [Akowitz et al., 1994; Sun et al., 2008].
In contrast to PK, NAP digestions that left greater amounts of residual PrP in brain, and a very different set of non-PrP proteins, reduced infectivity by >2.5 logs. This complementary finding adds to the likely importance of the infection-associated PK resistant proteins. Purification of virtually all starting infectious particles from cells without degenerative changes also showed these particles had reduced PrP and contained a highly select group of minor proteins [Sun et al., 2008]. A systematic comparative and direct molecular sequence analysis should help to resolve the fundamental importance of such minor proteins in the spread of infectivity and/or the intrinsic structure of the TSE infectious particle. At the current time we do not know why infected cells require more extensive PK digestions than brain to achieve >99.5% reductions in PrP. One possibility is that the intracellular location of PrP amyloid in GT1 cells has associated molecules that make it less accessible than the extracellular PrP amyloid of degenerating brain [Manuelidis et al., 1997; Manuelidis et al., 2007]. The extreme PK digestions used for cells gave only a 1.5 log titer reduction. It will be of interest to find if a shorter 2hr PK incubation of FU-CJD cells will preserve complete infectivity, as in brain. Less extensive PK digestion of cells may also help to resolve molecules of 25nm viruslike particles in-situ, and may also be used for enhanced particle purification [Manuelidis et al., 2007]. On a practical level, since GdnSCN has been shown to reduce infectivity by >4logs in brain [Manuelidis, 1997], practical and complete sterilization of precious instruments should be further effected by a subsequent digestion with NAP, and probably other keratinases.
The keratinase we used (NAP) is reasonably specific and, like the standard PK, was chromatographically pure. Several recent experiments have reported limited digestions with other proteases, but these proteases are often impure mixtures, and experiments have been done so that >10% of the PrP is retained. For example, the Sigma thermolysin that contains many other enzymes was used to digest centrifuged subcellular brain fractions precipitated with different chemicals [Cronier et al., 2008]. The percent of starting infectivity in these precipitated fractions was not reported. Using a neuroblastoma PK1 cell based assay, small differences in titer of ~2-fold appeared to be statistically significant, unlike previous statistics with the same cell assay [Mahal et al., 2007]. It is interesting, however, that the simplified precipitated brain fraction without any proteolytic digestion, but with intact PrP, showed a significant 1 log reduction in titer with only a short 1.5hr incubation at 70°C. This data further undermines the common belief that TSE agents require extremely high temperatures to achieve significant loss of titer. Other simplified infectious particle preparations may reveal conventional viral temperature inactivations for additional TSE agent strains. Pronase E, a mixture of three proteases, has also been used for limited digestion and enrichment of sensitive forms of PrP, and it was noted that “understanding the ratio of PrP molecules to infectivity is highly convoluted” [D'Castro et al., 2010].
Our current results demonstrating virtually complete digestion of all PrP with preservation of infectivity lead most simply to the conclusion that no form of prion protein is infectious. The use of whole homogenates and straightforward digestions here can be reproduced very easily, but whether such experiments will be done, or published, is less certain given the dominance and huge investment in the prion hypothesis. Many key prion experiments have never been repeated, and several, including spontaneous generation of infectivity from a 102L PrP mutation, have failed independent tests [Barron and Manson, 2003]. Even this extensive independent 102L transgenic work, in addition to an abundance of other experimental data that do not fit the prion hypothesis, or that contradict its many broad as well as specific conclusions [Manuelidis, 2007; Manuelidis et al., 2009a; Miyazawa et al., 2010] are rarely cited [Colby and Prusiner, 2011; Caughey et al., 2009]. The current experiments also call into question the multiple speculative forms of misfolded PrP required to encode the many unique TSE agent strains. The non-existent “Abwehrfermente” (defense enzymes) have several parallels to prions, and those irreproducible findings, but not the corrected data, continue to be cited [Deichmann and Benno, 1998; Kutschera, 2007]
Acknowledgments
Contract grant sponsor: NIH grant NS R01 012674-32
References
- Akowitz A, Sklaviadis T, Manuelidis L. Endogenous viral complexes with long RNA cosediment with the agent of Creutzfeldt-Jakob Disease. Nucleic Acids Res. 1994;22:1101–1107. doi: 10.1093/nar/22.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A, Simarro L, Islinger F, Nishida N, Manuelidis L. Two Creutzfeldt-Jakob disease agents reproduce prion protein-independent identities in cell cultures. Proc Natl Acad Sci USA. 2004;101:8768–8773. doi: 10.1073/pnas.0400158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007;4:645–50. doi: 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- Baker CA, Martin D, Manuelidis L. Microglia from CJD brain are infectious and show specific mRNA activation profiles. J. Virol. 2002;76:10905–10913. doi: 10.1128/JVI.76.21.10905-10913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron R, Manson J. A gene-targeted mouse model of P102L Gerstmann-Straussler-Scheinker syndrome. Clin Lab Med. 2003;1:161–73. doi: 10.1016/s0272-2712(02)00067-7. [DOI] [PubMed] [Google Scholar]
- Caughey B, Baron G, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby D, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a006833. epub a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronier S, Gros N, Tattum MH, Jackson GS, Clarke AR, Collinge J, Wadsworth JDF. Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin. Biochem J. 2008;416:297–305. doi: 10.1042/BJ20081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Castro L, Wenborn A, Gros N, Joiner S, Cronier S, Collinge J, Wadsworth J. Isolation of proteinase K-sensitive prions using pronase E and phosphotungstic acid. PLoS One. 2010;5:e15679. doi: 10.1371/journal.pone.0015679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann U, Benno M. The fraud of Abderhalden's enzymes. Nature. 1998;393:109–111. doi: 10.1038/30090. [DOI] [PubMed] [Google Scholar]
- Diringer H, Gelderblom H, Hilmert H, Ozel M, Edelbluth C, Kimberlin RH. Scrapie infectivity, fibrils and low molecular weight protein. Nature. 1983;306:476–478. doi: 10.1038/306476a0. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Gonzalez L, Espenes A, Press C, Martin S, Chaplin M, Davis L, Landsverk T, MacAldowie C, Eaton S, McGovern G. Transportation of prion protein across the intestinal mucosa of scrapie-susceptible and scrapie-resistant sheep. J. Pathol. 2006;209:4–14. doi: 10.1002/path.1962. [DOI] [PubMed] [Google Scholar]
- Kimberlin RH, Walker CA, Fraser H. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J. Gen. Virol. 1989;70:2017–2025. doi: 10.1099/0022-1317-70-8-2017. [DOI] [PubMed] [Google Scholar]
- Kutschera U. Abderhalden's fraud still wins him some supporters. Nature. 2007;446:136. doi: 10.1038/446136a. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sun R, Chakrabarty T, Manuelidis L. A rapid accurate culture assay for infectivity in transmissible encephalopathies. J. NeuroVirol. 2008;14:352–361. doi: 10.1080/13550280802105283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Baker C, Manuelidis L. New molecular markers of early and progressive CJD brain infection. J. Cellular Biochem. 2004;93:644–652. doi: 10.1002/jcb.20220. [DOI] [PubMed] [Google Scholar]
- Mahal S, Baker C, Demczyk C, Smith E, Julius C, Weissmann C. Prion strain discrimination in cell culture: the cell panel assay. Proc Natl Acad Sci U S A. 2007;104:20908–13. doi: 10.1073/pnas.0710054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Decontamination of Creutzfeldt-Jakob Disease and other transmissible agents. J. NeuroVirol. 1997;3:62–65. doi: 10.3109/13550289709015793. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Vaccination with an attenuated CJD strain prevents expression of a virulent agent. Proc. Natl. Acad. Sci. USA. 1998;95:2520–2525. doi: 10.1073/pnas.95.5.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Transmissible encephalopathies: speculations and realities. Viral Immunology. 2003;16:123–39. doi: 10.1089/088282403322017875. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A 25 nm Virion Is the Likely Cause of Transmissible Spongiform Encephalopathies. J Cell Biochem. 2007;100:897–915. doi: 10.1002/jcb.21090. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Nuclease resistant circular DNAs copurify with infectivity in scrapie and CJD. J Neurovirol. 2010;2:131–45. doi: 10.1007/s13365-010-0007-0. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Chakrabarty T, Miyazawa K, Nduom N-A, Emmerling K. The kuru infectious agent is a unique geographic isolate distinct from Creutzfeldt–Jakob disease and scrapie agents. Proc Natl Acad Sci U S A. 2009a;106:13529–13534. doi: 10.1073/pnas.0905825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Fritch W. Infectivity and host responses in Creutzfeldt-Jakob disease. Virology. 1996;216:46–59. doi: 10.1006/viro.1996.0033. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Fritch W, Xi YG. Evolution of a strain of CJD that induces BSE-like plaques. Science. 1997;277:94–98. doi: 10.1126/science.277.5322.94. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Liu Y, Mullins B. Strain-specific viral properties of variant Creutzfeldt–Jakob Disease (vCJD) are encoded by the agent and not by host prion protein. J Cell Biochem. 2009b;106:220–231. doi: 10.1002/jcb.21988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Sklaviadis T, Akowitz A, Fritch W. Viral particles are required for infection in neurodegenerative Creutzfeldt-Jakob disease. Proc. Natl. Acad. Sci. (USA) 1995;92:5124–5128. doi: 10.1073/pnas.92.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Sklaviadis T, Manuelidis E. On the origin and significance of scrapie associated fibrils. In: Court Lea., editor. Unconventional Virus Diseases of the Central Nervous System, Masson. Abbaye de Melleray; Bretagne: 1989. pp. 489–507. [Google Scholar]
- Manuelidis L, Yu Z-X, Barquero N, Mullins B. Cells infected with scrapie and Creutzfeldt-Jakob disease agents produce intracellular 25-nm virus-like particles. Proc Natl Acad Sci USA. 2007;104:1965–1970. doi: 10.1073/pnas.0610999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley MP, Bolton DC, Prusiner SB. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- Merz PA, Somerville RA, Wisniewski HM, Manuelidis L, Manuelidis EE. Scrapie associated fibrils in Creutzfeldt-Jakob disease. Nature. 1983;306:474–476. doi: 10.1038/306474a0. [DOI] [PubMed] [Google Scholar]
- Mitsuiki S, Hui Z, Matsumoto D, Sakai M, Moriyama Y, Furukawa K, Kanouchi H, Oka T. Degradation of PrP(Sc) by keratinolytic protease from Nocardiopsis sp. TOA-1. Biosci Biotechnol Biochem. 2006;70:1246–8. doi: 10.1271/bbb.70.1246. [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Emmerling K, Manuelidis L. Proliferative arrest of neural cells induces prion protein synthesis, nanotube formation, and cell-to-cell contacts. J Cell Biochem. 2010;111:239–47. doi: 10.1002/jcb.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa K, Emmerling K, Manuelidis L. Replication and spread of CJD, kuru and scrapie in vivo and in cell culture. Virulence. 2011;2 doi: 10.4161/viru.2.3.15880. epub PMID:21527829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N, Katamine S, Manuelidis L. Reciprocal interference between specific CJD and scrapie agents in neural cell cultures. Science. 2005;310:493–6. doi: 10.1126/science.1118155. [DOI] [PubMed] [Google Scholar]
- Prusiner S. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbel C, Pichner R, Groschup MH, Mueller-Hellwig S, Scherer S, Dietrich R, Maertlbauer E, Gareis M. Infectivity of Scrapie Prion Protein (PrPSc) Following In vitro Digestion with Bovine Gastrointestinal Microbiota. Zoonoses and Public Health. 2007;54:185–90. doi: 10.1111/j.1863-2378.2007.01040.x. [DOI] [PubMed] [Google Scholar]
- Sethi S, Lipford G, Wagner H, Kretzschmar H. Postexposure prophylaxis against prion disease with a stimulator of innate immunity. The Lancet. 2002;360:229–230. doi: 10.1016/S0140-6736(02)09513-2. [DOI] [PubMed] [Google Scholar]
- Sklaviadis TL, Manuelidis EE, Manuelidis L. Physical properties of the Creutzfeldt-Jakob disease agent. J. Virol. 1989;63:1212–1222. doi: 10.1128/jvi.63.3.1212-1222.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville R, Jul GN. Characterization of the effect of heat on agent strains of the transmissible spongiform encephalopathies. J Gen Virol. 2011;92:1738–48. doi: 10.1099/vir.0.030452-0. 7):1738-48. P. [DOI] [PubMed] [Google Scholar]
- Sun R, Liu Y, Zhang H, Manuelidis L. Quantitative recovery of scrapie agent with minimal protein from highly infectious cultures. Viral Immunol. 2008;21:293–302. doi: 10.1089/vim.2008.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SY, Takata M, Teruya K, Shinagawa M, Mohri S, Yokoyama T. Conformational change in hamster scrapie prion protein (PrP27-30) associated with proteinase K resistance and prion infectivity. J Vet Med Sci. 2008;70:159–65. doi: 10.1292/jvms.70.159. [DOI] [PubMed] [Google Scholar]
- Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen FE, DeArmond SJ, Prusiner SB. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan C-G, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1095–9203. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka M, Miwa T, Horii H, Takata M, Yokoyama T, Nishizawa K, Watanabe M, Shinagawa M, Murayama Y. Characterization of a proteolytic enzyme derived from a Bacillus strain that effectively degrades prion protein. Journal of Applied Microbiology. 2007;102:1365–2672. doi: 10.1111/j.1365-2672.2006.03080.x. [DOI] [PubMed] [Google Scholar]