Abstract
Bone marrow-derived mesenchymal stem cells (MSCs) are multipotent progenitors that can commit to osteoblast, chondrocyte, adipocyte, and several other lineages. The proper utilization of stem cells for clinical applications requires an integrated understanding of multiple signal inputs that control maintenance of stemness, proliferation, commitment, and differentiation. Various signaling pathways have been implicated in the regulation of MSC differentiation; however, complexities of pathway interactions, as well as seemingly contradictory results in the literature, create an often confusing and disjointed knowledge base. Several recent publications explore the integration of signaling pathways such as BMP, Wnt, Notch, Hedgehog, and Fibroblast Growth Factors in MSC osteoblast differentiation. The transcription factor Cbfa1/Runx2 has been implicated in these pathways as a potential focal point for signaling integration. This review will outline the current understanding of these pathways and indicate where both spatiotemporal effects during differentiation and comparable experimental conditions need to be considered in order to clarify the outcome(s) of differing regulatory levels of these signaling pathways.
Keywords: Mesenchymal Stem Cell (MSC), Bone Morphogenetic Protein (BMP), Wnt, Notch, Osteogenesis, Osteoblast differentiation, Runx2, Hedgehog, Fibroblast growth factor
Introduction
Stem cells are capable of self-renewal and differentiate to progeny cells which give rise to organized, functional networks of cells and extracellular matrix. These organized systems arise through embryonic development and are maintained in the adult as mature tissues and organs. Mesenchymal stem cells (MSCs) contribute to various musculoskeletal tissues as diverse as bone, cartilage, fat, muscle, ligament, and tendon [Pittenger et al., 1999]. There is increasing interest in developing clinical applications for MSCs, which are most commonly obtained from adult bone marrow. Indeed, MSCs have already been used in preclinical models for tissue engineering [Caplan, 2007], and allogeneic bone marrow transplants in children with osteogenesis imperfecta have demonstrated improved bone formation and function [Horwitz et al., 1999]. To enable the rational design of cell therapies to replace damaged or aged skeletal tissues, it is crucial to understand signaling pathways that guide MSC differentiation and how these signal inputs are integrated to initiate lineage commitment at the molecular level.
Bone development and homeostasis require several cell types that can be derived from MSCs. Osteoblasts may form bone directly via intramembranous bone formation or from a chondrocyte-derived cartilage template during endochondral bone formation [Lian et al., 2006]. The coordinated activities of chondrocytes and osteoblasts, both of MSC origin, together with the hematopoietic-derived osteoclasts that resorb bone, exist in a dynamic equilibrium to maintain bone structure and function. If this balance of bone formation and resorption is disturbed, the result is pathological osteoporosis (loss of bone) or osteopetrosis (increased, abnormal bone). Thus, bone homeostasis is carefully regulated by various signaling pathways. Notable paracrine signaling pathways include bone morphogenetic protein (BMP), Wnt, and Notch. An increased mechanistic knowledge of the significance of these pathways in MSC osteoblastogenesis is essential to develop methods to control cell fate in model systems and the eventual translation of that knowledge into novel therapeutics.
Mesenchymal stem cells in osteoblastogenesis
MSCs comprise less than 0.01% of the bone marrow cell population and may be isolated from hematopoietic cells in the marrow using density gradient centrifugation followed by adherence to tissue culture plastic. The undifferentiated cells appear fibroblast-like in morphology. Transplanted MSCs can migrate to sites of injury in animal models [Chamberlain et al., 2007], an appealing trait for tissue engineering purposes. MSCs can be differentiated into osteoblasts in vitro and require ascorbic acid and β-glycerophosphate to form a hydroxyapatite-rich mineralized matrix [Pittenger et al., 1999]. Kuznetsov et al. showed that human MSCs expanded from a single colony-forming unit precursor cell differentiated into osteoblasts in vivo by subcutaneously implanting MSC-containing scaffolds in mice and confirming the human origin of formed bone 45 weeks post-transplantation [Kuznetsov et al., 1997]. Although MSCs are classically derived from bone marrow, MSC-like multipotency has been described for cells derived from other tissue sources, including adipose tissue [Zuk et al., 2001], muscle [Qu-Petersen et al., 2002], and periosteum [De Bari et al., 2006]. More recently, perivascular mesenchymal cells (or pericytes) have also been shown to have MSC-like characteristics in culture and in fact may at least partially account for “MSCs” isolated from the aforementioned tissue sources. In a groundbreaking study, Sacchetti et al. identified CD146high pericytes surrounding bone marrow vascular sinusoids as self-renewing osteoprogenitor cells capable of ectopic bone formation [Sacchetti et al., 2007]. With confirmation of the mesenchymal progenitor cell and functional osteoblast progeny in vivo, identifying the molecular switch that instructs MSCs to commit to the osteoblastic lineage is of tremendous scientific interest.
Runx2 is a key transcriptional regulator of osteoblast differentiation
Early studies described Cbfa1, also called Runx2, as a transcription factor required for osteoblastogenesis. Ducy et al. reported that Runx2 bound the osteocalcin (OCN) promoter and was expressed in osteochondral progenitors as well as in early stages of osteoblastic differentiation. Forced expression of Runx2 in non-osteoblastic fibroblasts was sufficient to induce expression of osteoblastic markers such as type I collagen, bone sialoprotein, OCN, and osteopontin [Ducy et al., 1997]. Meanwhile, it was shown that Runx2 knockout mice exhibited no intramembranous or endochondral ossification [Komori et al., 1997; Otto et al., 1997], and the heterozygous phenotype mirrored the clavicular and cranial deformities of the human genetic disease cleidocranial dysplasia [Otto et al., 1997]. Runx2 thus serves as an essential regulator of bone formation by osteoblasts and is at the intersection of many signaling pathways regulating osteoblast differentiation.
Key signaling pathways in osteoblastogenesis
Bone Morphogenetic Protein (BMP)
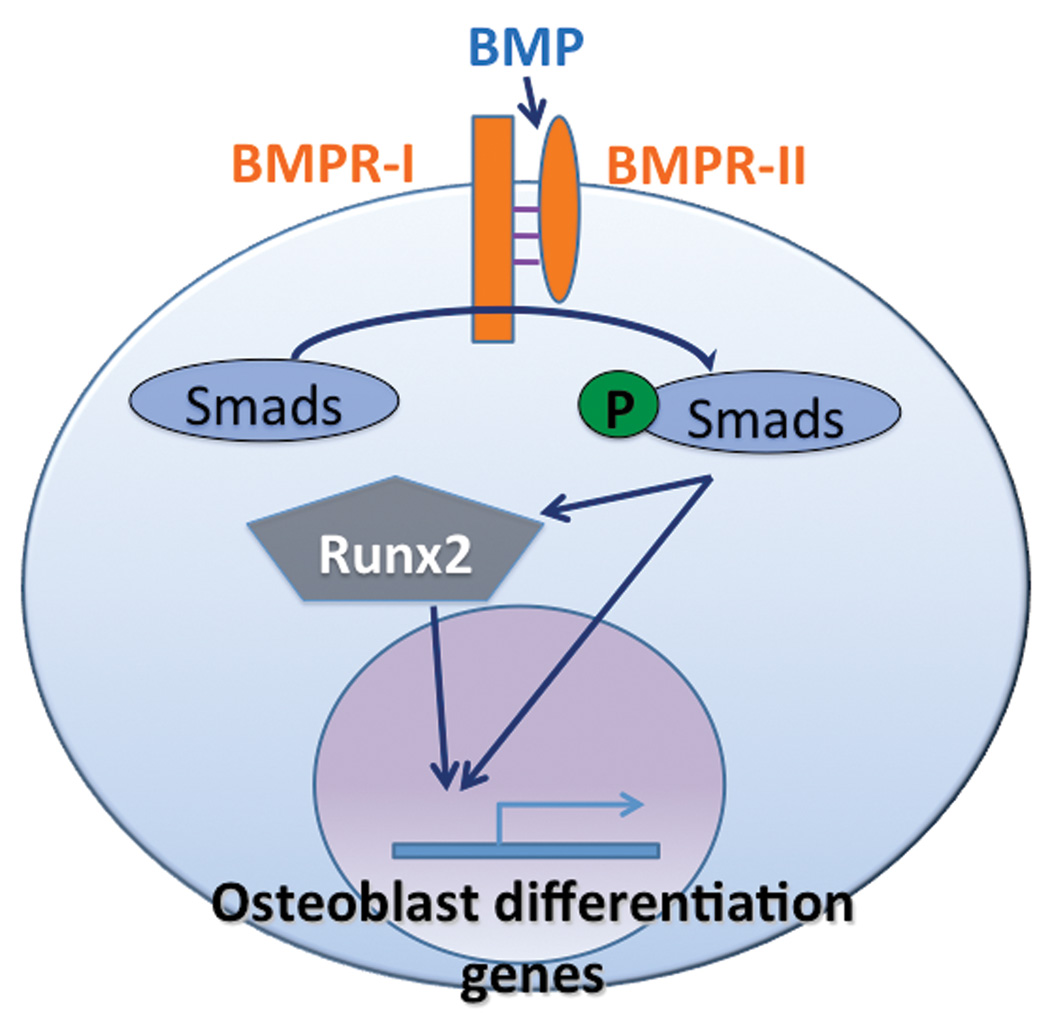
Expression and activity of transcription factors such as Runx2 are regulated by multiple signaling pathways. BMP, Wnt, and Notch signaling pathways all play important roles in osteoblast differentiation. BMPs were first purified from bovine bone and shown to induce ectopic bone formation in mice two decades ago [Sampath et al., 1987; Wang et al., 1988]. BMPs were subsequently identified as members of the transforming growth factor-β (TGF-β) superfamily of multifunctional cytokines [Celeste et al., 1990]. There are two subtypes of BMP receptors, BMPR-I and BMPR-II, which are serine-threonine kinase receptors. Upon BMP binding to the BMPR-II, BMPR-I is recruited to form an activated quaternary complex, which then phosphorylates and activates intracellular Smad proteins (Figure 1). Receptor Smads bind to a co-Smad and translocate to the nucleus to serve as transcription factors [Fujii et al., 1999; Kawabata et al., 1998].
Figure 1.
BMP signaling pathway: BMP binds heterodimeric receptor to activate Smad proteins, which transactivate osteoblastogenic genes either directly or via Runx2. Adapted from Kawabata et al 1998.
One of the BMP-Smad target genes is Runx2. In vitro, BMP-2 treatment increases Runx2 gene expression and alkaline phosphatase (ALP) protein levels in a human marrow stroma-derived cell line [Gori et al., 1999], and Runx2 overexpression suppressed myogenesis but required Smads to induce ALP expression and activity in C2C12 myoblasts [Lee et al., 2000]. Conversely, induced mutations in Runx2 disrupt Runx2-Smad interactions and early osteoblast differentiation [Javed et al., 2008]. In vivo, BMPs are essential for organogenesis in early and late development. Since BMP-2 and BMP-4 null mice are embryonic lethal [Winnier et al., 1995; Zhang and Bradley, 1996], studies have used conditional knockout alleles to delineate the physiological function of BMPs in skeletogenesis. Deletion of BMP ligands [Bandyopadhyay et al., 2006] or BMP receptors [Yoon et al., 2005] from the limb bud mesenchyme impairs chondrogenic or osteogenic differentiation and induces skeletal patterning defects.
Wnt
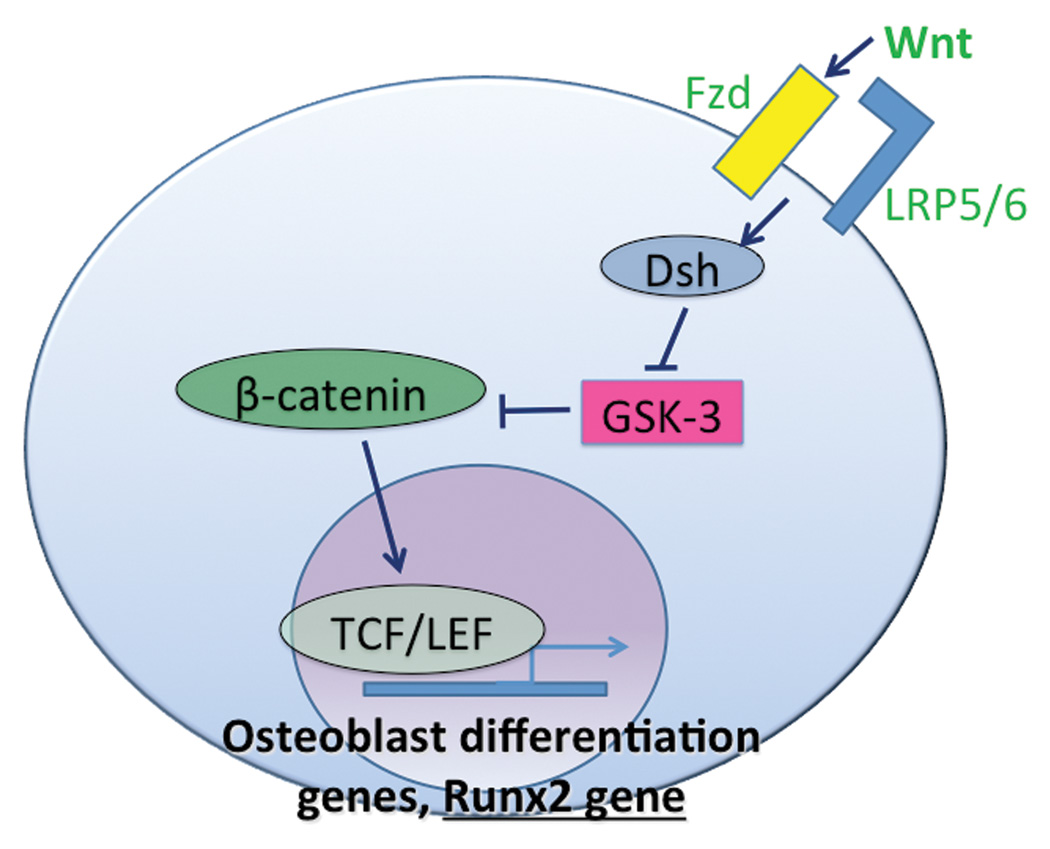
A second major molecular pathway involved in osteoblastogenesis is Wnt signaling. Wnts are historically categorized by whether they signal in a canonical or non-canonical manner. Canonical Wnts signal through β-catenin, whereas non-canonical Wnt signaling does not require β-catenin [Davis and Zur Nieden, 2008]. In the canonical pathway, Wnt binds to the transmembrane receptor Frizzled (Fzd) and co-receptor LRP5/6 (Figure 2). This event leads to intracellular accumulation of β-catenin, a signaling protein which can then enter the nucleus and interact with the transcription factor T-cell factor/lymphoid enhancer factor (TCF/LEF) to activate transcription of target genes. Cytoplasmic β-catenin is phosphorylated by glycogen synthase kinase-3 (GSK3) and thus is targeted for ubiquitination and proteasome-mediated degradation. The phosphorylation activity of GSK3 is inhibited by the cytoplasmic protein Dishevelled (Dsh) which is recruited by Fzd-Wnt binding events [Huelsken and Behrens, 2002]. Non-canonical Wnt pathways also use Fzd receptors but utilize unique co-receptors, including but not limited to Knypek, Ror2, and Cripto. Non-canonical Wnts signal through proteins such as phospholipase (PLC) and phosphokinase C (PKC) which regulate intracellular calcium release (Wnt/Ca2+ pathway) [Kohn and Moon, 2005]. Alternatively, Fzd can phosphorylate Dsh without co-receptor recruitment and activate small GTPases such as Rac and Rho (planar cell polarity pathway). Interestingly, non-canonical Wnt signaling may inhibit β-catenin transcriptional activity [Ishitani et al., 2003].
Figure 2.
Wnt/β-catenin signaling: Wnt-Fzd-LRP5/6 binding events recruit Dsh to stabilize intracellular β-catenin, which activates TCF/LEF for osteoblastogenic gene and Runx2 transcription. In the absence of Wnt, β-catenin is phosphorylated and marked for degradation by GSK-3. Adapted from Davis and Zur Nieden, 2008.
Extensive in vivo studies have demonstrated a role for canonical Wnt signaling in regulating osteoblastogenesis. Activating mutations in LRP5 in humans result in a high bone mass (HBM) phenotype, in which bone biopsies show increased trabecular bone volume and decreased fat within the marrow [Boyden et al., 2002; Little et al., 2002; Qiu et al., 2007]. Conversely, LRP5 inactivating mutations cause osteoporosis characterized by decreased bone and increased intramedullary fat, essentially the opposite phenotype of HBM [Gong et al., 2001; Qiu et al., 2007]. Murine studies have replicated these findings [Cui et al., 2011; Holmen et al., 2004]. Additionally, a conditional β-catenin knockout in skeletogenic mesenchyme caused ectopic formation of chondrocytes at the expense of osteoblasts [Day et al., 2005]. β-catenin is also required for postnatal bone maintenance in mice since an osteoblast-specific β-catenin mutation led to osteopenia and increased numbers of osteoclasts in mice [Holmen et al., 2005]. Furthermore, overexpression of Frzb, a secreted Wnt antagonist, abrogated ectopic bone formation by human periosteal cells in mice [Eyckmans et al., 2010].
In vitro studies on the effects of canonical and non-canonical Wnt signaling on osteoblastogenesis are inconclusive. Studies have shown that canonical Wnt signaling either through Wnt10b or GSK3 inhibition promotes osteoblastogenesis [Bennett et al., 2005; Bennett et al., 2007]. Similarly, human MSCs transduced with activated LRP5 and treated with Wnt3a in vitro were found to have increased ALP expression and activity with decreased lipid droplet formation, as well as the formation of ectopic mineralized bone when implanted in a scaffold subcutaneously in mice; the opposite phenotype was observed in hMSCs transduced with inactivated LRP5 and treated with Wnt3a [Qiu et al., 2007]. Meanwhile, other studies have suggested that canonical Wnt signaling maintains an undifferentiated proliferative state of MSCs, and non-canonical Wnt signaling promotes osteogenesis [Baksh et al., 2007; Boland et al., 2004].
Similar to BMP signaling, canonical Wnt signaling in osteoblastogenesis has been linked to Runx2. The Runx2 gene promoter contains a Wnt-responsive TCF regulatory element, and both β-catenin and TCF1 are recruited to the Runx2 locus [Gaur et al., 2005]. Additionally, transient activation of Wnt/β-catenin signaling in MSCs in vitro suppresses transcription of adipogenic transcription factor peroxisome proliferator-activated receptor-γ (PPAR-γ) and induces expression of bone lineage genes such as Dlx5 and Osterix [Kang et al., 2007]. Conversely, PPAR-γ inhibits OCN expression by repressing both the expression and transactivation ability of Runx2 [Jeon et al., 2003].
Notch
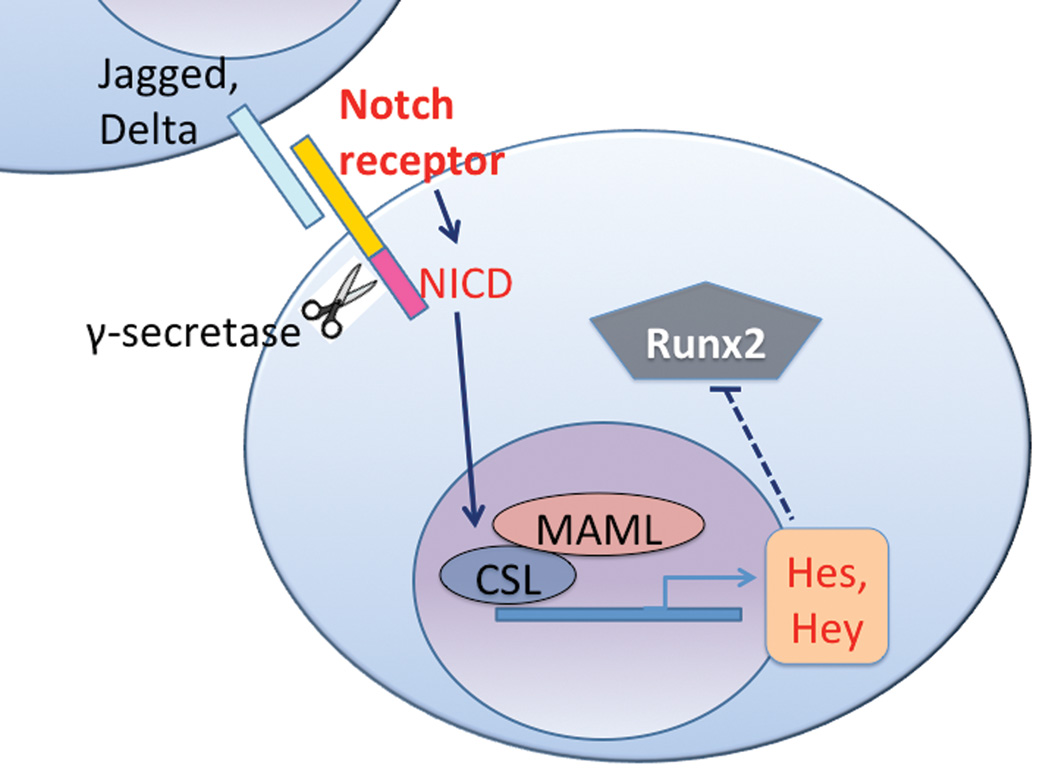
Notch signaling is a third pathway that is important for MSC differentiation into osteoblasts. Notch itself is a transmembrane receptor. When Notch interacts with membrane-bound ligands Delta or Jagged on the surface of neighboring cells, the Notch intracellular domain (NICD) is proteolytically cleaved from the membrane by γ-secretase (Figure 3) and translocates to the nucleus to complex with and activate the transcription factor CSL (CBF1 in humans, Suppressor of Hairless in Drosophila, LAG in Caenorhabditis elegans; also called RBP-Jkappa in mice). CSL then recruits the co-activator Mastermind-like (MAML) and initiates transcription of target genes such as Hes and Hey [Bolos et al., 2007; Watt et al., 2008].
Figure 3.
Notch signaling: Ligand expressed on neighboring cells binds to Notch receptor, initiating γ-secretase-mediated cleavage of the Notch receptor liberating the NICD which then binds to CSL and MAML for transcription of Hey/Hes which inhibit Runx2. Adapted from Watt et al. 2008.
Notch signaling may improve osteoblastogenesis but not necessarily enhance bone formation. Osteoblast-specific gain of Notch function in a NICD-overexpressing transgenic mouse results in abnormally dense or osteosclerotic bone, whereas loss of Notch signaling via γ-secretase mutations leads to late-onset, age-related osteoporosis [Engin et al., 2008]. The osteosclerotic phenotype is caused by enhanced proliferation of immature osteoblasts and not due to decreased osteoclastic activity. Conversely, the osteoporotic phenotype in loss-of-Notch-function mice displayed increased osteoclast numbers as a result of decreased Notch signaling in osteoblasts [Engin et al., 2008].
As with BMP and Wnt signaling in osteogenesis, Runx2 function is also influenced by Notch signaling. Runx2 transcriptional activity is physically antagonized by the protein encoded by Notch target gene Hey1 [Zamurovic et al., 2004]. Transient transfection of MSCs with NICD, Hes, or Hey all decreased Runx2 transactivity, and co-immunoprecipitation demonstrated direct binding of Hes or Hey to Runx2 [Hilton et al., 2008]. In addition, NICD can interact directly with Runx2 protein to repress terminal osteoblastic differentiation in vitro [Engin et al., 2008].
Interactions of BMP, Wnt, and Notch signaling pathways in osteoblastogenesis
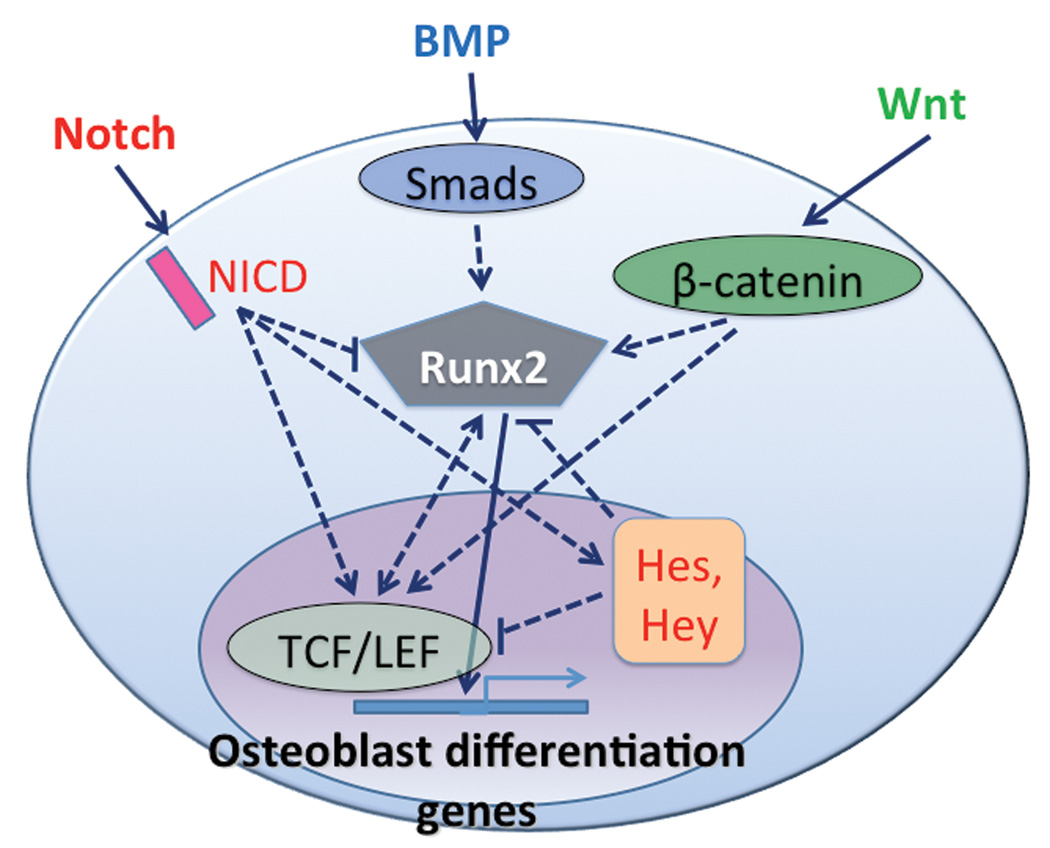
Clearly, Runx2 is a prime candidate for the intersection and integration of BMP, Wnt, and Notch signaling pathways in osteoblast differentiation. All three pathways directly regulate Runx2 transcriptional activity (Figure 4) but only a few studies address the integrated effects of these three signaling pathways on osteoblastogenesis. In addition, several other growth factors have been implicated in osteoblastogenesis, including the hedgehog and fibroblast growth factor families. Due to the complexity of this subject matter, we review the literature with individual sections focusing on different combinations of growth factor pathways.
Figure 4.
Integration of BMP, Wnt, and Notch signaling: a simplified Runx2-centric model.
Wnt, BMP, and Notch
To identify the effects of Notch on the BMP and Wnt pathways during osteoblast differentiation, one study used NICD overexpression in conjunction with BMP or Wnt treatment. NICD overexpression inhibited the positive effects of BMP-2 and Wnt3a on ALP activity in ST2 marrow stromal cells. Although NICD overexpression did not alter the level of phosphorylated Smad proteins with or without BMP, it did decrease Wnt-dependent reporter activity and levels of β-catenin [Deregowski et al., 2006]. In another study, Notch target gene Hey1 was induced by BMP-2 in the pre-osteoblastic MC3T3 cell line [Zamurovic et al., 2004]. Since Hey1 inhibits Runx2 transcriptional activity on osteoblastic genes, BMP-induced Hey1 expression may serve as a negative feedback regulator of BMP/Runx2-mediated osteoblastogenesis. These studies suggest that Notch signaling can regulate either BMP or Wnt-induced osteogenesis.
Wnt and BMP
Interactions between Wnt and BMP pathways in osteoblastogenesis have been explored the most extensively. One approach to examine BMP/Wnt interactions is by overexpressing or introducing the ligands or antagonists of both pathways into in vitro or in vivo experimental systems. For instance, addition of Wnt inhibitors DKK-1 and DKK-2 to the culture media of C2C12 cells, along with either Wnt3a or BMP-2, resulted in attenuated Wnt3a-induced ALP expression and enhanced BMP-2-induced ALP expression [Fujita and Janz, 2007]. In sharp contrast, BMP-2-induced ectopic bone formation in vivo was reduced by blocking β-catenin signaling, either by an adenovirus carrying DKK-1 or a conditional floxed β-catenin [Chen et al., 2007]. Furthermore, retroviral transduction of Wnt inhibitory factor-1 (WIF-1) into C3H10T1/2 cells inhibited BMP-2-induced ALP activity [Cho et al., 2009]. Although apparently conflicting, these studies suggest a more refined regulation downstream of antagonist/ligand/receptor interactions.
Indeed, different mutant stabilized forms of β-catenin used to mimic canonical Wnt signaling can antagonize or synergize with BMP-2-induced osteogenesis. Retrovirally expressed, phosphorylation-resistant β-catenin mutant protein (containing serine-to-alanine substitutions) in C3H10T1/2 cells increased ALP activity induced by Wnt signaling alone that was decreased when simultaneously treating the cells with BMP-2 [Bain et al., 2003]. In contrast, an N-terminal truncated β-catenin (deleting the GSK3 phosphorylation sites) did not enhance ALP activity on its own, but in presence of BMP-2 resulted in synergistically increased ALP activity, OCN expression, matrix mineralization, and new bone formation when injected into mouse calvaria in vivo [Mbalaviele et al., 2005]. These studies indicate that stabilization of β-catenin is not sufficient for Wnt regulation of BMP-induced ALP activity. One clear difference between these two studies is the physical size of the β-catenin mutant (point mutations at GSK3 phosphorylation sites versus truncation removing those sites). The truncated form of β-catenin may have other cellular effects besides inhibition of GSK3 phosphorylation, such as interfering with the association of β-catenin with other binding partners. For example, the truncated β-catenin was unable to bind α-catenin, which is thought to play a role in cell adhesion [Mbalaviele et al., 2005]. Such “off-target” effects may thus confound interpretations of experiments and potentially explain the contradictory reports of whether Wnt antagonizes or synergizes with BMP in osteogenesis. β-catenin also likely serves at the crossroads of Wnt and BMP signaling; depending on its binding partners, β-catenin may or may not inhibit BMP signaling.
With respect to whether BMP signaling regulates Wnt signal transduction, there is evidence that Smad1 sequesters the GSK3 inhibitor Dishevelled (Dsh) in the absence of Wnt signaling. When Wnt3a is present without BMP-2, Dsh is activated, dissociates from Smad1, and inhibits GSK3-mediated degradation of β-catenin. Addition of BMP-2 diminishes Wnt3A-induced transcriptional activity in ST2 cells, perhaps through BMP-induced formation of Smad1-Dsh complexes [Liu et al., 2006]. In contrast, it is suggested in primary calvarial osteoblasts that BMP-2 enhances Wnt signaling via upregulation of several Wnt ligands and receptors [Chen et al., 2007]. Canonical Wnt signaling may also stabilize BMP/Smad signaling by preventing GSK3-mediated degradation of Smad1 [Fuentealba et al., 2007].
Thus, integration of BMP and Wnt signaling pathways occurs at several different molecular steps, including Runx2, β-catenin, Dsh, and GSK3. Various manipulations of BMP and Wnt pathway components and antagonists have demonstrated complex regulation that depends on factors such as binding partner interactions, cellular context, and stabilization against degradation. Generally, BMP enhances MSC commitment to osteogenic differentiation while counteracting proliferative cues promoted by canonical Wnt. It is possible that Wnt signaling may sensitize uncommitted MSCs to respond to BMP, but overall induces complex effects on differentiation due to coincident stimulation of a number of different signals through promiscuous binding by β-catenin and other Wnt transducers.
BMP and Notch
At first glance, studies of the integrated effects of Notch and BMP signaling on osteoblastogenesis have produced dichotomous results. Several reports propose that Notch signaling enhances BMP-induced osteoblastogenesis. For instance, activating Notch signaling in MC3T3-E1 cells with adenovirally-overexpressed NICD was found to stimulate BMP-2-induced osteoblastogenesis, as measured by increased numbers of ALP-expressing cells and formation of calcified nodules in vitro [Tezuka et al., 2002]. To a similar end, enhanced ALP activity and in vivo ectopic bone formation were described in MC3T3-E1 cells that were simultaneously overexpressing Notch ligands and being treated with BMP-2. Without concomitant BMP-2 treatment, overexpression of Delta1 and Jagged1 had no effect on cell differentiation. In addition, inhibition of Notch signaling by gamma-secretase inhibitor, siRNA against Notch1, or a dominant-negative extracellular domain of Notch1, all resulted in decreased ALP activity and decreased promoter activity of BMP target genes [Nobta et al., 2005]. These findings suggest that Notch and BMP work synergistically to promote osteogenic differentiation.
Conversely, another study showed that in MC3T3-E1 cells overexpressing BMP-2, stimulation of Notch targets suppressed BMP-2-induced osteoblastogenesis. The authors suggest that Notch targets like Hey1 can be activated by CCN3/NOV, a regulator of cell differentiation. Co-transduction with adeno-CCN3/NOV and adeno-BMP-2 did not suppress BMP-2-induced Smad phosphorylation in Hey1-deficient cells, but recovery of Hey1 expression via adeno-Hey1 resulted in decreased phospho-Smad levels [Minamizato et al., 2007]. It is interesting that introducing adeno-CCN3/NOV alone did not cause inhibition of BMP targets like Runx2 and ALP expression, but only did so when adeno-BMP-2 was co-transduced into cells; perhaps excess BMP-2 signaling leads to feedback inhibition mediated by the Notch pathway. Similarly, BMP-2 treatment induced Hey1 expression in MC3T3 cells [Zamurovic et al., 2004], further suggesting that BMP osteogenic signaling is self-regulated through Hey1 inhibition of Runx2 transcriptional activity. Taken together with the previous results, this may indicate a negative feedback loop in which Notch ligands stimulate BMP-2-induced osteogenesis, but Notch target genes may inhibit the latter through inhibition of phospho-Smad. Hence Notch and BMP could simultaneously exert opposing controls on osteogenesis, underscoring the organizational complexity of signaling integration and the need for careful, spatiotemporally-controlled studies.
Hedgehog, BMP, and Wnt
Thus far, this review has focused on BMP, Wnt, and Notch signaling in osteoblastogenesis; however there are other signaling pathways that influence osteoblastogenesis and bone in vivo. For example, hedgehog (Hh) signaling is essential for skeletal patterning during development and several studies suggest that hedgehog-induced osteoblastogenesis occurs through Runx2 [Mak et al., 2006; St-Jacques et al., 1999]. Of the three Hh homologues, Sonic hedgehog (Shh) and Indian hedgehog (Ihh) have been implicated in osteoblast development. In general, Hh binds its cell surface receptor Patched, which relieves Patched-mediated suppression of a transmembrane protein called Smoothened (Smo). Then, Smo can activate an intracellular signaling cascade that eventually stabilizes the transcription factor Gli2, which induces transcription of Gli1 and other Hh target genes [Plaisant et al., 2009].
The removal of Hh signaling results in developmental defects in bone. Deletion of Shh caused vertebrae and distal limb skeletal malformations in mice [Chiang et al., 1996]. Ihh was shown to regulate Runx2 expression and hence osteoblastogenesis in the perichondrium [St-Jacques et al., 1999]. In Ihh null mouse embryos, the perichondrium lacked typical expression of multiple osteogenic markers including Runx2, Osterix, type I collagen, and ALP [Hu et al., 2005], suggesting that Ihh signaling is required for early osteoblastogenesis, likely through modulation of Runx2. These observations implicate Runx2 as a nexus linking Hh signaling together with other osteoblastogenic signaling pathways. The literature proposes both synergy and antagonism of Hh signaling with BMP or Wnt in osteogenic differentiation, as will be discussed below.
In conjunction with BMP-2, Hh appears to have a synergistic effect on osteogenesis in mesenchymal cell types, specifically in mice. Either Ihh or Shh treatment stimulated ALP activity synergistically with BMP-2 in C3H10T1/2 cells and MC3T3-E1 cells [Nakamura et al., 1997]. Similarly, recombinant N-terminal Shh led to higher percentages of cells responding to BMP-2 in terms of ALP activity; this was demonstrated in various murine mesenchymal cell types including C3H10T1/2, ST2, and primary mouse calvaria cells [Spinella-Jaegle et al., 2001]. The same study found that Shh treatment upregulated expression of ALP, Runx2, and OCN, at the expense of adipogenic commitment as indicated by decreased levels of adipogenic transcription factors C/EBPα and PPARγ in C3H10T1/2 cells, whether or not BMP-2 was also present [Spinella-Jaegle et al., 2001; Yuasa et al., 2002]. Intriguingly, the ability of mesenchymal cells to respond to Shh treatment may be dependent on the extent of terminal differentiation; i.e., cells essentially committed to the osteogenic lineage, such as pre-osteoblastic MC3T3-E1 cells and osteoblastic cell lines, did not respond to Shh in the presence of BMP-2, whereas the more pluripotent C3H10T1/2 cells did respond [Spinella-Jaegle et al., 2001]. Hence the contribution of Shh to osteoblastogenesis is likely more important in early stages of differentiation, e.g. for increasing osteogenic commitment while inhibiting adipogenic differentiation. In support of this conjecture, the removal of Ihh signaling by floxed Smo alleles caused mouse perichondrial osteoblast progenitors to undergo chondrogenesis instead of osteoblastogenesis [Long et al., 2004].
Moreover, the literature suggests that the effect of Hh on osteoblast differentiation, with or without BMP, may differ between species. Whereas Hh appears to promote osteoblastogenesis synergistically with BMP in mice, Hh may have non-synergistic or antagonistic activity in other species. In a neonatal rat metatarsal ex vivo culture, Shh supported endochondral ossification through Runx2 but did not synergistically increase ossification when simultaneously administered with low amounts of BMP-4, whereas higher amounts of BMP-4 actually diminished Shh-induced ossification [Krishnan et al., 2001]. In human adipose-derived stem cells treated with an osteogenic cocktail, Shh actually inhibited osteogenesis as measured by decreased ALP and Runx2 expression. Osteoinduction of these human cells led to a decreased Gli1 and Smo expression; however, expression levels of Ihh, Patched, and Gli2 did not change significantly [Plaisant et al., 2009], so it is unclear if osteoblastogenesis triggers a substantial decline in Hh signaling.
Hh signaling is also thought to function upstream of Wnt in osteoblast differentiation. In Ihh null mouse embryos, the normal accumulation of nuclear β-catenin in perichondrial cells was absent, suggesting that the lack of Ihh signaling disrupted canonical Wnt signaling [Hu et al., 2005]. Further evidence of Hh and Wnt crosstalk during osteogenesis was demonstrated by the co-expression of constitutively active Smo, to activate Hh signaling, and either Dkk1 or dominant-negative Tcf4, to antagonize Wnt signaling, which led to partial reduction in ALP and bone sialoprotein gene expression in C3H10T1/2 cells [Hu et al., 2005]. Similarly, overexpression of Wnt inhibitors significantly reduced both Shh-induced and BMP-2-induced ALP activity in C3H10T1/2 cells [Rawadi et al., 2003]. In contrast, BMP or Shh inhibitors decreased only BMP-2-mediated or Shh-mediated ALP activity, respectively, while having no effect on Wnt3a-stimulated ALP activity [Rawadi et al., 2003]. Thus while Hh signaling may regulate Wnt signaling, it appears that Wnt signaling is critical for Hh-induced osteogenesis.
The important role of Wnt signaling in Hh-mediated osteogenesis has also been shown in vivo. Mutant mice with inactivated canonical Wnt and upregulated Ihh signaling were generated via perichondrium-specific deletion of β-catenin and Patched. The mutant embryos displayed inhibited ossification and a severe decrease in the number of mature osteoblasts in limb sections. Despite diminished osteoblastogenesis, the expression of Ihh-induced Runx2 and Ihh signaling targets Gli1 and Hip1 was intact, suggesting that canonical Wnt signaling is not required for the Ihh signaling pathway itself but permits osteoblastogenesis downstream of Ihh signaling [Mak et al., 2006]. These in vitro and in vivo findings are all consistent with a model whereby Hh promotes Runx2 and early osteoblastic commitment of mesenchymal progenitors, but the latter process requires functional Wnt downstream.
FGF, BMP, and Wnt
Fibroblast growth factors (FGF) stimulate the proliferation and differentiation of a number of different cell types, including osteoblastic precursors and MSCs from various species [Tsutsumi et al., 2001]. FGF-2 has been shown to increase ALP protein levels without changing OCN mRNA levels in human MSCs [Ito et al., 2008], suggesting that the effect of FGF-2 on osteogenesis may be limited to supporting cell expansion, lineage commitment, and early stages of differentiation. In vivo data seem to support this idea, as FGF-2 knockout mice displayed less bone mass and increased marrow fat compared to wildtype mice. Bone marrow-derived MSCs cultured from FGF-2 knockout mice correspondingly tended to undergo adipogenesis at the expense of osteogenesis, mirroring the in vivo phenotype. Finally, exogenous FGF-2 blocked adipogenesis and rescued ALP activity and bone nodule formation in vitro [Montero et al., 2000; Xiao et al., 2010]. On the contrary, in a mouse model of achondroplasia, a gain-of-function mutation in FGF receptor-3 (FGFR-3) also led to decreased bone mass in mice. It was ascertained that activated FGFR-3 regulates both bone formation and resorption, as both osteoblastic and osteoclastic gene expression and activity were increased in marrow stromal cells from these mice [Su et al., 2010].
Interestingly, FGF may influence osteoblastogenesis at least partially through Runx2 modulation. Runx2 mRNA was found to be reduced in bone marrow-derived cell cultures from FGF-2 knockout mice and increased after FGF-2 treatment [Naganawa et al., 2006]. Other studies have indicated that Runx2 is phosphorylated and activated by FGF-2 [Kim et al., 2003; Naganawa et al., 2006]. Hence Runx2 is a potential conduit for FGF signaling crosstalk with other osteogenic signaling pathways. For instance, osteoblasts from FGF-2 null mice show impaired BMP function, i.e., BMP-2-induced bone formation and ALP activity is abrogated in FGF-2 null mice and marrow stromal culture, respectively [Naganawa et al., 2008]. Co-treatment of rat marrow-derived MSCs with both BMP-2 and basic FGF synergistically promoted higher osteoblastic gene expression and bone nodule formation than treatment with either factor alone [Hanada et al., 1997]. Moreover, FGF-2 also seems to regulate endogenous BMP-2 expression, as BMP-2 mRNA levels are increased by exogenous FGF-2 in calvarial osteoblasts and MC3T3-E1 cells [Choi et al., 2005] and reduced in calvariae from FGF-2 knockout mice which could be rescued by BMP-2 treatment [Naganawa et al., 2008]. Thus, FGF-2 and BMP-2 appear to be reciprocally regulated in osteoblasts, contributing to the balance of interacting signaling pathways in bone development and homeostasis.
Contrary to the case of FGF and BMP synergy, FGF signaling may negatively impact Wnt-induced osteogenesis. Studies of activating FGFR mutations in osteoblasts revealed downregulation of Wnt target genes [Ambrosetti et al., 2008; Mansukhani et al., 2005]. Recombinant FGF-1 has been shown to inhibit Wnt3a-induced osteoblastic transcription and ALP activity [Ambrosetti et al., 2008]. One proposed mechanism is through FGF-induced expression of Sox2 in osteoblasts, which has been shown to co-immunoprecipitate with β-catenin [Mansukhani et al., 2005]. Transfection with Sox2 also blocked ALP activity and mineralization in osteoblasts cultured in osteogenic media [Mansukhani et al., 2005], suggesting that FGF may regulate canonical Wnt-induced osteogenesis by sequestering β-catenin via Sox2 and thus preventing β-catenin from binding TCF/LEF. We conclude that FGF signaling antagonizes Wnt-mediated osteogenesis but also synergistically promotes BMP-mediated osteogenesis, potentially through Runx2 activation.
The complexity of interpreting signaling pathway interactions
Thus far, this review has described many examples of how multiple signaling pathways impact mesenchymal cell fate decisions. To understand the interplay of various signal inputs, models must be simplified to isolate experimental variables. Special attention is needed to evaluate whether experimental designs even permit logical comparisons when results from different studies are compared. Conflicting results in the literature may be due to fundamental differences in experimental conditions such as cell lines, culture conditions, or transfection methods. For example, rodent and human MSCs have been shown to be very different in their in vitro growth and differentiation conditions, a phenomenon which may limit cross-species comparison of cell behavior. While human MSCs can be isolated from bone marrow using flow cytometry and cultivated with reasonable purity, there are no reliable markers for murine MSC purification [Baddoo et al., 2003]. If it cannot be reasonably confirmed that cells of different species are functionally equivalent, then the implications of experimental results in animal models may not necessarily transfer to humans. Additionally, MSCs can be obtained from a variety of fetal or adult tissue sites besides bone marrow and there is considerable variability in the in vitro function of cells derived from different locations [Javazon et al., 2004]. Thus, consideration must also be given to the origin and differentiation state of cells.
Even within the same cell line, differing experimental designs between studies may show variable effects on signal transduction pathways that are sensitive to temporal or spatial modulation. Such modulation may result in diverse cellular responses including proliferation, differentiation, apoptosis, etc. [Engin et al., 2008]. For example, temporal molecular switching of homeodomain transcription factors Msx2, Dlx3, and Dlx5 occurs on the OCN promoter at different stages of osteoblast differentiation [Hassan et al., 2004]. If separate research teams choose to examine the behavior of the OCN promoter at different stages of osteoblast differentiation, they will report dissimilar transactivation events. Similarly, sequential stimulation of signaling pathways, rather than continuous overexpression or growth factor exposure, may more closely resemble physiological developmental cascade of events.
Another way that researchers studying the same pathway may arrive at disparate results is through different methodologies for experimental manipulations. Introducing point mutations at phosphorylation sites may be less detrimental to the native function of a signaling protein than truncating the protein to remove those sites, as the physical size of the native protein or the truncated region may be important for other molecular interactions. As another example, the use of adenoviral rather than retroviral vectors in inducing Notch signaling would cause a transient as opposed to a continuous effect, respectively [Deregowski et al., 2006], and levels of expression are difficult to control. Temporary Notch suppression enhanced in vivo bone formation, but over the longer term led to depletion of the mesenchymal progenitor pool and hence late-onset osteoporosis [Hilton et al., 2008]. One way to avoid supraphysiologic NICD overexpression while still achieving maximum physiologic Notch signal would be to expose cells to excess ligand for the Notch receptor. To this end, data shows that plating murine MSCs on recombinant Jagged-1 prevents osteoblast differentiation and increases progenitor proliferation (unpublished data, Shitaye and Hankenson).
Signaling proteins may also have compensatory homologues and/or pleiotropic effects, such that an experiment intended to target one pathway can in fact activate other cellular responses. Osteopenia secondary to LRP5 mutations is more severe in mice than in humans, which may be due to partial compensation by LRP6 and/or non-Wnt functions of β-catenin such as interactions with E-cadherin. On a similar note, experiments that combine growth factors may be confuse the effects of one factor for another. Hilton et al. described inhibition of osteoblastogenesis by Notch, and they cited a source [Tezuka et al., 2002] that argued a stimulatory role of Notch as being confounded by an interaction with BMP used in that study [Hilton et al., 2008]. Examples of such discrepancies are plentiful in the literature, and it takes critical evaluation to identify the source of confusion and to design experiments that truly clarify the pieces of the mechanistic puzzle. This discussion only describes a few of the complicated aspects of interpreting results from multiple signaling pathways; ultimately, understanding pathway integration will require working in a more unified, defined system.
Embryonic stem cells: integrated signaling approaches that could be applied to MSC
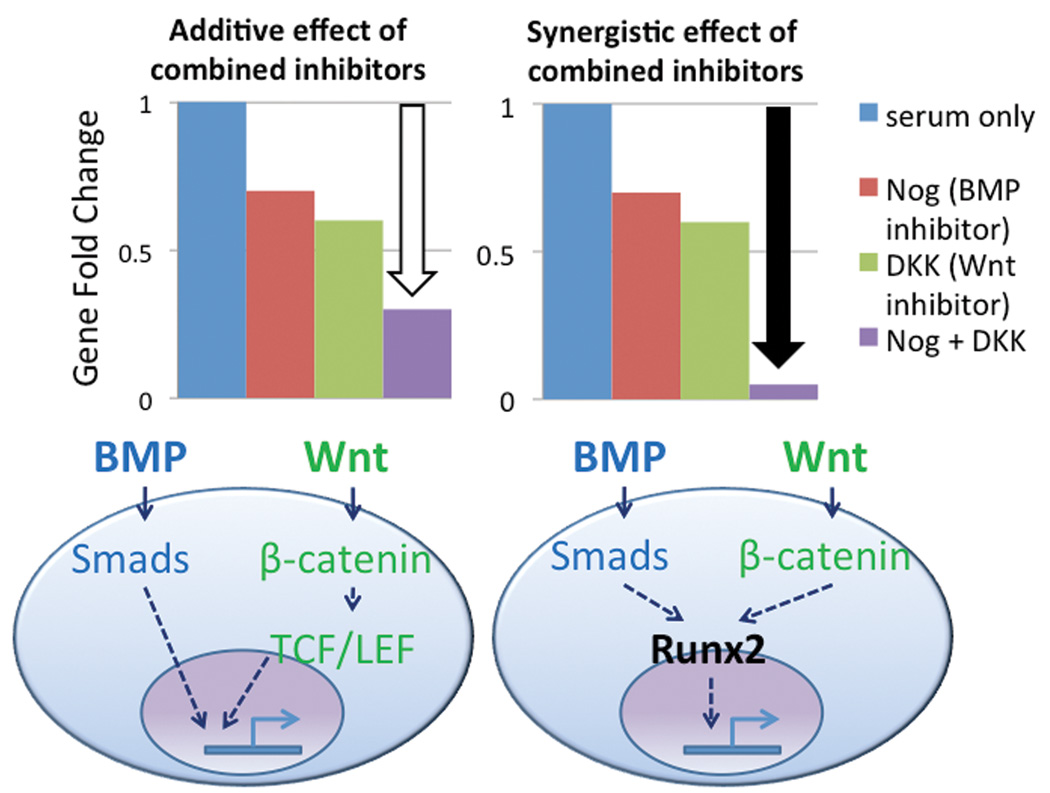
Various approaches used in embryonic stem cell (ESC) studies of signaling pathway integration could be applied to MSC differentiation. One study used inhibitors of each signaling pathway alone and in various combinations to illustrate how each pathway may be involved in commitment to blood vessel development [Lee et al., 2008]. Whether the effect of combined inhibitors is additive or synergistic affects mechanistic interpretations. For instance, an additive result suggests that the BMP and Wnt pathways may work in parallel to activate vertical signaling cascades converging on independent transactivating domains of the same gene. On the other hand, a synergistic effect of inhibiting BMP and Wnt pathways implies that the pathways could act through a common signal transducer (Figure 5).
Figure 5.
Additive (white arrow) vs. synergistic (black arrow) effects of combined inhibitors of different pathways suggest parallel/separate vs. integrated signaling transduction, respectively.
One method currently being used to study ESC differentiation involves flow cytometry to sort cells into groups at different stages of differentiation, followed by re-aggregation of those cells in culture and then gene expression analysis for undifferentiated vs. osteogenic or chondrogenic markers [Nostro et al., 2008]. This approach permits study of the regulation of distinct developmental stages in cell fate specification and therefore elucidates stage-specific roles for different signaling pathways. Another study utilized a stabilized conditional mutant β-catenin in ESCs and observed induction of different cell lineage markers when the cells were treated with inhibitors of other pathways such as BMP [Sumi et al., 2008]. These experiments were done in ESCs but could be translated to MSC and osteoblast progenitors.
Conclusions
Osteoblastogenesis is regulated by multiple signaling pathways. As described above, there are many factors to consider when comparing results from differing studies. In addition to being required for bone formation, Runx2 plays a central role in osteogenic differentiation studies of BMP, Wnt, Notch, Hh, and FGF pathways. Canonical and non-canonical Wnt signaling probably play different, interconnected roles in supporting undifferentiated, proliferative MSCs and stimulating those cells to respond to osteoinductive conditions. BMP is important for commitment to osteogenesis and may oppose proliferative pathways promoted by Wnt. Meanwhile, achieving maximal BMP expression and osteogenic function may depend on FGF. Early osteoblast expansion and commitment is supported by FGF and Wnt-dependent Hh signaling. Notch signaling drives proliferation of immature osteoblasts, leading to osteosclerosis in gain-of-function mutations and osteoporosis in loss-of-function mutations. Notch represses osteoblastic signaling induced by canonical Wnt and perhaps BMP. In all of these cases, the ability of mesenchymal progenitor cells to respond to signaling cues is most likely contextual, dependent upon the stage of osteoblast differentiation and the species-specific, tissue-specific origin of cells.
In order to harness the therapeutic potential of MSCs, a more precise molecular understanding of the complex process of differentiation is needed. This will involve learning how major signaling pathways, such as BMP, Wnt, and Notch, contribute to and integrate their signal inputs to influence cell fate. Advances in scientific technology for research applications promise to enable complicated analyses at the genomic level and to help organize methodologies to interpret results. The ability to manipulate MSC osteoblastogenesis has far-reaching clinical potential for pathological conditions such as osteoporosis, osteogenesis imperfecta, osteolytic lesions in metastatic cancers, primary bone tumors, high bone mass, and bone regeneration. At the same time, there is clinical relevance to understanding MSC differentiation in other lineages as well, such as chondrogenesis for arthritis and joint disorders, or adipogenesis for reconstructive or cosmetic surgery. Research in ESC differentiation has developed methods for systematically separating spatiotemporal relationships of key signaling factors and these approaches can and should be used to help guide research in MSC osteoblastogenesis.
Acknowledgements
The authors gratefully thank Jeroen Eyckmans for fruitful discussions and critically reading the manuscript, as well as the Medical Scientist Training Program for supporting this work. KDH is supported by NIH Grants R01 AR-054714 and R01 DE-017471.
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contributions
GLL drafted the manuscript and prepared the figures. GLL and KDH contributed to the conceptual design, edited, and approved the final manuscript.
References
- Ambrosetti D, Holmes G, Mansukhani A, Basilico C. Fibroblast growth factor signaling uses multiple mechanisms to inhibit Wnt-induced transcription in osteoblasts. Mol Cell Biol. 2008;28:4759–4771. doi: 10.1128/MCB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, Phinney DG. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–1249. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- Bain G, Muller T, Wang X, Papkoff J. Activated beta-catenin induces osteoblast differentiation of C3H10T1/2 cells and participates in BMP2 mediated signal transduction. Biochem Biophys Res Commun. 2003;301:84–91. doi: 10.1016/s0006-291x(02)02951-0. [DOI] [PubMed] [Google Scholar]
- Baksh D, Boland GM, Tuan RS. Cross-talk between Wnt signaling pathways in human mesenchymal stem cells leads to functional antagonism during osteogenic differentiation. J Cell Biochem. 2007;101:1109–1124. doi: 10.1002/jcb.21097. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CN, Ouyang H, Ma YL, Zeng Q, Gerin I, Sousa KM, Lane TF, Krishnan V, Hankenson KD, MacDougald OA. Wnt10b increases postnatal bone formation by enhancing osteoblast differentiation. J Bone Miner Res. 2007;22:1924–1932. doi: 10.1359/jbmr.070810. [DOI] [PubMed] [Google Scholar]
- Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- Celeste AJ, Iannazzi JA, Taylor RC, Hewick RM, Rosen V, Wang EA, Wozney JM. Identification of transforming growth factor beta family members present in bone-inductive protein purified from bovine bone. Proc Natl Acad Sci U S A. 1990;87:9843–9847. doi: 10.1073/pnas.87.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- Chen Y, Whetstone HC, Youn A, Nadesan P, Chow EC, Lin AC, Alman BA. Beta-catenin signaling pathway is crucial for bone morphogenetic protein 2 to induce new bone formation. J Biol Chem. 2007;282:526–533. doi: 10.1074/jbc.M602700200. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Cho SW, Yang JY, Sun HJ, Jung JY, Her SJ, Cho HY, Choi HJ, Kim SW, Kim SY, Shin CS. Wnt inhibitory factor (WIF)-1 inhibits osteoblastic differentiation in mouse embryonic mesenchymal cells. Bone. 2009;44:1069–1077. doi: 10.1016/j.bone.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Choi KY, Kim HJ, Lee MH, Kwon TG, Nah HD, Furuichi T, Komori T, Nam SH, Kim YJ, Ryoo HM. Runx2 regulates FGF2-induced Bmp2 expression during cranial bone development. Dev Dyn. 2005;233:115–121. doi: 10.1002/dvdy.20323. [DOI] [PubMed] [Google Scholar]
- Cui Y, Niziolek PJ, MacDonald BT, Zylstra CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon RA, Brommage R, Liu Q, Mseeh F, Powell DR, Yang QM, Zambrowicz B, Gerrits H, Gossen JA, He X, Bader M, Williams BO, Warman ML, Robling AG. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LA, Zur Nieden NI. Mesodermal fate decisions of a stem cell: the Wnt switch. Cell Mol Life Sci. 2008;65:2658–2674. doi: 10.1007/s00018-008-8042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- De Bari C, Dell'Accio F, Vanlauwe J, Eyckmans J, Khan IM, Archer CW, Jones EA, McGonagle D, Mitsiadis TA, Pitzalis C, Luyten FP. Mesenchymal multipotency of adult human periosteal cells demonstrated by single-cell lineage analysis. Arthritis Rheum. 2006;54:1209–1221. doi: 10.1002/art.21753. [DOI] [PubMed] [Google Scholar]
- Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J Biol Chem. 2006;281:6203–6210. doi: 10.1074/jbc.M508370200. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyckmans J, Roberts SJ, Schrooten J, Luyten FP. A clinically relevant model of osteoinduction: a process requiring calcium phosphate and BMP/Wnt signalling. J Cell Mol Med. 2010;14:1845–1856. doi: 10.1111/j.1582-4934.2009.00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Takeda K, Imamura T, Aoki H, Sampath TK, Enomoto S, Kawabata M, Kato M, Ichijo H, Miyazono K. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell. 1999;10:3801–3813. doi: 10.1091/mbc.10.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Janz S. Attenuation of WNT signaling by DKK-1 and -2 regulates BMP2-induced osteoblast differentiation and expression of OPG, RANKL and M-CSF. Mol Cancer. 2007;6:71. doi: 10.1186/1476-4598-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Gori F, Thomas T, Hicok KC, Spelsberg TC, Riggs BL. Differentiation of human marrow stromal precursor cells: bone morphogenetic protein-2 increases OSF2/CBFA1, enhances osteoblast commitment, and inhibits late adipocyte maturation. J Bone Miner Res. 1999;14:1522–1535. doi: 10.1359/jbmr.1999.14.9.1522. [DOI] [PubMed] [Google Scholar]
- Hanada K, Dennis JE, Caplan AI. Stimulatory effects of basic fibroblast growth factor and bone morphogenetic protein-2 on osteogenic differentiation of rat bone marrow-derived mesenchymal stem cells. J Bone Miner Res. 1997;12:1606–1614. doi: 10.1359/jbmr.1997.12.10.1606. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF, Glatt V, Bouxsein ML, Ai M, Warman ML, Williams BO. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res. 2004;19:2033–2040. doi: 10.1359/JBMR.040907. [DOI] [PubMed] [Google Scholar]
- Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL, Williams BO. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- Horwitz EM, Prockop DJ, Fitzpatrick LA, Koo WW, Gordon PL, Neel M, Sussman M, Orchard P, Marx JC, Pyeritz RE, Brenner MK. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci. 2002;115:3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J, Matsumoto K. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Sawada R, Fujiwara Y, Tsuchiya T. FGF-2 increases osteogenic and chondrogenic differentiation potentials of human mesenchymal stem cells by inactivation of TGF-beta signaling. Cytotechnology. 2008;56:1–7. doi: 10.1007/s10616-007-9092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javazon EH, Beggs KJ, Flake AW. Mesenchymal stem cells: paradoxes of passaging. Exp Hematol. 2004;32:414–425. doi: 10.1016/j.exphem.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon MJ, Kim JA, Kwon SH, Kim SW, Park KS, Park SW, Kim SY, Shin CS. Activation of peroxisome proliferator-activated receptor-gamma inhibits the Runx2-mediated transcription of osteocalcin in osteoblasts. J Biol Chem. 2003;278:23270–23277. doi: 10.1074/jbc.M211610200. [DOI] [PubMed] [Google Scholar]
- Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9:49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Bae SC, Choi JY, Ryoo HM. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J Biol Chem. 2003;278:319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Ma Y, Moseley J, Geiser A, Friant S, Frolik C. Bone anabolic effects of sonic/indian hedgehog are mediated by bmp-2/4-dependent pathways in the neonatal rat metatarsal model. Endocrinology. 2001;142:940–947. doi: 10.1210/endo.142.2.7922. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Krebsbach PH, Satomura K, Kerr J, Riminucci M, Benayahu D, Robey PG. Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J Bone Miner Res. 1997;12:1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, Lappe MM, Spitzer L, Zweier S, Braunschweiger K, Benchekroun Y, Hu X, Adair R, Chee L, FitzGerald MG, Tulig C, Caruso A, Tzellas N, Bawa A, Franklin B, McGuire S, Nogues X, Gong G, Allen KM, Anisowicz A, Morales AJ, Lomedico PT, Recker SM, Van Eerdewegh P, Recker RR, Johnson ML. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tang Y, Qiu T, Cao X, Clemens TL. A dishevelled-1/Smad1 interaction couples WNT and bone morphogenetic protein signaling pathways in uncommitted bone marrow stromal cells. J Biol Chem. 2006;281:17156–17163. doi: 10.1074/jbc.M513812200. [DOI] [PubMed] [Google Scholar]
- Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, McMahon AP. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131:1309–1318. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- Mak KK, Chen MH, Day TF, Chuang PT, Yang Y. Wnt/beta-catenin signaling interacts differentially with Ihh signaling in controlling endochondral bone and synovial joint formation. Development. 2006;133:3695–3707. doi: 10.1242/dev.02546. [DOI] [PubMed] [Google Scholar]
- Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbalaviele G, Sheikh S, Stains JP, Salazar VS, Cheng SL, Chen D, Civitelli R. Beta-catenin and BMP-2 synergize to promote osteoblast differentiation and new bone formation. J Cell Biochem. 2005;94:403–418. doi: 10.1002/jcb.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamizato T, Sakamoto K, Liu T, Kokubo H, Katsube K, Perbal B, Nakamura S, Yamaguchi A. CCN3/NOV inhibits BMP-2-induced osteoblast differentiation by interacting with BMP and Notch signaling pathways. Biochem Biophys Res Commun. 2007;354:567–573. doi: 10.1016/j.bbrc.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganawa T, Xiao L, Abogunde E, Sobue T, Kalajzic I, Sabbieti M, Agas D, Hurley MM. In vivo and in vitro comparison of the effects of FGF-2 null and haplo-insufficiency on bone formation in mice. Biochem Biophys Res Commun. 2006;339:490–498. doi: 10.1016/j.bbrc.2005.10.215. [DOI] [PubMed] [Google Scholar]
- Naganawa T, Xiao L, Coffin JD, Doetschman T, Sabbieti MG, Agas D, Hurley MM. Reduced expression and function of bone morphogenetic protein-2 in bones of Fgf2 null mice. J Cell Biochem. 2008;103:1975–1988. doi: 10.1002/jcb.21589. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Aikawa T, Iwamoto-Enomoto M, Iwamoto M, Higuchi Y, Pacifici M, Kinto N, Yamaguchi A, Noji S, Kurisu K, Matsuya T. Induction of osteogenic differentiation by hedgehog proteins. Biochem Biophys Res Commun. 1997;237:465–469. doi: 10.1006/bbrc.1997.7156. [DOI] [PubMed] [Google Scholar]
- Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem. 2005;280:15842–15848. doi: 10.1074/jbc.M412891200. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Plaisant M, Fontaine C, Cousin W, Rochet N, Dani C, Peraldi P. Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells. 2009;27:703–713. doi: 10.1634/stemcells.2008-0888. [DOI] [PubMed] [Google Scholar]
- Qiu W, Andersen TE, Bollerslev J, Mandrup S, Abdallah BM, Kassem M. Patients with high bone mass phenotype exhibit enhanced osteoblast differentiation and inhibition of adipogenesis of human mesenchymal stem cells. J Bone Miner Res. 2007;22:1720–1731. doi: 10.1359/jbmr.070721. [DOI] [PubMed] [Google Scholar]
- Qu-Petersen Z, Deasy B, Jankowski R, Ikezawa M, Cummins J, Pruchnic R, Mytinger J, Cao B, Gates C, Wernig A, Huard J. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. J Cell Biol. 2002;157:851–864. doi: 10.1083/jcb.200108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawadi G, Vayssiere B, Dunn F, Baron R, Roman-Roman S. BMP-2 controls alkaline phosphatase expression and osteoblast mineralization by a Wnt autocrine loop. J Bone Miner Res. 2003;18:1842–1853. doi: 10.1359/jbmr.2003.18.10.1842. [DOI] [PubMed] [Google Scholar]
- Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, Bianco P. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Sampath TK, Muthukumaran N, Reddi AH. Isolation of osteogenin, an extracellular matrix-associated, bone-inductive protein, by heparin affinity chromatography. Proc Natl Acad Sci U S A. 1987;84:7109–7113. doi: 10.1073/pnas.84.20.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitaye H, Hankenson KD. Immobilized Jagged-1 promotes proliferation of primary marrow stromal cells and inhibits osteogenic differentiationeditor^editors [Google Scholar]
- Spinella-Jaegle S, Rawadi G, Kawai S, Gallea S, Faucheu C, Mollat P, Courtois B, Bergaud B, Ramez V, Blanchet AM, Adelmant G, Baron R, Roman-Roman S. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. J Cell Sci. 2001;114:2085–2094. doi: 10.1242/jcs.114.11.2085. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Sun Q, Li C, Lu X, Qi H, Chen S, Yang J, Du X, Zhao L, He Q, Jin M, Shen Y, Chen D, Chen L. Gain-of-function mutation in FGFR3 in mice leads to decreased bone mass by affecting both osteoblastogenesis and osteoclastogenesis. Hum Mol Genet. 2010;19:1199–1210. doi: 10.1093/hmg/ddp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Tezuka K, Yasuda M, Watanabe N, Morimura N, Kuroda K, Miyatani S, Hozumi N. Stimulation of osteoblastic cell differentiation by Notch. J Bone Miner Res. 2002;17:231–239. doi: 10.1359/jbmr.2002.17.2.231. [DOI] [PubMed] [Google Scholar]
- Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferation in response to FGF. Biochem Biophys Res Commun. 2001;288:413–419. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- Wang EA, Rosen V, Cordes P, Hewick RM, Kriz MJ, Luxenberg DP, Sibley BS, Wozney JM. Purification and characterization of other distinct bone-inducing factors. Proc Natl Acad Sci U S A. 1988;85:9484–9488. doi: 10.1073/pnas.85.24.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol. 2008;20:171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Xiao L, Sobue T, Eisliger A, Kronenberg MS, Coffin JD, Doetschman T, Hurley MM. Disruption of the Fgf2 Gene Activates the Adipogenic and Suppresses the Osteogenic Program in Mesenchymal Marrow Stromal Stem Cells. Bone. doi: 10.1016/j.bone.2010.05.021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa T, Kataoka H, Kinto N, Iwamoto M, Enomoto-Iwamoto M, Iemura S, Ueno N, Shibata Y, Kurosawa H, Yamaguchi A. Sonic hedgehog is involved in osteoblast differentiation by cooperating with BMP-2. J Cell Physiol. 2002;193:225–232. doi: 10.1002/jcp.10166. [DOI] [PubMed] [Google Scholar]
- Zamurovic N, Cappellen D, Rohner D, Susa M. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 2004;279:37704–37715. doi: 10.1074/jbc.M403813200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]