Abstract
Protein-based markers that classify tumor subtypes and predict therapeutic response would be clinically useful in guiding patient treatment. We investigated the LC-MS/MS-identified protein biosignatures in 39 baseline breast cancer specimens including 28 HER2-positive and 11 triple-negative (TNBC) tumors. Twenty proteins were found to correctly classify all HER2 positive and 7 of the 11 TNBC tumors. Among them, galectin-3-binding protein and ALDH1A1 were found preferentially elevated in TNBC, whereas CK19, transferrin, transketolase, and thymosin β4 and β10 were elevated in HER2-positive cancers. In addition, several proteins such as enolase, vimentin, peroxiredoxin 5, Hsp 70, periostin precursor, RhoA, cathepsin D preproprotein, and annexin 1 were found to be associated with the tumor responses to treatment within each subtype. The MS-based proteomic findings appear promising in guiding tumor classification and predicting response. When sufficiently validated, some of these candidate protein markers could have great potential in improving breast cancer treatment.
1. Introduction
Chemotherapy has long been used to treat all types of cancer. Although survival benefits from adjuvant systemic chemotherapy in breast cancer have been thoroughly documented [1], success is not uniform with many still dying after the initial chemotherapy. The unpredictable tumor response to chemotherapy in any given patient and the significant toxicity manifested in all demand a better strategy for delivering cancer therapy.
In selective subtypes of breast cancer, therapies targeting specific signal transduction and/or metabolic pathways have been successful. For example, Herceptin for HER2/neu positive breast cancer [2, 3] and poly(ADP ribose) polymerase (PARP) inhibitors for triple-negative breast cancer with defective DNA-repair [4, 5] are among the recent successes of targeted therapy. The success of target therapy has led to an explosion of interest in developing tailored systemic therapy.
Breast cancer is a heterogeneous disease molecularly, histologically, and clinically. Clinical outcomes from the same treatment vary widely even among patients with tumors of identical stage and histology. Breast cancers developed from an accumulation of genetic alterations may partially explain the differences observed including tumor responses to anticancer agents [6]. Recently gene expression analysis has identified five subtypes of breast cancer which overlaps with clinical tumor classification according to the expression of three biomarkers, estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor 2 (HER2). Clinically these three markers are prognostically and therapeutically important in guiding treatment selection [7–10]; however, they do not fully reflect the complexity and heterogeneity of breast cancer and do not always predict the outcome of the treatment. For example, Herceptin as a single agent or in combination with chemotherapy has been shown to reduce recurrent disease and to save lives in patients with HER2-positive breast cancer, yet a significant number of HER2 overexpression tumors do not respond to the treatment [11]. Additional molecular targets are expected to improve tailored treatment in the future.
Proteomics has been employed in recent years to identify new disease-related biomarkers for cancer diagnosis and implementation of tailored treatment [12–15]. The tumor proteomes representing a global protein expression of cancer may provide new insights into the molecules that govern the dynamic cellular activities of tumor cells. Therefore, we choose to study breast tumor protein signatures in breast cancer classification and in predicting tumor response to treatment.
Previously we used SELDI mass spectrometry to profile tumor response to neoadjuvant treatment and found that significant m/z profile differences existed between cancers of nonresponders (tumor regression rate ≤25%) and others (tumor regression rate >25%) [16]. In this current study we have applied the LC-MS/MS technology to study the breast cancer proteomes in human tissues and identify unique proteins that may have the potential to separate two subtypes of breast cancer (TNBC versus HER2+) and to predict drug responses within each subtype.
2. Materials and Methods
2.1. Collection of Breast Tumor Tissues and Classification of Response
Breast tumors were collected, processed, and banked as previously described [16]. This study was approved by the UCLA institutional review board (IRB). Tumors from 39 consented patients with locally advanced breast cancer were collected from a neoadjuvant clinical trial [17]. Eleven were triple-negative breast tumors (TNBC, ER-/PR-/HER2-), and 28 were HER2-positive tumors (HER2+). The tumor specimens were uniformly collected according to a standard operating procedure established in our laboratory. Baseline tumor specimens were obtained by either core needle biopsy or surgical biopsy before starting the neoadjuvant Taxotere/Carboplatin/±Herceptin treatment (TC ± H). Evaluation of tumor response to the treatment was measured both by pathologic examination of surgically removed tissue and by clinical assessment including physical examination and/or imaging studies. The pathological response of the tumor was reported as either pathologically complete response (pCR) or having residual tumor. Because a baseline tumor size by pathologic evaluation was not possible in patients receiving neoadjuvant treatment, the clinically or imaging-measured tumor size prior to chemotherapy was used as the baseline tumor size. Pathological assessment after chemotherapy including tumor size, lymph node staging, and tumor biomarkers was performed on the specimen obtained from the definitive breast cancer surgery [16]. The tumor regression rate (TRR) was used to evaluate tumor response induced by neoadjuvant therapy, and it was calculated as follows: (baseline tumor size − residual tumor size)/baseline tumor size × 100%, where the baseline tumor size was measured clinically, and the postchemotherapy residual invasive tumor size was measured pathologically. The tumor response was categorized into three groups: responders (TRR > 75%, R), intermediate responders (25% < TRR ≤ 75%, IR), and nonresponders (TRR ≤ 25%, NR).
2.2. Protein Extraction and Abundant Protein Depletion
Protein extraction from tumors and depletion of abundant proteins from tumor lysates were performed as previously described [16]. Briefly, frozen tumors were homogenized in liquid nitrogen and suspended in 1% Triton X-100. The samples were refrozen at −80°C and thawed on ice twice. Following centrifugation (10,000 g, 10 min, 4°C), the supernatants were subjected to albumin and immunoglobulin depletion using an albumin and IgG removal kit (Amersham) as well as hemoglobin depletion using Ni-NTA magnetic agarose beads (Qiagen). Protein concentrations of each preparation were determined by the BioRad protein assays.
Because the blood proteins in the breast cancer tissue can cause significant ion suppression of lower abundance cancer-related proteins/peptides which may mask ion signals of less abundant peptides with similar M/Z ratios and retention times. In addition, the over presentation of serum proteins in the specimen may lower the amount of the cancer-related proteins available for LC-MS/MS analysis [18]. As a result, selected abundant serum proteins were depleted from the tissue extracts. Our preliminary test has shown more than 95% albumin, IgG and hemoglobin were removed by the described method, and more meaningful proteins have been detected.
2.3. Trypsin Digestion
The dried protein samples were dissolved in 6 M guanidine HCl, reduced with DTT (5 mM–15 mM), and alkylated using 10 mM iodoacetamide. Samples were then diluted with NH4HCO3 to lower guanidine HCl concentration (1 M), mixed with trypsin (1 : 50 w/w ratio, sequencing grade, Promega) containing 50 mM ammonium bicarbonate, and incubated at 37°C overnight. Samples were desalted by C18 Microspin columns (The Nest Group), and the eluates were dried in a vacuum centrifuge.
2.4. LC-MS/MS Analysis
Each digested and dried sample was prepared for LC-MS/MS analysis as previously reported [19]. Briefly the samples were redissolved in Buffer A (H2O/acetonitrile/formic acid, 98.9/1/0.1, typically at 0.7 μg protein/uL), and aliquots were injected (5 μL) onto an in-house-prepared C18 trap. The retained materials were placed onto a reverse phase column (New Objective C18, 15 cm, 75 μM diameter, 5 μm particle size equilibrated in Buffer A) and eluted (300 nL/min, Eksigent Nanolc-2D) with an increasing concentration of Buffer B (acetonitrile/water/formic acid, 98.9/1/0.1; min/%B: 0/5, 10/10, 112/40, 130/60, 135/90, 140/90). Eluted peptides were analyzed by MS and data-dependent MS/MS (collision-induced dissociation) using online data-dependent tandem mass spectrometry (LTQ Orbitrap, Thermo Fisher Scientific) in which the seven most abundant precursor ions were selected for MS/MS. Before testing the experimental specimens, the reproducibility of LC-MS/MS analysis was confirmed by examining the triplicates of two different tissue samples, and similar proteins were identified from the triplicates of each sample with more than 90% overlapping.
2.5. Database Searching and Analysis
BioWorks software (version 3.3.1, Thermo Fisher Sceintific), based on the SEQUEST algorithm (SRF v.5, Thermo Fisher Scientific), was used to search the mass spectra against a human trypsin indexed database (human.fasta.hdr database, Version 12.2, 227246 entries) as described by Whelan et al. [19]. SEQUEST was searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 50 PPM. The search tolerated up to two missed trypsin cleavages with variable modifications for carboxyamidomethylation (57.02146 Da) and methionine oxidation (15.99492 Da). Scaffold (version 3.0.3, Proteome Software, Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet Algorithm [20]. Protein identifications were accepted if they could be established with a greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet Algorithm [21].
From the resulting MS/MS protein identifications, a list of proteins was generated for each sample. A list of semi-quantitative protein abundances in the different samples was developed using the normalized spectrum counts of the identified tryptic peptides from each protein, as compiled by the Scaffold program. The protein lists and their relative abundances were then compared to find differentially expressed proteins between two groups.
2.6. Statistic Analysis
The data files exported from Scaffold were further processed as Excel files. The top 60% (180) abundant proteins of the 315 identified proteins were further selected for hierarchical clustering and supervised classification studies. Those proteins with at least a 2-fold difference in mean spectral counts between any two groups were selected for analysis in the web-based Gene Expression Profile Analysis Suite (GEPAS, version 4.0, http://www.gepas.org). Five different classification algorithms were tested to select candidate markers in the GEPAS software: Support Vector Machines (SVM), K-Nearest Neighbor Clustering (KNN), Diagonal Linear Discriminant Analysis (DLDA), Prediction Analysis with Microarrays (PAM), and Self-Organizing Map (SOM).
2.7. Immunohistochemistry Staining
A small portion of each of the baseline tumors was embedded in OCT and stored at −80°C. Endogenous peroxidase activity was quenched with 0.6% hydrogen peroxide in methanol for 10 minutes, and endogenous biotin was eliminated by Biotin Blocking System (DAKO, x0590). After blocking with 1 : 5 diluted normal goat serum or fetal bovine serum, slides were incubated for 1 hour with primary antibody (CK19, mouse IgG, ready to use, DAKO; galectin-3-binding protein, Goat IgG, 1 : 200 dilution, R&D) and 30 minutes with biotinylated secondary antibody (biotinylated anti-mouse Ig, 1 : 800 dilution, DAKO; biotinylated anti-goat Ig, 1 : 200 dilution, Vector Labs). Antigen-antibody complexes were then detected by the StreptABComplex/HRP method (DAKO) using diaminobenzidine as a chromogenic substrate (DAKO). Immunostained slides were lightly counterstained with hematoxylin. For negative controls, primary antibodies were replaced by mouse IgG or goat IgG.
3. Results
3.1. Patient Characteristics
The reported thirty-nine baseline tumor specimens included 28 HER2-positive breast cancers and 11 TNBC with HER2 status determined by fluorescence in situ hybridization (FISH) assay. Fifteen of the 28 HER2+ patients were randomized to receive TC, and the remaining 13 received TC and Herceptin (TCH) before surgery. All eleven patients with TNBC received neoadjuvant TC. Following the neoadjuvant treatment, 28 patients with HER2+ tumors showed 12 responders (R), including 7 with pathological complete response (pCR) and 5 with a tumor regression rate >75%, 12 intermediate responders (IR), and 4 nonresponders (NR). In the TNBC group, there were 7 responders including 6 pCR and 1 with tumor regression rate >75%, 3 IR, and 1 NR. The clinical characteristics and pathologic features of the 11 TNBC and 28 HER2+ cases are summarized in Tables 1(a) and 1(b).
Table 1.
(a) Clinical characteristics of 11 TNBC tumors
| LTQ Orbitrap sample ID | Patient age | Ethnicity | TR % | Response | T stage | Histological type | ER | PR | FISH R/G ratio | Neoadjuvant |
|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 61 | White | 80 | R | T3 | IDC | − | − | 1.10 | TC |
| #2 | 29 | Hispanic | −60 | NR | T3 | IDC | − | − | 0.92 | TC |
| #5 | 55 | Hispanic | 100 | R (pCR) | T3 | IDC | − | − | 0.92 | TC |
| #6 | 54 | Hispanic | 100 | R (pCR) | T3 | IDC | − | − | 1.01 | TC |
| #7 | 40 | Asian | 45 | IR | T3 | IDC | − | − | 1.17 | TC |
| #8 | 44 | White | 100a | R (pCR) | T3 | IDC | − | − | 1.00 | TC |
| #9 | 49 | Hispanic | 48 | IR | T4 | IDC | − | − | 1.10 | TC |
| #10 | 53 | White | 100 | R (pCR) | T3 | IDC | − | − | 1.20 | TC |
| #11 | 84 | Asian | 100 | R (pCR) | T4 | IDC | − | − | 1.03 | TC |
| #36 | 45 | Hispanic | 30 | IR | T3 | IDC | − | − | 1.27 | TC |
| #37 | 38 | White | 100 | R (pCR) | T2 | IDC | − | − | 1.10 | TC |
aLN positive without residual primary cancer.
(b) Clinical characteristics of 28 HER2+ tumors
| LTQ Orbitrap sample ID | Patient age | Ethnicity | TRR % | Response | T stage | Histological type | ER | PR | FISH R/G ratio | Neoadjuvant |
|---|---|---|---|---|---|---|---|---|---|---|
| #17 | 38 | White | 40 | IR | T3 | IDC | + | − | 12.4 | TC |
| #18 | 63 | Asian | 100 | R (pCR) | T3 | IDC | + | − | 12.7 | TCH |
| #19 | 57 | White | 100 | R (pCR) | T3 | IDC | − | − | 4.6 | TC |
| #20 | 56 | Asian | 78.2 | R | T4 | IDC | + | − | 10.71 | TC |
| #21 | 51 | Black | 56 | IR | T3 | IDC | − | − | 19.97 | TCH |
| #22 | 31 | White | 45.5 | IR | T3 | IDC | + | + | 2.2 | TC |
| #23 | 55 | White | 80 | R | T4 | IDC | + | + | 3.8 | TC |
| #24 | 45 | Asian | 75 | IR | T4 | IDC | + | + | 2.7 | TCH |
| #25 | 42 | Hispanic | 63.5 | IR | T4 | IDC | + | − | 2.5 | TC |
| #26 | 50 | White | 67.1 | IR | T3 | IDC | − | − | 2.41 | TCH |
| #27 | 33 | White | 82.9 | R | T3 | IDC | + | + | 3.03 | TCH |
| #28 | 40 | White | 66.7 | IR | T3 | IDC | − | − | 8.1 | TC |
| #29 | 35 | Hispanic | 100a | R (pCR) | T3 | IDC | − | − | 42.2 | TCH |
| #30 | 44 | White | 97.3 | R | T4 | IDC | − | − | 4.2 | TC |
| #31 | 30 | White | −7.7 | NR | T3 | IDC | + | − | 5 | TC |
| #32 | 57 | White | 25% | NR | T4 | IDC | + | − | >4 | TCH |
| #33 | 37 | White | 33.3 | IR | T2 | IDC | + | + | 9.49 | TC |
| #34 | 36 | Black | 25 | NR | T3 | IDC | − | − | 5.1 | TC |
| #35 | 42 | White | 60 | IR | T2 | IDC | + | + | 4.5 | TCH |
| #38 | 55 | White | 42.3 | IR | T4 | IDC | − | − | 3.9 | TCH |
| #39 | 47 | White | 100b | R (pCR) | T3 | IDC | + | + | >20 | TCH |
| #40 | 50 | Asian | 100b | R (pCR) | T3 | IDC | + | + | 4.19 | TCH |
| #41 | 58 | White | 50 | IR | T4 | IDC | + | + | 2.1 | TCH |
| #42 | 40 | Asian | 60 | IR | T2 | IDC | − | − | 16 | TC |
| #43 | 37 | White | −85.6 | NR | T3 | IDC | + | + | 3.1 | TC |
| #44 | 49 | White | 100b | R (pCR) | T2 | IDC | + | − | 7.7 | TCH |
| #45 | 55 | Asian | 92.6 | R | T3 | IDC | − | − | 9.9 | TC |
| #46 | 41 | Hispanic | 100 | R (pCR) | T2 | IDC | − | − | 9.2 | TC |
aLN positive and residual DCIS; bresidual DCIS only.
3.2. Protein Comparison between HER2+ and TNBC Groups
Proteins identified by MS/MS from the 39 tumors showed that 48 proteins were only found in HER2+ tumors, 24 were only seen in TNBC, and 243 proteins were shared by both, but the quantity of the shared proteins differed widely in the two tumor types. In this study, we focused the analysis on the top 60% abundant proteins (180/315) detected in the 39 tumors.
The 20 most abundant shared proteins by both subtypes of cancer were summarized in Table 2. Among them apolipoprotein A-I and D, enolase 1, tumor rejection antigen (gp96) 1, transgelin 2, cofilin 1, profilin, heat shock proteins 70, and annexins 5 were found to be present in significant quantity in both types of breast cancer. Some of these shared proteins found in sufficient amount may be useful for breast cancer detection.
Table 2.
The 20 most abundant proteins shared by both HER2-positive and TNBC tumors.
| Identified proteins | Accession no. | MW |
|---|---|---|
| Apolipoprotein A-I | gi∣90108664 | 28 kDa |
| Vimentin | gi∣62414289 | 54 kDa |
| Enolase 1 | gi∣4503571 | 47 kDa |
| Alpha-1 antitrypsin | gi∣157086955 | 45 kDa |
| Triosephosphate isomerase 1 | gi∣4507645 (+2) | 27 kDa |
| Cyclophilin A | gi∣1633054 | 18 kDa |
| Apolipoprotein D | gi∣619383 | 28 kDa |
| Cofilin 1 | gi∣5031635 | 19 kDa |
| Chaperonin | gi∣31542947 | 61 kDa |
| Transgelin 2 | gi∣4507357 | 22 kDa |
| Heat shock 70 kDa protein 5 | gi∣16507237 | 72 kDa |
| Tumor rejection antigen (gp96) 1 | gi∣4507677 | 92 kDa |
| S100 calcium-binding protein A11 | gi∣5032057 | 12 kDa |
| Lumican precursor | gi∣4505047 | 38 kDa |
| Tropomyosin 4 | gi∣4507651 | 29 kDa |
| ATP synthase, H+ transporting, mitochondrial F1 complex | gi∣32189394 | 57 kDa |
| Prosaposin isoform a preproprotein | gi∣11386147 | 58 kDa |
| Profilin | gi∣157838211 (+4) | 15 kDa |
| Heat shock 70 kDa protein 8 isoform 1 | gi∣5729877 | 71 kDa |
| Annexin 5 | gi∣4502107 | 36 kDa |
Of the 180 top abundant proteins observed in the 39 breast cancer specimens, 61 were found to have a ≥2-fold difference of spectrum counts between the two subtypes of breast cancer (HER2+ versus TNBC). Because some of these proteins were not detected in every sample, we further refined the list of differential proteins by selecting only those detected in ≥50% of the cases in either group. The selected 44 differentially expressed proteins were tested by hierarchical clustering to classify HER2+ breast cancer versus TNBC. These differentially expressed proteins correctly classified all 28 HER2+ tumors and 8 of the 11 TNBC by unweighted pair-group method using arithmetic average (UPGMA) (Figure 1).
Figure 1.
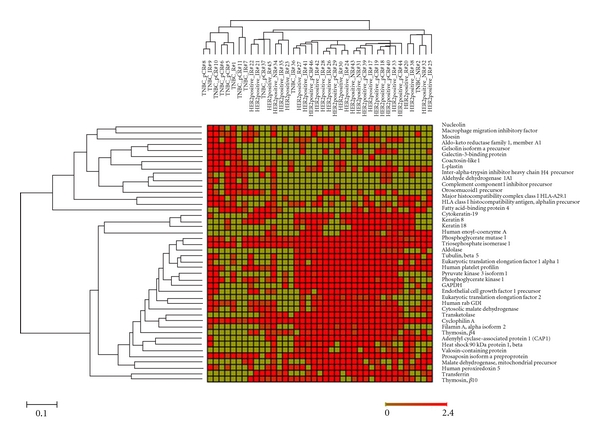
Heat map displaying the expression of 44 proteins in 28 HER2-positive and 11 TNBC tumors. Classification of 39 breast cancer cases into 2 groups based on tumor subtypes (HER-positive tumors and TNBC tumors) by the hierarchical clustering using GEPAS software. Each column represents a case as labeled on top, the short labeling cases are “TNBC” with sample ID, and long labeling cases are “HER2-positive” with sample ID. Each row represents a protein ID as indicated at the right. 44 proteins were expressed by ≥2-fold differences and detected in ≥50% of the cases in either group.
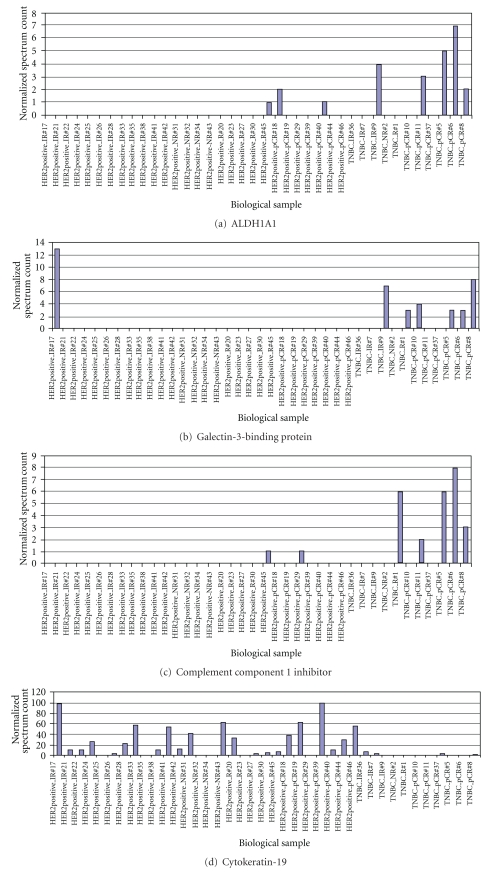
Self-validation of selected proteins in tumor classification was tested using a supervised classification. The 44 differentially expressed proteins were used to build a model separating subtypes of the tumors by Prophet, a web interface from the Gene Expression Profile Analysis Suite. Error rates were calculated as the number of misclassified tumors divided by total tumor cases tested. The error rates using various numbers of proteins by different models (SVM, KNN, DLDA, PAM, and SOM) were estimated by leaving-one-out tests (see File A in Supplementary Material available online at doi:10.1155/2011/896476). SVM had the lowest error rate (10%, 4/39) with 90% accuracy in tumor classification. The top 20 protein candidates (Table 3) selected by SVM model successfully classified all 28 HER2+ tumors and 7 of the 11 TNBC. Among the 20 differentially expressed proteins, G3BP, ALDH1A1, and complement component 1 inhibitor overexpression were found to be associated with TNBC subtype, whereas overexpression of CK19, transferrin, transketolase, and thymosin β4 and β10 were associated with HER2+ tumors (Figure 2).
Table 3.
Top 20 differentially expressed proteins selected by supervised classification methods for classifying two tumor subtypes.
| Rank | Accession no. | Protein name | MW | HER2+/TNBC mean | Subcellular location | Function |
|---|---|---|---|---|---|---|
| 1 | gi∣10946578 | Thymosin β4 | 5 kDa | 2.99 | Cytoplasm, cytoskeleton | For cytoskeletal binding, involved in cell growth and maintenance |
| 2 | gi∣4507521 | Transketolase | 68 kDa | 4.20 | Cytosol | Involved in metabolism. Associated with cell proliferation of uterine and cervical cancer. |
| 3 | gi∣1633054 | Cyclophilin A | 18 kDa | 2.45 | Cytoplasma | Involved in accelerate the folding of proteins |
| 4 | gi∣73858568 | Complement component 1 inhibitor | 55 kDa | 0.33 | Secreted | Regulating the complement cascade |
| 5 | gi∣4557871 | Transferrin | 77 kDa | 16.38 | Secreted | Essential for cell growth and iron-dependent metabolic processes |
| 6 | gi∣90111766 | Keratin type I cytoskeletal 19 | 44 kDa | 11.29 | Cytoskeleton | Involved in metastatic progression of breast cancer |
| 7 | gi∣10863895 | Thymosin β10 | 5 kDa | 2.25 | Cytoplasm, cytoskeleton | For cytoskeletal binding, involved in cell growth and maintenance |
| 8 | gi∣5031863 | Galectin-3-binding protein | 65 kDa | 0.41 | Secreted | Modulating cell-cell and cell-matrix interactions |
| 9 | gi∣4505753 (+1) | Phosphoglycerate mutase 1 | 29 kDa | 2.51 | Cytosol | Involved in glycolysis |
| 10 | gi∣5174391 | Aldo-keto reductase family 1, member A1 | 37 kDa | 0.30 | Cytosol | Involved in the reduction of biogenic and xenobiotic aldehydes |
| 11 | gi∣21361176 | Aldehyde dehydrogenase 1A1 | 55 kDa | 0.39 | Cytoplasm | Detoxifying enzyme responsible for oxidating of intracellular aldehydes. A marker for cancer stem cells |
| 12 | gi∣4505185 | Macrophage migration inhibitory factor | 12 kDa | 0.36 | Secreted, cytoplasm | Involved in integrin signaling pathways |
| 13 | gi∣4507645 (+2) | Triosephosphate isomerase 1 | 27 kDa | 2.49 | Cytosol, nucleus | Fatty acid biosynthesis, gluconeogenesis, glycolysis, lipid synthesis |
| 14 | gi∣4930167 | Aldolase A | 39 kDa | 6.41 | Extracellular, cytoskeleton | Involved in glycolysis |
| 15 | gi∣116241280 | Adenylyl cyclase-associated protein 1 (CAP 1) | 52 kDa | 3.03 | Membrane | Regulating filament dynamics, cell polarity and signal transduction, |
| 16 | gi∣21624607 (+5) | Coactosin-like 1 | 16 kDa | 0.42 | Cytoplasm, cytoskeleton | Regulating the actin cytoskeleton |
| 17 | gi∣160420317 | Filamin A, alpha isoform 2 | 281 kDa | 3.10 | Cytoplasm | Anchoring transmembrane proteins to the actin cytoskeleton, scaffold for cytoplasmic signaling proteins |
| 18 | gi∣6005942 | Valosin-containing protein | 89 kDa | 3.26 | Cytosol, nucleus | Fragmentation of Golgi stacks during mitosis and reassembly |
| 19 | gi∣5174539 | Cytosolic malate dehydrogenase | 36 kDa | 2.52 | Cytoplasm | Involved glycolysis, oxidation reduction, and tricarboxylic acid cycle |
| 20 | gi∣33286418 (+2) | Pyruvate kinase 3 | 58 kDa | 6 | Cytoplasm, nucleus | Involved in glycolysis |
Figure 2.
Representative proteins differentially expressed by HER2+ and TNBC tumors. (a)–(c): proteins preferentially expressed in TNBC. (d)-(e): proteins preferentially expressed in HER2+ tumors.
3.3. Proteins Correlated with Different Tumor Response to Neoadjuvant Treatment among HER2+ Tumors
Of the 28 HER2+ tumors, there were 12 R (including 7 with pCR), 12 IR, and 4 NR. We compared proteomic differences between the two groups with extreme tumor response (pCR and NR) and found that 48 of the 180 proteins had an expressional difference ≥2-fold between 7 pCR versus 4 NR tumors. Self-validation of these potential marker proteins by five supervised classification methods suggested that the KNN had the lowest error rate (9%, 1/11) in predicting tumor response (Files B and C). By using KNN = 1 method, 100% (4/4) NR and 85.7% (6/7) pCR were correctly grouped by 20 selected proteins (Table 4). Of the 20 proteins, overexpressions of enolase1, vimentin, and L-plastin in HER2-positive tumors were associated with pCR, whereas high level of heat shock proteins 70 (Hsp70) and peroxiredoxin 5 (Prx V) were found only in the NR cases.
Table 4.
Top 20 proteins predicting tumor response to neoadjuvant treatment in HER2-positive tumors.
| Rank | Protein name | Accession no. | pCR/NR mean | Subcellular location | Function |
|---|---|---|---|---|---|
| 1 | Enolase 1 | gi∣4503571 | 2.59 | Cytoplasm, cell membrane | Multifunctional enzyme |
| 2 | Heterogeneous nuclear ribonucleoprotein A2/B1 isoform B1 | gi∣14043072 | 3.51 | Nucleus, cytoplasm | Pre-mRNA processing |
| 3 | Heat shock 70 kDa protein 1 | gi∣75061728 | 0.24 | Cytoplasm | Stress response |
| 4 | Vimentin | gi∣62414289 | 9.94 | Cytosol | Class III intermediate filaments |
| 5 | Vesicle amine transport protein 1 | gi∣18379349 | 0.50 | Cytoplasmic vesicle membrane | Neurotransmitter transport |
| 6 | Coronin, actin-binding protein, 1A | gi∣5902134 | 2.00 | Cytoplasm | Component of the cytoskeleton of highly motile cells |
| 7 | Fatty acid-binding protein 4 | gi∣4557579 (+1) | 0.23 | Cytoplasm, nucleus | Lipid transport protein |
| 8 | Peroxiredoxin 5 | gi∣15826629 | 0.37 | Mitochondrion, cytoplasm, peroxisome | Antioxidant, oxidoreductase peroxidase |
| 9 | Heat shock 70 kDa protein 9 | gi∣24234688 | 0.15 | Mitochondrion | Control of cell proliferation and cellular aging |
| 10 | Leucine aminopeptidase 3 | gi∣41393561 | 2.94 | Cell membrane, secreted | Cell-cell signaling |
| 11 | Apolipoprotein D | gi∣619383 | 2.90 | Secreted | Lipid metabolic process |
| 12 | L-plastin | gi∣4504965 | 3.14 | Cytoplasm, cell membrane | Activation of T cells, intracellular protein transport |
| 13 | Anterior gradient protein 2 homolog precursor | gi∣5453541 | 0.11 | Secreted, endoplasmic reticulum | Mucus secretion |
| 14 | Heat shock 10 kDa protein 1 | gi∣4504523 | 0.37 | Mitochondrion | Stress response |
| 15 | ATP synthase, H+ transporting, mitochondrial F1 complex | gi∣4757810 | 0.41 | Mitochondrion | Proton-transporting ATP synthase complex assembly |
| 16 | Glutathione transferase | gi∣20664358 (+5) | 3.29 | Cytoplasm | Glutathione metabolic process |
| 17 | Chaperonin | gi∣31542947 | 0.33 | Mitochondrion | Stress response |
| 18 | Complement component 3 precursor | gi∣115298678 | 3.00 | Secreted | Activation of the complement system |
| 19 | Heterogeneous nuclear ribonucleoprotein D isoform a | gi∣14110420 | 2.19 | Nucleus, cytoplasm | Transcription regulation |
| 20 | Malate dehydrogenase | gi∣6648067 (+1) | 0.22 | Cytoplasm | Tricarboxylic acid cycle |
3.4. Proteins Predicting TNBC Tumor Response to Neoadjuvant Treatment
Among the 11 TNBC cases, there were 7 R (including 6 pCR), 3 IR, and 1 NR. Due to the small sample size, the proteins of responders' tumor (R) were compared to all the remaining tumors with less response (IR + NR). Sixty-three of 180 proteins had a ≥2-fold mean differences between the two groups of TNBC with different response to the same treatment. Self-validation of these proteins by five supervised classification methods was used to compare the accuracy in predicting a tumor response. Using DLDA method, 6 of 7 tumors in the R group and 3 of 4 tumors in IR/NR group were correctly classified by the 30 selected proteins (error rate 18%) (Files D and E). Of these 30 proteins, the increased heat shock 70 kDa protein 8, periostin, Ras homolog gene family member A (RhoA), actinin alpha 4, cathepsin D preproprotein, annexin 1, and several other proteins were associated with drug resistance in TNBC (Table 5).
Table 5.
Top 30 proteins predicting tumor response to neoadjuvant chemotherapy in TNBC tumors.
| Rank | Protein name | Accession no. | R/IR + NR mean | Subcellular location | Function |
|---|---|---|---|---|---|
| 1 | Heat shock 70 kDa protein 8 isoform 1 | gi∣5729877 | 0.32 | Stress response | |
| 2 | Periostin precursor (PN) (osteoblast-specific factor 2) | gi∣93138709 | 0.31 | Nucleus | Transcription regulation |
| 3 | Cyclophilin A | gi∣1633054 | 0.41 | Secreted | Cell attachment adhesion and spreading |
| 4 | Tyrosine 3/tryptophan 5-monooxygenase activation protein | gi∣5803225 (+1) | 3.71 | Nucleus | Protein binding |
| 5 | Profilin | gi∣157838211 (+4) | 0.32 | Cytoplasm, cytoskeleton | Actin cytoskeleton organization |
| 6 | Cardiac muscle alpha actin 1 proprotein | gi∣4885049 | 0.08 | Cytoplasm, cytokeleton | actin filament-based movement, apoptosis |
| 7 | Beta actin | gi∣4501885 | 0.22 | Cytoplasm, cytokeleton | Cell motility |
| 8 | Caldesmon (CDM) | gi∣2498204 | 0.42 | Cytoplasm, cytokeleton | Actin- and myosin-binding protein |
| 9 | Tubulin β5 | gi∣7106439 | 0.19 | Cytosol | Major constituent of microtubules |
| 10 | Tropomyosin 2 (beta) isoform 1 | gi∣42476296 | 0.11 | Cytoplasm, cytokeleton | Binding to actin filaments |
| 11 | Actinin, α4 | gi∣12025678 | 0.11 | Nucleus, cytoplasm | Protein transport |
| 12 | Ras homolog gene family, member A (RhoA) | gi∣10835049 (+4) | 0.33 | Cytoplasm, cell membrane | Regulating a signal transduction pathway |
| 13 | Heterogeneous nuclear ribonucleoprotein K | gi∣13384620 | 0.33 | Cytoplasm, nucleus | Pre-mRNA-binding proteins |
| 14 | Tubulin α1 | gi∣6755901 | 0.36 | Cytosol | Major constituent of microtubules |
| 15 | Tropomyosin 4 | gi∣4507651 | 0.35 | Cytoplasm, cytokeleton | Binds to actin filaments |
| 16 | Complement component 1 inhibitor precursor | gi∣73858568 | 4.57 | Secreted | Complement pathway |
| 17 | ATP synthase, H+ transporting, mitochondrial F1 complex | gi∣4757810 | 0.43 | Mitochondrion | Proton-transporting ATP synthase complex assembly |
| 18 | Calnexin precursor | gi∣10716563 | 0.42 | Endoplasmic reticulum membrane, cell membrane | Calcium-binding protein |
| 19 | Eukaryotic translation elongation factor 1 alpha 1 | gi∣4503471 | 0.29 | Cytoplasm | Protein biosynthesis |
| 20 | Annexin I | gi∣4502101 | 0.25 | Nucleus, cytoplasm, membrane | Calcium/phospholipid-binding protein |
| 21 | Triosephosphate isomerase 1 | gi∣4507645 (+2) | 0.35 | Cytosol, nucleus | Fatty acid biosynthesis, gluconeogenesis, glycolysis, lipid synthesis |
| 22 | Cathepsin D preproprotein | gi∣4503143 | 0.35 | Lysosome | proteolysis |
| 23 | Alpha glucosidase II alpha subunit isoform 2 | gi∣38202257 | 0.19 | Cytosol | Glycan metabolism, N-glycan metabolism |
| 24 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | gi∣4507949 (+1) | 0.42 | Nucleus | Protein binding |
| 25 | Thymosin β10 | gi∣10863895 | 0.42 | Cytoplasm, cytokeleton | cytoskeleton organization |
| 26 | Aconitase 2 precursor | gi∣4501867 | 0.44 | Mitochondrion | Carbohydrate metabolism, tricarboxylic acid cycle |
| 27 | Heterogeneous nuclear ribonucleoprotein D isoform a | gi∣14110420 | 0.48 | Nucleus, cytoplasm | Transcription regulation |
| 28 | Serine (or cysteine) proteinase inhibitor | gi∣32454741 | 0.25 | Secreted | Inhibits activated protein C, plasminogen activator |
| 29 | Lumican precursor | gi∣4505047 | 0.28 | Secreted | Binds to laminin |
| 30 | Apolipoprotein D | gi∣619383 | 2.86 | Secreted | Transport |
3.5. Evaluation of CK19 and G3BP Expression in TNBC and HER2+ Frozen Tumors
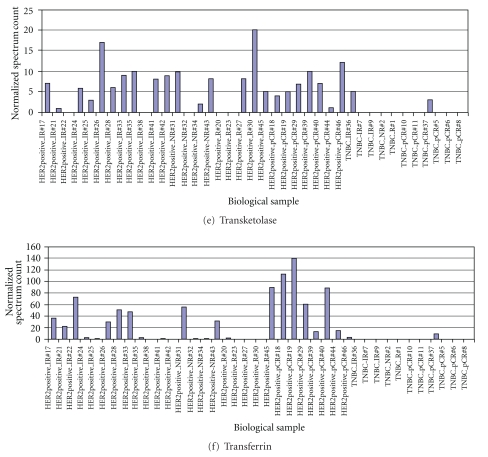
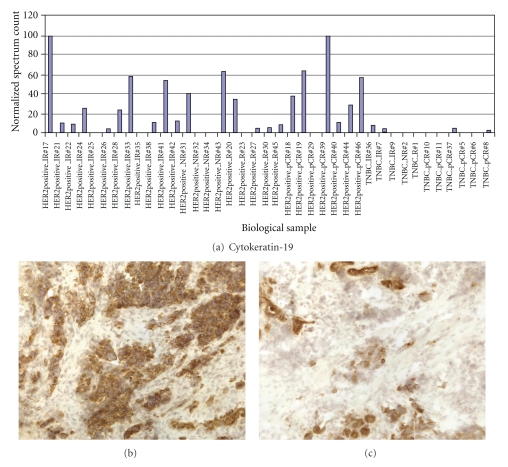
CK19 and G3BP protein expressions were tested in breast cancer tumors by immunohistochemistry. The CK19 and G3BP staining showed cytoplasmic/membrane staining pattern in breast cancer cells. The overexpression of CK19 was found in HER2+ breast tumors (Figure 3) while the expression of G3BP was found to be upregulated in most TNBC (Figure 4). The concordance findings between the mass spectrometry analysis and immunohistochemical staining of the same tumor suggested that high-throughput mass spectrometry may be used as a screening tool to discover disease-related biomarkers.
Figure 3.
CK19 expressions detected by LC-MS/MS and immunohistochemical (IHC) staining. Elevated CK19 expressions found in HER+ tumor group by LC-MS/MS and confirmed by IHC in most of the frozen HER2+ tumors. (a) Normalized spectrum count of CK19 detected in 39 breast cancer tissues. (b) Immunohistochemical staining of CK19 in a HER2+ frozen tumor (power 200x). (c) Immunohistochemical staining of CK19 in a TNBC frozen tumor (power 200x).
Figure 4.
G3BP expressions detected by LC-MS/MS and immunohistochemical (IHC) staining. Elevated G3BP expressions found in TNBC group by LC-MS/MS and confirmed by IHC in most of the frozen TNBC tumors. (a) Normalized spectrum count of G3BP detected from 39 breast cancer tissues. (b) Immunohistochemical staining of G3BP in a TNBC frozen tumor (power 200x). (c) Immunohistochemical staining of G3BP in a HER2+ frozen tumor (power 200x).
4. Discussion
In this discovery study, the MS-detected proteomic differences between two subtypes of breast cancer (HER2+ versus TNBC tumors) were explored, and proteomic prediction of tumor response to neoadjuvant chemotherapy was investigated. LC-MS/MS data sets of proteins from the 39 tumors analyzed allowed us to identify several candidate proteins that could classify tumor subtypes and predict tumor response to neoadjuvant chemotherapy.
Two clinical subtypes of breast cancer, HER2-positive and triple-negative breast cancers, defined by immunohistochemical staining and fluorescence in situ hybridization of three biomarkers of breast cancer have also been confirmed by gene analysis as two distinctive types of breast cancer. In this study, we reported that proteomic analysis could also separate the two subtypes by the unique biosignature associated with each type of breast cancer (Table 3). We also reported the potential of proteomic analysis in classifying drug-resistant TNBC and HER2+ breast cancer (Tables 4 and 5).
Through an extensive literature review, some of the identified proteins have reported roles that are relevant to cancer biology and treatment. In TNBC tumors, we observed that the levels of G3BP, ALDH1A1, and complement component 1 inhibitor protein were preferentially elevated. All of them have been reported to have important biological properties in cancer progression. G3BP, also known as 90-kDa Mac-2-binding protein, is a member of the beta-galactoside-binding protein family and has a role in modulating cell-cell and cell-matrix interactions. It has been shown that G3BP is overexpressed in a variety of cancer cells such as colon, gastric, and breast cancer, and its overexpression appears to correlate with tumor progression, and metastasis [22–26]. Our report is the first to describe G3BP overexpression in human TNBC by both mass spectrometry analysis and immunohistochemical staining method.
One protein correlated with triple-negative breast cancer meriting a discussion is ALDH1A1, a detoxifying enzyme responsible for oxidizing intracellular aldehydes. This process is important in early differentiation of stem cells through conversion of retinol to retinoic acid [27]. ALDH1 is considered to be a breast cancer stem cell marker and also a predictor for poor prognosis [27]. Because breast cancer stem cells have been implicated in radiation and chemotherapy resistance, as well as increasing the potential for metastasis, our finding of ALDH1A1 in TNBC may explain the more frequent relapse in TNBC patients. Previously, we have observed an overexpression of ALDH1 in TNBC when compared with hormone-receptor-positive and HER2-negative breast cancer [28]. In this paper, we also found a preferential overexpression of ALDH1 in TNBC over HER2-positive tumors. The unique overexpression of ALDH1 in TNBC tumors may point out an important population as the origin of some TNBC whereby notch signaling-dependent stem cell targets may be leveraged for target therapy development [29].
A different set of proteins was found preferentially elevated in HER2+ tumors. This list included CK19, transferrin, transketolase, and thymosin β4 and β10, and the biological significance of some of them will be discussed.
Cytokeratins are known to be important in cellular motility, signaling, and division. While CK8/CK18 were similarly detected in both HER2+ and TNBC tumors, elevated CK19 was more commonly found in HER2+ tumors. Our observation coincides with the finding reported by Schulz et al. using a combination of 2D-DIGE/mass spectrometry and western blot [30]. Both our current and previous papers suggest that CK19 is low in TNBC when compared with either HER2-positive or HER2-negative but hormone-receptor-positive breast cancer [31]. Although the biological significance and the mechanism of reduced CK19 in TNBC are not clear, it could be related to the frequent recurrence and poor overall survival rate seen in TNBC patients [32].
Transferrin, another protein associated with HER2-positive cancer, is essential for cell growth and iron-dependent metabolic activities including DNA synthesis, electron transport, and mitogenic signaling pathways [33]. The elevation of transferrin receptor (CD71) was reported to be a marker of poor outcome [33]. Vyhlidal et al. reported that transferrin is regulated by estrogen hormone [34], and tamoxifen was shown to be ineffective in ER-positive breast cancer with transferrin overexpression which coincides with the tamoxifen resistance observed in HER2-positive hormone-receptor-positive breast cancer. Taken together, the ineffectiveness of tamoxifen in women with HER2-positive and hormone-receptor-positive cancer may be related to the prevalent expression of transferrin in these tumors.
Thymosin β4 and β10 are members of a family of highly conserved small acid peptides that control the growth and differentiation of many cell types. They act as major actin-sequestering factor and play a role in cancer cell motility, invasion and metastasis [35, 36]. Thymosin β4 stimulates tumor metastasis by activating cell migration and angiogenesis in lung cancer and is associated with poor prognosis [37–39]. Elevations of thymosin β4 and β10 have also been reported in a number of other cancers including melanoma and breast cancer [40]. In the same tumor, the level of thymosin β10 was significantly higher in the cancer cells than in the normal breast parenchymal cells of the uninvolved area [41]. Its association with high-grade and poorly differentiated cancer cells is consistent with our findings of thymosin β10 overexpression in HER2-positive breast cancer. Further studies are required to confirm its overexpression and to determine its role in HER2-positive breast cancer.
In this study, we also reported the MS-identified protein signature predicting drug-induced tumor response in HER2-positive tumors. We found that enolase 1, vimentin, L-plastin, and ApoD predicted a favorable response of HER2-positive tumors. In contrast, elevated peroxiredoxin 5 and heat shock proteins 70 were found in nonresponding HER2-positive tumors.
Enolase 1(ENO1), a phosphopyruvate dehydratase, is a key glycolytic enzyme involved in anaerobic metabolism under hypoxic conditions of cancer growth, and a cell surface plasminogen receptor for tumor invasion. Overexpression of ENO1 in breast, and lung cancers is associated with tumor progression and rapid tumor growth [42]. While our study did not specifically studying the prognostic value of ENO1, the observation of ENO1 in HER2-positive tumors supports the prognostic importance of this molecule. While it was seen in the more aggressive subtype of breast cancer, our study showed ENO1 elevation in HER2-positive tumors seemed to indicate a better tumor response to chemotherapy.
Vimentin is a member of the intermediate filament family. Along with microtubules and actin microfilaments, vimentin is an integral component of the cell cytoskeleton. In cancer, altered vimentin level is associated with a dedifferentiated phenotype, increased motility, invasiveness, and poor clinical prognosis [43, 44]. Vimentin overexpression was found in 90.5% of grade III breast carcinomas [45] which may explain its presence in both HER2-positive breast cancer and in TNBC. Our study found that vimentin, although an aggressive marker for breast cancer growth, is another indicator for a favorable tumor response to chemotherapy. L-plastin is an actin-binding protein involved in cancer cell migration, invasion, and metastasis, and its expression in breast cancer cell lines correlates with the degree of invasiveness [30, 46]. In this paper, L-plastin overexpression in HER2-positive breast cancer was associated with a likelihood of pCR.
In contrast to those molecules associated with favorable tumor response to neoadjuvant therapy, high levels of Prx V in HER2-positive breast cancers were found to be associated with poor response to the same chemotherapy regimen. Peroxiredoxins (Prxs) represent a novel group of peroxidases containing high antioxidant activity involved in cell differentiation and apoptosis [47], and Prx V is particularly effective in reducing reactive oxygen species (ROS). Moreover, Prx V is found in peroxisomes and mitochondria where protection against ROS is mostly needed. The antioxidant activity of Prx V may be associated with drug resistance of the tumor cells.
While some molecules are unique to the characteristics of individual subtype of breast cancer, Hsp70 overexpression was found by us to be associated with drug resistance in both HER2-positive and TNBC tumors. Heat shock proteins are overexpressed in a wide range of human cancers and are implicated in tumor cell proliferation, differentiation, death, invasion, metastasis, and immune recognition [48]. Consistent with the cellular functions of Hsp70, clinically it has been correlated with poor prognosis in breast, endometrial, cervical, and bladder cancers. Others have also reported that Hsp70 mediated drug resistance through its inhibitory effect on chemotherapy-induced tumor cell apoptosis [48–50].
In TNBC tumors, a list of different proteins was found to be overexpressed in tumors resistant to neoadjuvant chemotherapy. In addition to Hsp70, proteins such as periostin precursor (OSF-2), RhoA, actinin α4, cathepsin D preproprotein, and annexin 1 predicted a poor response of TNBC to treatment. Although all of them were known to have important cancer biological properties, they have not been linked to chemotherapy susceptibility until now.
Periostin was originally identified in a mouse osteoblastic cell line as an extracellular matrix adhesion protein for pre-osteoblasts. In addition to forming bones, teeth, and heart, periostin was recently found to be overexpressed in various types of human cancer. Periostin interacts with multiple cell-surface receptors (most notable integrins) and signals via the PI3-K/Akt and other pathways to promote cancer cell survival, epithelial-mesenchymal transition, invasion, and metastasis [51]. In breast cancer, periostin was found upregulated at both the mRNA and protein levels [51–55]. Activation of the Akt/PKB cellular survival pathway with consequential protection of tumor cells and endothelial cells from stress-induced cell death [51, 56] may contribute to the periostin-mediated drug resistance in cancer. To our knowledge, this is the first paper to link periostin to drug resistance in TNBC.
RhoA is a member of the Ras superfamily. It is involved in the regulation and timing of cell division. It is a small GTPase protein known to regulate the actin cytoskeleton in the formation of stress fibers. RhoA protein levels were significantly increased in breast cancer compared with the matched normal tissue. It has been reported by Fritz et al. that an elevated RhoA protein level correlated with increasing breast tumor grade and poor prognosis [57].
Actinin α4 is another interesting protein that we found to indicate a poor tumor response to neoadjuvant therapy. It is thought that the actinin α4 cross-links actin filaments and connects the actin cytoskeleton to the cell membrane. The accumulation of actinin α4 in the cytoplasm is related to tumor invasiveness and metastasis, probably by enhancing cell motility, and was suggested to be a novel prognosticator in patients with ovarian and breast cancer [58].
Cathepsin D, an acid protease, is active in intracellular protein breakdown and is involved in the pathogenesis of several diseases. Its preproprotein secreted by cancer cells, acting as a mitogen on both cancer and stromal cells, stimulates both proinvasive and prometastatic properties of cancer cells. Many studies found that cathepsin D preproprotein/cathepsin D level represents an independent prognostic factor in a variety of cancers and is, therefore, considered to be a potential target for anticancer therapy [59]. Others have also shown that overexpression of cathepsin D in human breast cancers is associated with a higher risk of relapse and metastasis [59–61]. In our study, cathepsin D preproprotein appeared to be a drug-resistant marker in TNBC.
Although many proteins identified in this pilot study are interesting with promising potential, this study has several limitations. First, the tumors used in this study were collected from a clinical trial which provided many controlled clinical data; however, the sample size available for proteomic analysis was small. As a result, the findings derived from a small sample size always warrant a cautious interpretation. Second, the HER2-positive group consisted of tumors with different ER and PR status which might interfere with the conclusion. The potential false associations with HER2 might be solved by stratifying the HER2-positive tumors according to hormonal receptor status in a larger study. Lastly, the HER2-positive patients in this study were randomized to receive either chemotherapy alone or chemotherapy with Herceptin. The selected drug-resistant markers may represent the resistance not only to the chemotherapy but also to Herceptin.
In summary, our study has led to the identification of a list of important breast cancer proteins. The study also suggests that MS-based protein profiling may be an important tool in discovery of cancer biosignatures for tumor subtyping and prediction of treatment outcome. When sufficiently validated, some of these candidate protein markers could be used to improve breast cancer care. In addition, due to the heterogeneous and complex nature of the breast cancer tissue specimens, more refined methods need to be developed to maximize the protein identification to allow the capture of the best protein candidate markers for clinical use.
Supplementary Material
The error rates of tumor classification predicted by different sets of proteins in each model (SVM, KNN, DLDA, PAM, and SOM) were estimated by leaving-one-out test (GEPAS, version 4.0, http://www.gepas.org).
File A: SVM had the lowest error rate (10%, 4/39) in tumor classification using 20 proteins listed in Table 3.
File B and C: KNN had the lowest error rate (9%, 1/11) in predicting HER2-positive tumor response using 20 proteins listed in Table 4. By using KNN = 1 method, 100% (4/4) tumors in NR and 85.7% (6/7) tumors in pCR were correctly grouped.
File D and E: DLDA had the lowest error rate (18%, 2/11) in predicting TNBC tumor response using 30 proteins listed in Table 5. 85.7% (6/7) tumors in the R group and 75% (3/4) tumors in IR/NR group were correctly classified.
Conflict of Interests
The authors declare that they have no conflict of interests.
Funding
Grant support: The present study was supported by grant funds from NIH (NCI no. 1RO1 CA 093736-01A1), the Gonda Foundation, the Entertainment Industry Foundation (EIF)/Women's Cancer Research Fund, and the Friends of the Breast Program at UCLA.
Acknowledgments
The authors thank Jeffrey A. Gornbein (Department of Biomathematics, UCLA) for providing critical advice on statistical analysis.
Abbreviations
- DLDA:
Diagonal linear discriminant analysis
- G3BP:
Galectin-3-binding protein
- GEPAS:
Gene expression pattern analysis suite
- IR:
Intermediate responders (>25% but ≤75% tumor regression)
- KNN:
K nearest neighbor
- MS:
Mass spectrometry
- NR:
Nonresponders, chemoresistant tumors (≤25% tumor regression)
- PAM:
Prediction analysis with microarrays
- pCR:
Pathological complete response, no residual cancer found at primary tumor site
- R:
Responders (>75% of tumor regression)
- SOM:
Self-organizing map
- SVM:
Support vector machines
- TC ± H:
Taxotere/Carboplatin/±Herceptin treatment
- TNBC:
Triple negative breast tumors
- TRR:
Tumor regression rate.
References
- 1.Carlson RW, Brown E, Burstein HJ, et al. NCCN task force report: adjuvant therapy for breast cancer. Journal of the National Comprehensive Cancer Network. 2006;4(supplement 1):S1–S26. [PubMed] [Google Scholar]
- 2.Baselga J, Gianni L, Geyer C, Perez EA, Riva A, Jackisch C. Future options with trastuzumab for primary systemic and adjuvant therapy. Seminars in Oncology. 2004;31(5, supplement 10):51–57. doi: 10.1053/j.seminoncol.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 3.Ross JS, Schenkein DP, Pietrusko R, et al. Targeted therapies for cancer 2004. American Journal of Clinical Pathology. 2004;122(4):598–609. doi: 10.1309/5CWP-U41A-FR1V-YM3F. [DOI] [PubMed] [Google Scholar]
- 4.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England Journal of Medicine. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 5.Anders CK, Winer EP, Ford JM, et al. Poly(ADP-ribose) polymerase inhibition: “targeted” therapy for triple-negative breast cancer. Clinical Cancer Research. 2010;16(19):4702–4710. doi: 10.1158/1078-0432.CCR-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radpour R, Barekati Z, Kohler C, Holzgreve W, Zhong XY. New trends in molecular biomarker discovery for breast cancer. Genetic Testing and Molecular Biomarkers. 2009;13(5):565–571. doi: 10.1089/gtmb.2009.0060. [DOI] [PubMed] [Google Scholar]
- 7.Reis-Filho JS, Tutt ANJ. Triple negative tumours: a critical review. Histopathology. 2008;52(1):108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 8.Perou CM, Sørile T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 9.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sørlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang HR. Trastuzumab-based neoadjuvant therapy in patients with HER2-positive breast cancer. Cancer. 2010;116(12):2856–2867. doi: 10.1002/cncr.25120. [DOI] [PubMed] [Google Scholar]
- 12.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nature Clinical Practice Oncology. 2008;5(10):588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 13.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Molecular & Cellular Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Hondermarck H, Tastet C, Yazidi-Belkoura IE, Toillon RA, Bourhis XL. Proteomics of breast cancer: the quest for markers and therapeutic targets. Journal of Proteome Research. 2008;7(4):1403–1411. doi: 10.1021/pr700870c. [DOI] [PubMed] [Google Scholar]
- 15.Röwer C, Vissers JPC, Koy C, et al. Towards a proteome signature for invasive ductal breast carcinoma derived from label-free nanoscale LC-MS protein expression profiling of tumorous and glandular tissue. Analytical and Bioanalytical Chemistry. 2009;395(8):2443–2456. doi: 10.1007/s00216-009-3187-9. [DOI] [PubMed] [Google Scholar]
- 16.He J, Shen D, Chung DU, et al. Tumor proteomic profiling predicts the susceptibility of breast cancer to chemotherapy. International Journal of Oncology. 2009;35(4):683–692. doi: 10.3892/ijo_00000380. [DOI] [PubMed] [Google Scholar]
- 17.Chang HR, Glaspy J, Allison MA, et al. Differential response of triple-negative breast cancer to a docetaxel and carboplatin-based neoadjuvant treatment. Cancer. 2010;116(18):4227–4237. doi: 10.1002/cncr.25309. [DOI] [PubMed] [Google Scholar]
- 18.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnology. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 19.Whelan SA, Lu M, He J, et al. Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. Journal of Proteome Research. 2009;8(8):4151–4160. doi: 10.1021/pr900322g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 21.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 22.Ulmer TA, Keeler V, Loh L, et al. Tumor-associated antigen 90K/Mac-2-binding protein: possible role in colon cancer. Journal of Cellular Biochemistry. 2006;98(5):1351–1366. doi: 10.1002/jcb.20784. [DOI] [PubMed] [Google Scholar]
- 23.Park YP, Choi SC, Kim JH, et al. Up-regulation of Mac-2 binding protein by hTERT in gastric cancer. International Journal of Cancer. 2007;120(4):813–820. doi: 10.1002/ijc.22369. [DOI] [PubMed] [Google Scholar]
- 24.Mbeunkui F, Metge BJ, Shevde LA, Pannell LK. Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. Journal of Proteome Research. 2007;6(8):2993–3002. doi: 10.1021/pr060629m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Ao X, Vuong H, et al. Membrane glycoproteins associated with breast tumor cell progression identified by a lectin affinity approach. Journal of Proteome Research. 2008;7(10):4313–4325. doi: 10.1021/pr8002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Q, Guo X, Nash GB, et al. Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Research. 2009;69(17):6799–6806. doi: 10.1158/0008-5472.CAN-09-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M, Whitelegge JP, Whelan SA, et al. Hydrophobic fractionation enhances novel protein detection by mass spectrometry in triple negative breast cancer. Journal of Proteomics and Bioinformatics. 2010;3(2):1–10. doi: 10.4172/jpb.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Research. 2010;70(23):9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz DM, Böllner C, Thomas G, et al. Identification of differentially expressed proteins in triple-negative breast carcinomas using DIGE and mass spectrometry. Journal of Proteome Research. 2009;8(7):3430–3438. doi: 10.1021/pr900071h. [DOI] [PubMed] [Google Scholar]
- 31.Lu M, Whelan SA, He J, et al. Hydrophobic proteome analysis of triple negative and hormone-receptor- positive-her2-negative breast cancer by mass spectrometer. Clinical Proteomics. 2010;6(3):93–103. doi: 10.1007/s12014-010-9052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parikh RR, Yang Q, Higgins SA, Haffty BG. Outcomes in young women with breast cancer of triple-negative phenotype: the prognostic significance of CK19 expression. International Journal of Radiation Oncology Biology Physics. 2008;70(1):35–42. doi: 10.1016/j.ijrobp.2007.05.066. [DOI] [PubMed] [Google Scholar]
- 33.Habashy HO, Powe DG, Staka CM, et al. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Research and Treatment. 2010;119(2):283–293. doi: 10.1007/s10549-009-0345-x. [DOI] [PubMed] [Google Scholar]
- 34.Vyhlidal C, Li X, Safe S. Estrogen regulation of transferrin gene expression in MCF-7 human breast cancer cells. Journal of Molecular Endocrinology. 2002;29(3):305–317. doi: 10.1677/jme.0.0290305. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa T, Huang SK, Martinez SR, et al. Proteomic profiling of primary breast cancer predicts axillary lymph node metastasis. Cancer Research. 2006;66(24):11825–11830. doi: 10.1158/0008-5472.CAN-06-2337. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein AL. Thymosin β4: a new molecular target for antitumor strategies. Journal of the National Cancer Institute. 2003;95(22):1646–1647. doi: 10.1093/jnci/djg126. [DOI] [PubMed] [Google Scholar]
- 37.Cha HJ, Jeong MJ, Kleinman HK. Role of thymosin β4 in tumor metastasis and angiogenesis. Journal of the National Cancer Institute. 2003;95(22):1674–1680. doi: 10.1093/jnci/djg100. [DOI] [PubMed] [Google Scholar]
- 38.Rahman SMJ, Gonzalez AL, Li M, et al. Lung cancer diagnosis from proteomic analysis of preinvasive lesions. Cancer Research. 2011;71(8):3009–3017. doi: 10.1158/0008-5472.CAN-10-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu BJ, Gonzalez AL, Kikuchi T, et al. MALDI-MS derived prognostic protein markers for resected non-small cell lung cancer. Proteomics—Clinical Applications. 2008;2(10-11):1508–1517. doi: 10.1002/prca.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutton CW, Rustogi N, Gurkan C, et al. Quantitative proteomic profiling of matched normal and tumor breast tissues. Journal of Proteome Research. 2010;9(8):3891–3902. doi: 10.1021/pr100113a. [DOI] [PubMed] [Google Scholar]
- 41.Verghese-Nikolakaki S, Apostolikas N, Livaniou E, Ithakissios DS, Evangelatos GP. Preliminary findings on the expression of thymosin beta-10 in human breast cancer. British Journal of Cancer. 1996;74(9):1441–1444. doi: 10.1038/bjc.1996.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennipman A, van Oirschot BA, Smits J, Rijksen G, Staal GEJ. Glycolytic enzyme activities in breast cancer metastases. Tumor Biology. 1988;9(5):241–248. doi: 10.1159/000217568. [DOI] [PubMed] [Google Scholar]
- 43.Kokkinos MI, Wafai R, Wong MK, Newgreen DF, Thompson EW, Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer—observations in vitro and in vivo. Cells Tissues Organs. 2007;185(1–3):191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 44.Hendrix MJC, Seftor EA, Seftor REB, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. American Journal of Pathology. 1997;150(2):483–495. [PMC free article] [PubMed] [Google Scholar]
- 45.Korsching E, Packeisen J, Liedtke C, et al. The origin of vimentin expression in invasive breast cancer: epithelial- mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? Journal of Pathology. 2005;206(4):451–457. doi: 10.1002/path.1797. [DOI] [PubMed] [Google Scholar]
- 46.Lin CS, Chen ZP, Park T, Ghosh K, Leavitt J. Characterization of the human L-plastin gene promoter in normal and neoplastic cells. Journal of Biological Chemistry. 1993;268(4):2793–2801. [PubMed] [Google Scholar]
- 47.Karihtala P, Mäntyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clinical Cancer Research. 2003;9(9):3418–3424. [PubMed] [Google Scholar]
- 48.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress and Chaperones. 2005;10(2):86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jäättelä M. Escaping cell death: survival proteins in cancer. Experimental Cell Research. 1999;248(1):30–43. doi: 10.1006/excr.1999.4455. [DOI] [PubMed] [Google Scholar]
- 50.Mosser DD, Caron AW, Bourget L, et al. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Molecular and Cellular Biology. 2000;20(19):7146–7159. doi: 10.1128/mcb.20.19.7146-7159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan K, Bao S, Ouyang G. The multifaceted role of periostin in tumorigenesis. Cellular and Molecular Life Sciences. 2009;66(14):2219–2230. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao R, Bao S, Bai X, et al. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Molecular and Cellular Biology. 2004;24(9):3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang X, Zhao J, Hajivandi M, et al. Quantification of membrane and membrane-bound proteins in normal and malignant breast cancer cells isolated from the same patient with primary breast carcinoma. Journal of Proteome Research. 2006;5(10):2632–2641. doi: 10.1021/pr060125o. [DOI] [PubMed] [Google Scholar]
- 54.Liang X, Huuskonen J, Hajivandi M, et al. Identification and quantification of proteins differentially secreted by a pair of normal and malignant breast-cancer cell lines. Proteomics. 2009;9(1):182–193. doi: 10.1002/pmic.200700957. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Zhang G, Li J, Tao Q, Tang W. The expression analysis of periostin in human breast cancer. Journal of Surgical Research. 2010;160(1):102–106. doi: 10.1016/j.jss.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 56.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for αVβ3 and αVβ5 integrins and promotes cell motility. Cancer Research. 2002;62(18):5358–5364. [PubMed] [Google Scholar]
- 57.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. British Journal of Cancer. 2002;87(6):635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honda K, Yamada T, Endo R, et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. Journal of Cell Biology. 1998;140(6):1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benes P, Vetvicka V, Fusek M. Cathepsin D-many functions of one aspartic protease. Critical Reviews in Oncology/Hematology. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thorpe SM, Rochefort H, Garcia M, et al. Association between high concentrations of M(r) 52,000 cathepsin D and poor prognosis in primary human breast cancer. Cancer Research. 1989;49(21):6008–6014. [PubMed] [Google Scholar]
- 61.Spyratos F, Maudelonde T, Brouillet JP, et al. Cathepsin D: an independent prognostic factor for metastasis breast cancer. The Lancet. 1989;2(8672):1115–1118. doi: 10.1016/s0140-6736(89)91487-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The error rates of tumor classification predicted by different sets of proteins in each model (SVM, KNN, DLDA, PAM, and SOM) were estimated by leaving-one-out test (GEPAS, version 4.0, http://www.gepas.org).
File A: SVM had the lowest error rate (10%, 4/39) in tumor classification using 20 proteins listed in Table 3.
File B and C: KNN had the lowest error rate (9%, 1/11) in predicting HER2-positive tumor response using 20 proteins listed in Table 4. By using KNN = 1 method, 100% (4/4) tumors in NR and 85.7% (6/7) tumors in pCR were correctly grouped.
File D and E: DLDA had the lowest error rate (18%, 2/11) in predicting TNBC tumor response using 30 proteins listed in Table 5. 85.7% (6/7) tumors in the R group and 75% (3/4) tumors in IR/NR group were correctly classified.