Abstract
Determination of the prevalence of accumulated antiretroviral drug resistance among persons infected with human immunodeficiency virus (HIV) is complicated by the lack of routine measurement in clinical care. By using data from 8 clinic-based cohorts from the North American AIDS Cohort Collaboration on Research and Design, drug-resistance mutations from those with genotype tests were determined and scored using the Genotypic Resistance Interpretation Algorithm developed at Stanford University. For each year from 2000 through 2005, the prevalence was calculated using data from the tested subset, assumptions that incorporated clinical knowledge, and multiple imputation methods to yield a complete data set. A total of 9,289 patients contributed data to the analysis; 3,959 had at least 1 viral load above 1,000 copies/mL, of whom 2,962 (75%) had undergone at least 1 genotype test. Using these methods, the authors estimated that the prevalence of accumulated resistance to 2 or more antiretroviral drug classes had increased from 14% in 2000 to 17% in 2005 (P < 0.001). In contrast, the prevalence of resistance in the tested subset declined from 57% to 36% for 2 or more classes. The authors’ use of clinical knowledge and multiple imputation methods revealed trends in HIV drug resistance among patients in care that were markedly different from those observed using only data from patients who had undergone genotype tests.
Keywords: antiretroviral therapy, highly active; drug resistance; genotype; HIV
Missingness of data is a ubiquitous problem in epidemiologic studies and results from 2 primary causes. The first contributor is lack of response, which can result from refusal (nonresponse) or loss to follow-up (nonparticipation). Incomplete data may also occur structurally, such as when data collection is determined by clinical-care decisions. Those selected for a medical procedure, intervention, or test will contribute data, whereas those not selected or indicated for treatment will not. By definition, the selected sample will differ clinically from those who do not receive the treatment.
Human immunodeficiency virus (HIV) drug resistance is one area of research in which the amount of missing data is heavily influenced by selection. Combination antiretroviral therapy (ART) has been highly successful over the past decade in reducing the morbidity and mortality associated with HIV-1 infection (1) and restoring immunologic function (2). However, poor adherence (and subsequent virologic failure), as well as extensive treatment exposures (particularly to suboptimal treatment regimens), can result in acquired resistance to both specific HIV drugs and entire drug classes (3–5), limiting treatment options. Population trends in resistance could thus be a primary motivator for the development of new therapeutic agents and could be informative for estimating the burden of uncontrolled disease and cost of care. Furthermore, individuals with a resistant virus that is uncontrolled by their current regimens will contribute to part of a community viral load, one of the key metrics identified in the 2010 National HIV/AIDS Strategy (6).
Most data available on drug resistance status arise from genotype testing, that is, genotyping of sampled HIV strains to detect mutations that confer drug resistance. These tests have several characteristics that complicate analysis of even simple prevalence descriptions. First, standard laboratory tests measure only genotypic resistance among subjects with a high viral load. As a consequence, individuals who undergo genotype testing likely differ clinically from those who do not. Second, Department of Health and Human Services guidelines recommend that these resistance tests be used to guide the choice of ART for those who may be failing to respond to other therapies. Third, characterization of HIV drug resistance is complicated by the impact of changing drug regimens on circulating viral strains. Therapies will suppress but not eradicate an established viral strain. Such archived resistance cannot be detected by genotype testing with commercial assays but contributes to the overall burden of drug resistance because it influences treatment options and effectiveness (7).
To estimate of the prevalence of latent and circulating resistance among treated populations in clinical care, approaches should be used that address the noted selection and missing data issues. Unfortunately, the majority of studies that have assessed the burden of resistance made inferences using only data from individuals who had undergone genotype tests in the course of clinical care. It is well known that the complete case analysis approach can lead to bias and diminished precision (8–10).
In the present article, we describe methods to yield resistance-prevalence inferences for individuals who are in clinical care and engaged in treatment. Our approach centers around the idea of completing missing resistance status data by incorporating clinical knowledge of the influences of many factors (e.g., patient drug history, viral load trajectory, and past genotype testing information) on an individual’s resistance probability. Multiple-imputation methods (11) alone could be utilized to obtain an asymptotically unbiased estimate of the prevalence of resistance if missingness could be assumed random, conditional on factors measured and available to the investigator (11). However, it is unlikely that missingness of data will be conditionally independent of a strain’s being resistant, given what is known of HIV biology. For example, adherence measurements are not commonly available; however, a patient with an extensive history of poor adherence might be both less likely to have been tested (because modifying the existing regimen might not be beneficial until adherence is improved) and more likely to harbor resistance (unless adherence declines below the threshold at which there is no selective pressure on the virus to induce a resistant strain to emerge). Given that current clinical guidelines specify the consideration of factors such as drug and viral load history, as well as resistance testing in the optimization of therapy (12) (as this information best informs clinical judgment as to the potential for archived resistance and thus a therapy’s effectiveness), incorporating such clinical understanding into a data-completion algorithm could offer an alternative method for obtaining estimates of the burden of resistance.
MATERIALS AND METHODS
Study sample
The North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) is a consortium of clinical and interval HIV cohorts from Canada and the United States (13). It is 1 of 7 regional collaborations of the International Epidemiologic Databases to Evaluate AIDS that are supported by the National Institutes of Health. Genotype testing data were collected from 8 clinic-based cohorts within the NA-ACCORD during the period of interest (2000–2005), and those centers agreed to participate in the present analysis. We identified those individuals who had initiated ART before 2006 and had at least 1 clinic visit between 2000 and 2006. Participants contributed only to estimates in years in which they were actively followed (seen in the clinic at least once), as inferences were targeted at the population engaged in clinical care. Cohorts contributed data on specific genotypic resistance mutations identified by using tests conducted as part of clinical care. We included viral load information from participants with genotypic data if an HIV-1 RNA measurement was available in the 6 months before the genotype test.
Study definitions and design
There are currently 5 distinct classes of antiretroviral drugs available to HIV-infected individuals in developed countries. These drugs include nucleoside-analogue reverse transcriptase inhibitors (NRTIs), protease inhibitors (PIs), nonnucleoside reverse transcriptase inhibitors (NNRTIs), fusion inhibitors, and integrase inhibitors. Effective antiretroviral therapies rely on a combination of drugs, preferably from a mix of different classes. Here, we focus on the 3 major classes of drugs (PIs, NNRTs, and NRTIs) that have been available since the late 1990s. We defined ART as a regimen including 2 or more NRTIs with at least 1 PI or NNRTI. Triple NRTI regimens with abacavir or tenofovir were also included.
Genotype mutations were analyzed using the Genotypic Resistance Interpretation Algorithm, version 4.3.6 (HIVdb Program, Stanford University, Stanford, California), which assigns inferred levels of resistance to commonly used PIs and RTIs on the basis of user-submitted protease and RT gene sequences (14). Genotypic resistance to any single drug was considered present if the algorithm assigned a score of 30 (“intermediate resistance”) or higher for the occurrence of a given mutation pattern. We defined “class” resistance as intermediate or higher resistance to any single drug in any given therapeutic drug class available at the time of the genotype test. Because HIV-1 RNA undergoes reverse transcription into DNA and is inserted into the host cell’s DNA, various HIV mutations are considered to be archived into the latent reservoir (7). Therefore, resistance mutations were considered to accumulate over time and were carried forward, with new resistance tests maintaining or increasing resistance scores from previous years. If results from more than 1 test were available for an individual in a given year, the scores from the last test in that year (representing the cumulative scores from all past tests) were used. Participants were considered to have viremia in a given year if they had at least 1 HIV-1 RNA measurement higher than 1,000 copies/mL after 3 continuous months on an initial antiretroviral regimen. Because genotype testing requires a high viral load for reliable results, we also considered genotype testing of a marker of viremia in the year of the test. Covariate data for those tested were taken from the visit closest to the date of the test within the previous 6 months. For participants for whom we did not have testing data, covariate information was used from the last visit with viremia (in those with viremia in a given year) or the last visit in the year (in those without viremia in a given year). These time points were selected to provide the participant with the maximum opportunity to receive genotype testing (in the former case) or become viremic (in the latter case).
Statistical analysis
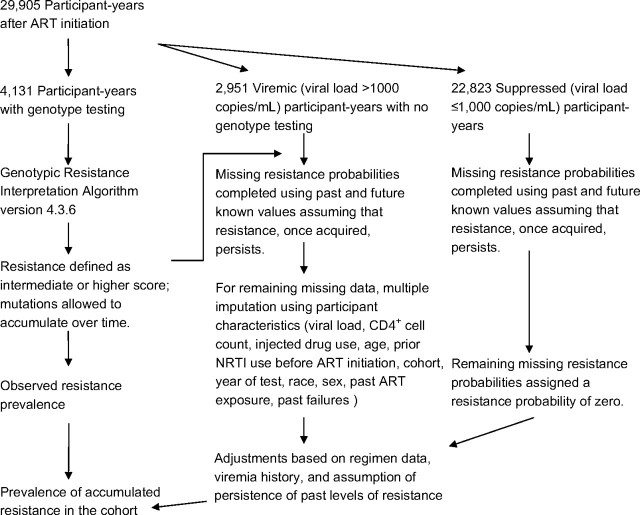
Estimates of the prevalences of accumulated 2- and 3-class resistance were obtained by creating a complete data set of individual resistance probabilities for each calendar year between 2000, when genotypic resistance testing became widely available in the clinic setting, and 2005. Current scientific understanding regarding the persistence of resistant viral strains and the clinical context for the development of resistance were applied to complete the missing data as follows. The accumulated 2- and 3-class-resistance data from the subset of the cohort for whom we had genotypic testing information were extended forward in time with the assumption that resistance was present indefinitely (because of the archival nature of the HIV virus). Thus, in years in which the participant was seen (contributed viral load data) but did not undergo a genotype resistance test, any past level of accumulated resistance was considered still to be present. Participants with no previous testing information who were virally suppressed (i.e., had HIV-1 RNA levels ≤1,000 copies/mL) in a given year were assumed to have no resistance in that year. Similarly, participants with viremia who later demonstrated suppression on an NNRTI or PI regimen were assumed to have accumulated no resistance to those drug classes. For the remaining participant-years with viremia but unknown resistance status, we used the multiple-imputation method (11) to estimate the probability of resistance. Five imputation data sets were created using a single-chain Markov chain Monte Carlo method with the following covariates as main terms: most recent HIV-1 RNA level and CD4+ cell count, history of injected drug use, age, prior NRTI use before ART initiation, cumulative exposure to each drug class, cumulative number of past regimen failures, cohort, year of genotype test, race, and sex. The model was fitted only in viremic participants because covariate associations were found to vary by viremia status. An expectation-maximization algorithm was used to find the posterior mode as the starting value for the chain. The final multiclass resistance prevalence estimates were determined as the average of the estimates resulting from the 5 imputation data sets. The process used to complete the data is illustrated schematically in Figure 1.
Figure 1.
Diagram illustrating the use of clinical knowledge in conjunction with the multiple-imputation method to complete missing data on human immunodeficiency virus drug resistance status, North American AIDS Cohort Collaboration on Research and Design, 2000–2005. ART, antiretroviral therapy; NRTI, nucleoside-analogue reverse transcriptase inhibitor.
The probability of having accumulated resistance (the prevalence of accumulated resistance in the cohort) was estimated for each year in all included participants who were seen in that year. The trend in the yearly estimates was assessed by regressing the individual resistance values on the year and evaluating the significance and direction of the slope. Confidence intervals were obtained by adding the within-imputation and between-imputation variability. All analyses were performed using SAS, version 9.2 (SAS Institute, Inc., Cary, North Carolina).
RESULTS
Cohort virologic response
The analysis included 9,289 patients who participated in 1 of 8 clinic-based cohorts and who initiated ART before the end of 2005 and underwent HIV-1 RNA testing in 2000–2005. Of these patients, 3,959 (43%) had at least 1 HIV-1 RNA level greater than 1,000 copies/mL after at least 3 months of an initial therapy, and 2,962 (75% of those with viremia) had at least 1 genotype resistance test performed.
In 2000, a total of 5,004 patients treated with antiviral drugs had at least 1 strain of HIV-1 RNA measured; of these, 1,480 (30%) had at least 1 HIV-1 RNA level above 1,000 copies/mL after at least 3 months of therapy and 817 (16% of the total participants and 55% of those with viremia) underwent genotype resistance testing (Table 1). In 2005, a total of 4,653 participants who had been treated with antiretroviral drugs had at least 1 HIV-1 RNA level measured; of these, 778 (17%) had at least 1 HIV-1 RNA level above 1,000 copies/mL after at least 3 months of therapy and 486 (10% of the total participants and 62% of those with viremia) underwent genotype testing (Table 1). The proportion of treated patients who had an HIV-1 RNA level above 1,000 copies/mL declined over time (P < 0.001), as has been previously noted (15–18). The percentage of individuals who had genotype resistance testing out of the total number of patients on ART declined from 2000 to 2005 (P < 0.001), whereas the percentage of those tested out of the total number with viremia increased (P < 0.001).
Table 1.
Frequency of Detectable Viremia and Genotype Testing Among Patients Treated With Antiretroviral Drugs, North American AIDS Cohort Collaboration on Research and Design, 2000–2005
| Year | No. of Patients on Antiretroviral Therapy | Protease Inhibitor-Based Regimen |
Nonnucleoside Reverse Transcriptase Inhibitor-Based Regimen |
No. of Patients Who Had >1,000 copies/mL During the Yeara |
No. of Patients Who Had a Genotypic Resistance Test During the Year |
||||
| No. of Cases | % | No. of Cases | % | No. of Cases | % | No. of Cases | % of Totala, % of No. With Viremiaa | ||
| 2000 | 5,004 | 1,920 | 38 | 1,387 | 28 | 1,480 | 30 | 817 | 16/55 |
| 2001 | 5,093 | 1,850 | 36 | 1,552 | 30 | 1,318 | 26 | 735 | 14/56 |
| 2002 | 5,063 | 1,767 | 35 | 1,627 | 32 | 1,360 | 27 | 781 | 15/57 |
| 2003 | 5,052 | 1,778 | 35 | 1,688 | 33 | 1,160 | 23 | 692 | 14/60 |
| 2004 | 5,040 | 2,035 | 40 | 1,614 | 32 | 986 | 20 | 620 | 12/63 |
| 2005 | 4,653 | 1,817 | 39 | 1,254 | 27 | 778 | 17 | 486 | 10/62 |
P < 0.05 for comparisons across calendar year by Cochran-Armitage trend test.
Missing resistance status
Comparing those participants who did and did not have genotype testing among the participants at risk for testing (HIV-1 RNA levels >1,000 copies/mL; Table 2), we found that those receiving genotype testing had lower median HIV-1 RNA levels at the time of the test than did those who were viremic but not tested in both 2000 and 2005. Those tested in 2000 were less likely to have received NRTIs before their first combination therapy regimen (57% vs. 69%) than were those who were viremic but not tested in that same year. In both 2000 and 2005, those not tested were more likely to report lapses in their ART regimen during the year. Median CD4+ cell counts and ages were approximately the same across testing groups. Race (white vs. nonwhite), sex, and injection drug use (yes vs. no) distributions were also similar between those tested and those not tested in a given year.
Table 2.
Characteristics of Individuals on Combination Antiretroviral Therapy, North American AIDS Cohort Collaboration on Research and Design, 2000–2005
| Characteristic | 2000 |
2005 |
||||||||||
| Viral Load >1,000 copies/mL (n = 1,480) |
Viral Load <1,000 copies/mL (n = 3,524) |
Viral Load >1,000 copies/mL (n = 778) |
Viral Load <1,000 copies/mL (n = 3,875) |
|||||||||
| With a Genotypic Test Within Year (n = 817) |
No Genotypic Test Within Year (n = 663) |
With a Genotypic Test Within Year (n = 486) |
No Genotypic Test Within Year (n = 292) |
|||||||||
| % | Median (IQR) | % | Median (IQR) | % | Median (IQR) | % | Median (IQR) | % | Median (IQR) | % | Median (IQR) | |
| Male sex | 80 | 81 | 82 | 74 | 79 | 80 | ||||||
| White | 43 | 42 | 54 | 34 | 34 | 42 | ||||||
| Use of nucleoside-analogue reverse transcriptase inhibitors prior to start of antiretroviral therapy | 57a | 69a | 44 | 46 | 39 | 28 | ||||||
| Lapse in antiretroviral therapy during the year | 25a | 33a | 16 | 26a | 50a | 25 | ||||||
| Injected drug use | 23 | 22 | 20 | 19 | 22 | 19 | ||||||
| Age, years | 40 (36–47) | 40 (36–46) | 41 (36–47) | 43 (37–49) | 45 (39–51) | 44 (39–51) | ||||||
| CD4+, cells/mm3 | 229 (90–380) | 221 (83–400) | 430 (269–635) | 229 (100–418) | 224 (103–380) | 440 (300–628) | ||||||
| Viral load, copies/mL | 13,991a (2,588–72,849) | 17,000a (3,854–72,756) | <50 | 8,770a (1,160–52,200) | 24,456a (5,575–100,010) | <50 | ||||||
Abbreviation: IQR, interquartile range.
P < 0.05 by χ2 test or Wilcoxon rank sum test comparing those with and without genotype tests among those at risk for testing (viral load >1,000 copies/mL).
Of 29,905 participant-years, we were missing genotype data for 25,774. However, only 11% were person-years with unsuppressed viral loads. Of this, 9%, 7%, 6%, 5%, 4%, and 5% of the data on the accumulation of 2 or more classes of resistance remained missing for years 2000 through 2005, respectively, after the algorithm based on clinical knowledge was applied to complete the data. Following the multiple-imputation method to complete the remaining missing resistance data, the relative increase in variance (as a result of the remaining missingness) was estimated to be 2% and the relative efficiency of the imputation estimator based on the 5 imputation data sets was 99%. Results were similar for 3-class resistance data.
Class resistance over time
When data from those in the cohort with missing genotype testing were ignored, the observed prevalence of class resistance to 2 or more antiretroviral drug classes decreased (P < 0.001 for trend; Table 3). The observed prevalences in the tested subgroup were 57%, 56%, 49%, 34%, 37%, and 36% for the years 2000–2005, respectively. Likewise, the observed prevalence of class resistance to 3 antiretroviral drug classes declined over time (22% and 17% in 2000 and 2005, respectively; P < 0.001 for trend). The level of observed wild-type (i.e., those without any detected drug resistance) remained stable, at 12% in 2000 and 13% in 2005.
Table 3.
Prevalence of 2- and 3-Class Resistance by Year Comparing Patients Who Underwent Human Immunodeficiency Virus Genotype Testing With the Entire Cohort, North American AIDS Cohort Collaboration on Research and Design, 2000–2005
| Year | Prevalence of Observed Resistance in Those With Resistance Testing |
Prevalence of Accumulated Resistance in Entire Cohort Estimated Using Multiple Imputation Methods |
||||||
| Accumulation of 2 or More Classesa of Resistance Mutationsb |
Accumulation of 3 Classesa of Resistance Mutationsb |
Accumulation of 2 or More Classesa of Resistance Mutationsc |
Accumulation of 3 Classesa of Resistance Mutationsc |
|||||
| % | 95% CI | % | 95% CI | % | 95% CI | % | 95% CI | |
| 2000 | 57 | 53, 60 | 22 | 19, 25 | 14 | 13, 15 | 6 | 5, 7 |
| 2001 | 56 | 52, 59 | 22 | 19, 25 | 17 | 16, 18 | 7 | 7, 8 |
| 2002 | 49 | 45, 53 | 17 | 14, 20 | 18 | 17, 19 | 8 | 7, 9 |
| 2003 | 34 | 30, 38 | 16 | 13, 18 | 17 | 16, 18 | 8 | 7, 9 |
| 2004 | 37 | 33, 41 | 17 | 14, 19 | 17 | 16, 18 | 7 | 7, 8 |
| 2005 | 36 | 32, 41 | 17 | 14, 21 | 17 | 16, 18 | 8 | 7, 9 |
Abbreviation: CI, confidence interval.
Classes refers to the 3 main antiretroviral therapy drug classes: nucleoside-analogue reverse transcriptase inhibitors, protease inhibitors, and nonnucleoside reverse transcriptase inhibitors.
P < 0.05 by Cochran-Armitage trend test.
P < 0.05 for parameter estimate from the linear regression of prevalence on year.
When combining estimated resistance (in those who were viremic but not tested) with observed resistance (in those who were tested), we found that the prevalence of accumulated resistance in the entire cohort was much lower than the observed prevalence in those with testing. There was a slight increase in the estimated accumulated prevalence over time, mostly from 2000 to 2001 (P < 0.001 for trend; Table 3). We estimated the prevalences of accumulated resistance to 2 or more classes to be 14%, 17%, 18%, 17%, 17%, and 17% for the years 2000 through 2005, respectively. The prevalence of accumulated 3-class resistance also slightly increased from 6% in 2000 to 7% in 2005 (P = 0.008).
Sensitivity analyses
Incorporation of best clinical knowledge accounted for the completion of 93% of the missing data and had the largest impact on estimates of the prevalence of resistance. When the algorithm for completing missing data using clinical knowledge was ignored and the multiple-imputation method was used as the sole means of completing the missing data, the estimates for the prevalence of accumulated resistance to 2 or more classes of drugs in the cohort were 51%, 61%, 64%, 64%, 65%, and 66% for 2000–2005, respectively. The estimates for the prevalence of accumulated 3-class resistance in the cohort were 32%, 41%, 45%, 46%, 48%, and 50%, respectively. In contrast, using only clinical knowledge to complete missing data and ignoring the remaining missingness resulted in lower estimates for the prevalence. For example, the prevalences of accumulated resistance to 2 or more classes of drugs were estimated to be 10%, 13%, 15%, 14%, 14%, and 14% and to 3 classes were estimated to be 4%, 5%, 5%, 5%, 5%, and 5% for 2000–2005, respectively.
Regimen and viral load data were used to inform individual resistance probability estimates as a surrogate for testing information. However, future suppression on a PI or NNRTI regimen does not fully exclude the possibility of resistance to those classes. Excluding the regimen information resulted in minor upward shifts of the estimated prevalences of 1%–2%.
DISCUSSION
In the present article, we used a modified multiple-imputation method that augmented the statistical approach with current clinical and scientific knowledge. The application of this method to the study of antiretroviral drug resistance enabled us to consider a large body of clinical knowledge concerning archiving of resistance, drug cross-class resistance, and future likelihood of viral suppression. The evolving epidemiology of antiretroviral drug resistance in clinical practice has not been fully described, as inferences have relied on data from resistance tests performed during the course of routine clinical practice. Thus, a large percentage of missing data has been routinely ignored in studies characterizing the magnitude and trends in resistance. By estimating the prevalence of accumulated resistance using data from a representative sample of North American patients who had been treated in clinical care, we found contrasting inferences regarding the trend in 2- and 3-class resistance compared with those in the genotype-tested subset. The prevalences of 2- and 3-class resistance in the entire cohort remained essentially stable except in 2000, which accounted for the significant trend test. This trend, compared with the dramatic decreases in class resistance observed among the tested subset, speaks to 2 factors: first, the inability of testing to capture the persistence of mutant viral strains in the latent reservoir, and second, selection factors that determine who is tested. Although the observed estimates do attempt to correct for the latent reservoir by summing past resistance scores over repeated tests, patients who were subsequently doing well on therapy (with a suppressed viral load) after a past failure did not undergo subsequent tests. Using the methods presented here, we were able to infer the likely resistance level for such individuals based on their past testing results (e.g., PI-class resistant in 2000), an assumption of archived resistance (e.g., PI-class resistant in all subsequent years), and knowledge of their viral load trajectory (e.g., suppressed viral load since 2000 and thus no additional acquired resistance). Such scientific knowledge regarding the development and persistence of resistance took precedence over a random draw from a multivariate normal distribution for completing the missing resistance data.
The results using data from the subset of individuals who underwent genotype resistance testing were generally consistent with trends observed in several recent European cohorts (13, 19–21). This apparent decline in resistance in the subset of genotyped patients could be attributed to several factors, including an earlier use of genotype testing among those failing to respond to ART (22), low adherence to the prescribed therapeutic regimen (23–25), or the increased use of regimens that are less likely to select for drug resistance (23, 26). All of these factors could affect the percentages of mutations detected in the circulating virus. Comparisons between those who did and did not undergo genotype testing revealed clear contrasts that suggested that inferences concerning resistance trends cannot be extrapolated from the tested subcohort to the clinical-care population. Further, testing results alone do not capture the reservoir of archived resistance in the larger cohort of HIV-infected patients (unless repeated frequent testing is used throughout follow-up). The multiple imputation method, along with assumptions about the structure of the resistance data, provided 1 potential methodological approach to quantifying the burden of resistance in the population over time in lieu of universal repeated testing data. It should be noted that this approach still requires conditional independence to hold in the partially completed data (after clinical knowledge-based imputation) (10, 27). The validity of the individual resistance probability estimates for those not tested relies upon the predictive strength of the covariates included in the multiple-imputation model, which may not comprise the optimal set of explanatory factors. In the present example, however, only approximately 6% of the data arose from multiple-imputation models as a result of our use of clinical knowledge to inform the data completion.
The greatest influence on the estimated prevalences and subsequent trend arose from the integration of current scientific knowledge about resistance. In particular, the assumption concerning the persistence of resistant strains in an individual over time defined a data structure that treated resistance as a monotonically increasing function. The assumption that despite a positive response to new regimens and an absence of detectable drug-resistant viremia, an individual continues to harbor resistant viral strains reflects current understanding and evidence (28) and was incorporated into methods used to compute both the observed prevalence and the estimated cohort prevalence, though it was much more influential on cohort inferences for which the assumed structure was used to complete missing data. Thus, the prevalence of resistance we observed in any year was likely higher than what would be expected from a cross-sectional survey of resistance. However, because the earliest genotype testing data we have are from 1997, the accumulation of resistance only pertains to resistance captured after this time. Further, resistant variants fade as selective pressure is removed, such as during a period of poor or interrupted adherence to therapeutic regimens. Therefore, our estimates of the prevalence of accumulated resistance likely represent a lower bound for the true prevalence.
A limitation of the analysis was the simplifying assumption that genotype testing results, when available, were an accurate reflection of phenotypic drug susceptibility. Genotype tests have a limited ability to detect resistant circulating strains at low viral loads and thus might result in falsely classifying an individual as not harboring resistant viral strains. Of additional concern are tests that are administered after therapy has been discontinued. With selective pressure no longer present, resistant HIV strains may be undetectable at the time of the test. Such misclassification of resistance status among the tested through either mechanism would have affected both the observed and accumulated prevalence estimates and resulted in underestimation of the true prevalence of multiclass resistance.
Of further interest is the generalizability of the estimates presented. The target population in this study was HIV-infected individuals engaged in clinical care. The source population was an open cohort of participants initiating ART treatment who were observed during the year for which they contributed to the prevalence estimate. We capture primarily the burden of acquired resistance arising from treatment exposure. The overall pool of resistance also includes community-acquired resistance, which could not be adequately addressed in the present study because of the limited numbers of pre-ART genotype tests. The prevalence of community-acquired multiclass resistance in North America is estimated to be 6%–17% (29, 30).
Patients who exhibit virologic failure (the result of either nonadherence or resistance) on antiretroviral therapy tend to have worse long-term outcomes than do those who achieve durable viral suppression (3, 31–36). The contrast in inferences presented here between the tested sample and the entire cohort highlights the value of appropriate handling of missing data in informing clinical practice and policy.
Acknowledgments
Author affiliations: Department of Epidemiology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Alison G. Abraham, Bryan Lau, Richard D. Moore, Jinbing Zhang, Lisa Jacobson, Greg Kirk, Jennifer Thorne, Stephen J. Gange, Kelly Gebo); Department of Medicine, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Bryan Lau); Division of Infectious Diseases, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Richard D. Moore); Division of Ocular Immunology, The Wilmer Eye Institute, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Jennifer Thorne); Division of Infectious Diseases, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Kelly Gebo); Departments of Epidemiology and Medicine, University of California, San Francisco, San Francisco, California (Steven Deeks, Jeff Martin); Division of Infectious Diseases, University of North Carolina, Chapel Hill, North Carolina (Joseph Eron); Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina (Sonia Napravnik); Laboratory Program, Drug Treatment Program, BC Centre for Excellence in HIV/AIDS, Vancouver, British Columbia, Canada (Richard Harrigan); Epidemiology and Population Health Program, BC Centre for Excellence in HIV/AIDS, Vancouver, British Columbia, Canada (Robert Hogg); Departments of Medicine, Microbiology, Immunology and Infectious Diseases, Pathology, and Laboratory Medicine, University of Calgary, Calgary, Alberta, Canada (John M. Gill); Department of Medicine, Division of Allergy and Infectious Diseases, University of Washington, Seattle, Washington (Mari Kitahata); Department of Medicine, Division of Infectious Diseases and Immunodeficiency Service, McGill University, Montreal, Quebec, Canada (Marina Klein); Department of Medicine, Division of Infectious Diseases, University of Toronto, Toronto, Ontario, Canada (Anita Rachlis); Department of Psychiatry, Ontario HIV Treatment Network, University of Toronto, Toronto, Ontario, Canada (Sean Rourke); Division of Infectious Diseases, Case Western Reserve University, Cleveland, Ohio (Benigno Rodriguez); Division of Infectious Diseases, School of Medicine, University of California, San Diego, San Diego, California (Constance Benson); Department of Biostatistics, School of Public Health, Harvard University, Boston, Massachusetts (Ron Bosch); Department of Medicine, Division of Allergy and Infectious Diseases, University of Washington, Seattle, Washington (Ann Collier); Division of Cancer Epidemiology and Genetics, Infections and Immunoepidemiology Branch, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (James Goedert); Epidemiology Branch, Office of Clinical Research Operations, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland (Rosemary McKaig); Division of Research, Kaiser Permanente Northern California, Oakland, California (Michael Horberg, Michael Silverberg); Department of Internal Medicine, VA Connecticut Healthcare System and Yale University School of Medicine, New Haven, Connecticut (Amy Justice); Department of Medicine, Division of Infectious Diseases, Vanderbilt University Medical Center, Nashville, Tennessee (Timothy Sterling); and Department of Medicine, Division of Infectious Diseases, University of Alabama at Birmingham, Birmingham, Alabama (James Willig).
This work was supported by the National Institutes of Health (grants U01-AI069918, U01-AA013566, U01-AI31834, U01-AI34989, U01-AI34993, U01-AI34994, U01-AI35004, U01-AI35039, U01-AI35040, U01-AI35041, U01-AI35042, U01-AI35043, U01-AI37613, U01-AI37984, U01-AI38855, U01-AI38858, U01-AI42590, U01-AI68634, U01-AI68636, U01-HD32632, U10-EY08057, U10-EY08052, U10- EY08067, UL1-RR024131, MO1-RR-00052, M01-RR00071, M01-RR00079, M01-RR00083, M01-RR00722, MM01- RR025747, P30-AI27757, P30-AI27767, P30-AI50410, P30-AI54999, R01-DA04334, R01-DA12568, R01-MH54907, R24-AI067039, Z01-CP010176, AHQ290-01-0012, N02-CP55504, R01-DA11602, AI-69432, AI-69434, K01-AI071725, K01-AI071754, R01-AA16893, K24-00432, K23-AI-61-0320, and K23 EY013707); the Centers for Disease Control and Prevention (grant CDC200-2006-18797); the Canadian Institutes for Health Research (grants TGF-96118, HCP-97105, CBR-86906, CBR-94036, KRS-86251, and 169621); and the Canadian Trials Network (project number 242).
North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) cohorts and representatives (*contributed to this study): AIDS Link to the IntraVenous Experience (Dr. Gregory D. Kirk); Adult AIDS Clinical Trials Group Longitudinal Linked Randomized Trials (Drs. Constance A. Benson, Ronald J. Bosch, and Ann C. Collier); *HAART Observational Medical Evaluation and Research (Drs. Robert S. Hogg, Richard Harrigan, Julio Montaner, and Angela Cescon); HIV Outpatient Study (Drs. John T. Brooks and Kate Buchacz); HIV Research Network (Dr. Kelly A. Gebo); *Johns Hopkins HIV Clinical Cohort (Dr. Richard D. Moore); *John T. Carey Special Immunology Unit Patient Care and Research Database, Case Western Reserve University (Dr. Benigno Rodriguez); *Kaiser Permanente Northern California (Drs. Michael A. Horberg and Michael J. Silverberg); Longitudinal Study of Ocular Complications of AIDS (Dr. Jennifer E. Thorne); Multicenter Hemophilia Cohort Study–II (Dr. James J. Goedert); Multicenter AIDS Cohort Study (Dr. Lisa P. Jacobson); *Montreal Chest Institute Immunodeficiency Service Cohort (Dr. Marina B. Klein); *Ontario HIV Treatment Network Cohort Study (Drs. Sean B. Rourke, Ann Burchell, and Anita R. Rachlis); *Southern Alberta Clinic Cohort (Dr. M. John Gill); Studies of the Consequences of the Protease Inhibitor Era (Drs. Steven G. Deeks and Jeffery N. Martin); University of Alabama at Birmingham 1917 Clinic Cohort (Drs. Michael S. Saag, Michael Mugavero, and James Willig); *University of North Carolina, Chapel Hill HIV Clinic Cohort (Drs. Joseph J. Eron and Sonia Napravnik); *University of Washington HIV Cohort (Drs. Mari M. Kitahata and Heidi M. Crane); Veterans Aging Cohort Study (Drs. Amy C. Justice, Robert Dubrow, and David Fiellin); Vanderbilt-Meharry CFAR Cohort (Drs. Timothy R. Sterling, Sam Stinette, and David Haas); Women’s Interagency HIV Study (Drs. Stephen J. Gange and Kathryn Anastos). NA-ACCORD Executive Committee: Drs. Richard D. Moore, Michael S. Saag, Stephen J. Gange, Mari M. Kitahata, and Rosemary G. McKaig, as well as Aimee M. Freeman. Epidemiology/Biostatistics Core: Dr. Stephen J. Gange, Dr. Alison G. Abraham, Dr. Bryan Lau, Dr. Keri N. Althoff, Jinbing Zhang, Jerry Jing, Dr. Elizabeth Golub, Dr. Shari Modur, David Hanna, Peter Rebeiro, Adell Mendes, and Aaron Platt. Data Management Core: Dr. Mari M. Kitahata, Dr. Stephen E. Van Rompaey, Dr. Heidi M. Crane, Eric Webster, Liz Morton, and Brenda Simon.
Conflict of interest: none declared.
Glossary
Abbreviations
- AIDS
acquired immunodeficiency syndrome
- ART
antiretroviral therapy
- HIV
human immunodeficiency virus
- NNRTI
nonnucleoside reverse transcriptase inhibitor
- NRTI
nucleoside-analogue reverse transcriptase inhibitor
- PI
protease inhibitor
References
- 1.Palella FJ, Jr, Delaney KM, Moorman AC, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG, Barbour JD, Martin JN, et al. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J Infect Dis. 2000;181(3):946–953. doi: 10.1086/315334. [DOI] [PubMed] [Google Scholar]
- 3.Lucas GM. Antiretroviral adherence, drug resistance, viral fitness and HIV disease progression: a tangled web is woven. J Antimicrob Chemother. 2005;55(4):413–416. doi: 10.1093/jac/dki042. [DOI] [PubMed] [Google Scholar]
- 4.Deeks SG. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet. 2003;362(9400):2002–2011. doi: 10.1016/S0140-6736(03)15022-2. [DOI] [PubMed] [Google Scholar]
- 5.Napravnik S, Keys JR, Quinlivan EB, et al. Triple-class antiretroviral drug resistance: risk and predictors among HIV-1-infected patients. AIDS. 2007;21(7):825–834. doi: 10.1097/QAD.0b013e32805e8764. [DOI] [PubMed] [Google Scholar]
- 6.Office of National AIDS Policy. 2010 National HIV/AIDS Strategy. Washington, DC: Office of National AIDS Policy; 2010. ( http://www.whitehouse.gov/administration/eop/onap/nhas/). (Accessed March 1, 2011) [Google Scholar]
- 7.Siliciano JD, Siliciano RF. A long-term latent reservoir for HIV-1: discovery and clinical implications. J Antimicrob Chemother. 2004;54(1):6–9. doi: 10.1093/jac/dkh292. [DOI] [PubMed] [Google Scholar]
- 8.Greenland S, Finkle WD. A critical look at methods for handling missing covariates in epidemiologic regression analyses. Am J Epidemiol. 1995;142(12):1255–1264. doi: 10.1093/oxfordjournals.aje.a117592. [DOI] [PubMed] [Google Scholar]
- 9.Vach W, Blettner M. Biased estimation of the odds ratio in case-control studies due to the use of ad hoc methods of correcting for missing values for confounding variables. Am J Epidemiol. 1991;134(8):895–907. doi: 10.1093/oxfordjournals.aje.a116164. [DOI] [PubMed] [Google Scholar]
- 10.Rubin DB. Multiple imputation after 18+ years. J Am Stat Assoc. 1996;91(434):473–489. [Google Scholar]
- 11.Little RJA, Rubin DB. Statistical Analysis With Missing Data. New York, NY: John Wiley & Sons, Inc; 2002. [Google Scholar]
- 12.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. Washington, DC: Department of Health and Human Services; 2009. ( http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf). (Accessed May 14, 2010) [Google Scholar]
- 13.Gange SJ, Kitahata MM, Saag MS, et al. Cohort profile: the North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) Int J Epidemiol. 2007;36(2):294–301. doi: 10.1093/ije/dyl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee SY, Gonzales MJ, Kantor R, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bannister WP, Kirk O, Gatell JM, et al. Regional changes over time in initial virologic response rates to combination antiretroviral therapy across Europe. J Acquir Immune Defic Syndr. 2006;42(2):229–237. doi: 10.1097/01.qai.0000214815.95786.31. [DOI] [PubMed] [Google Scholar]
- 16.Lampe FC, Gatell JM, Staszewski S, et al. Changes over time in risk of initial virological failure of combination antiretroviral therapy: a multicohort analysis, 1996 to 2002. Arch Intern Med. 2006;166(5):521–528. doi: 10.1001/archinte.166.5.521. [DOI] [PubMed] [Google Scholar]
- 17.Lohse N, Obel N, Kronborg G, et al. Declining risk of triple-class antiretroviral drug failure in Danish HIV-infected individuals. AIDS. 2005;19(8):815–822. doi: 10.1097/01.aids.0000168976.51843.9f. [DOI] [PubMed] [Google Scholar]
- 18.Deeks SG, Gange SJ, Kitahata MM, et al. Trends in multidrug treatment failure and subsequent mortality among antiretroviral therapy-experienced patients with HIV infection in North America. Clin Infect Dis. 2009;49(10):1582–1590. doi: 10.1086/644768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Mendoza C, Garrido C, Corral A, et al. Changing rates and patterns of drug resistance mutations in antiretroviral-experienced HIV-infected patients. AIDS Res Hum Retroviruses. 2007;23(7):879–885. doi: 10.1089/aid.2005.0072. [DOI] [PubMed] [Google Scholar]
- 20.Di Giambenedetto S, Bracciale L, Colafigli M, et al. Declining prevalence of HIV-1 drug resistance in treatment-failing patients: a clinical cohort study. Antivir Ther. 2007;12(5):835–839. [PubMed] [Google Scholar]
- 21.Vercauteren J, Deforche K, Theys K, et al. The incidence of multidrug and full class resistance in HIV-1 infected patients is decreasing over time (2001–2006) in Portugal. Retrovirology. 2008;5(1):12. doi: 10.1186/1742-4690-5-12. (doi:10.1186/1742-4690-5-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch MS, Günthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 23.Bangsberg DR, Acosta EP, Gupta R, et al. Adherence-resistance relationships for protease and non-nucleoside reverse transcriptase inhibitors explained by virological fitness. AIDS. 2006;20(2):223–231. doi: 10.1097/01.aids.0000199825.34241.49. [DOI] [PubMed] [Google Scholar]
- 24.Bangsberg DR, Kroetz DL, Deeks SG. Adherence-resistance relationships to combination HIV antiretroviral therapy. Curr HIV/AIDS Rep. 2007;4(2):65–72. doi: 10.1007/s11904-007-0010-0. [DOI] [PubMed] [Google Scholar]
- 25.Harrigan PR, Hogg RS, Dong WW, et al. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. J Infect Dis. 2005;191(3):339–347. doi: 10.1086/427192. [DOI] [PubMed] [Google Scholar]
- 26.von Wyl V, Yerly S, Böni J, et al. Emergence of HIV-1 drug resistance in previously untreated patients initiating combination antiretroviral treatment: a comparison of different regimen types. Arch Intern Med. 2007;167(16):1782–1790. doi: 10.1001/archinte.167.16.1782. [DOI] [PubMed] [Google Scholar]
- 27.Rubin DB, Stern HS, Vehovar V. Handling don’t know survey responses: the case of the Slovenian plebiscite. J Am Stat Assoc. 1995;90(431):822–828. [Google Scholar]
- 28.Hermankova M, Ray SC, Ruff C, et al. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/mL receiving combination therapy. JAMA. 2001;286(2):196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- 29.Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 30.Lima VD, Harrigan PR, Sénécal M, et al. Epidemiology of antiretroviral multiclass resistance. Am J Epidemiol. 2010;172(4):460–468. doi: 10.1093/aje/kwq101. [DOI] [PubMed] [Google Scholar]
- 31.Grover D, Copas A, Green H, et al. What is the risk of mortality following diagnosis of multidrug-resistant HIV-1? J Antimicrob Chemother. 2008;61(3):705–713. doi: 10.1093/jac/dkm522. [DOI] [PubMed] [Google Scholar]
- 32.Hogg RS, Bangsberg DR, Lima VD, et al. Emergence of drug resistance is associated with an increased risk of death among patients first starting HAART. PLoS Med. 2006;3(9):e356. doi: 10.1371/journal.pmed.0030356. (doi: 10.1371/journal.pmed.0030356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas GM, Gallant JE, Moore RD. Relationship between drug resistance and HIV-1 disease progression or death in patients undergoing resistance testing. AIDS. 2004;18(11):1539–1548. doi: 10.1097/01.aids.0000131339.68666.1a. [DOI] [PubMed] [Google Scholar]
- 34.Mocroft A, Ledergerber B, Viard JP, et al. Time to virological failure of 3 classes of antiretrovirals after initiation of highly active antiretroviral therapy: results from the EuroSIDA study group. J Infect Dis. 2004;190(11):1947–1956. doi: 10.1086/425424. [DOI] [PubMed] [Google Scholar]
- 35.Raffanti SP, Fusco JS, Sherrill BH, et al. Effect of persistent moderate viremia on disease progression during HIV therapy. J Acquir Immune Defic Syndr. 2004;37(1):1147–1154. doi: 10.1097/01.qai.0000136738.24090.d0. [DOI] [PubMed] [Google Scholar]
- 36.Recsky MA, Brumme ZL, Chan KJ, et al. Antiretroviral resistance among HIV-infected persons who have died in British Columbia, in the era of modern antiretroviral therapy. J Infect Dis. 2004;190(2):285–292. doi: 10.1086/422007. [DOI] [PubMed] [Google Scholar]