Abstract
Background
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal-dominant vascular disorder, characterized by recurrent epistaxis, mucocutaneous telangiectases, and arteriovenous malformations (AVMs) in various visceral organs. Endoglin (ENG) and activin receptor-like kinase 1 (ACVRL1; ALK1), receptors for transforming growth factor-β (TGF-β) superfamily, have been identified as the principal HHT-causing genes.
Methods
Three unrelated Korean HHT patients and their asymptomatic as well as symptomatic family members were genetically diagnosed by sequencing whole exons and their flanking regions of ENG and ACVRL1. Functionality of an aberrant translation start codon, which is created by a substitution mutation at the 5'-untranslated region (UTR) of ENG found in a HHT family, was tested by transient in vitro transfection assay. Decay of the mutant transcripts was also assessed by allele-specific expression analysis.
Results
Two ENG and one ACVRL1 mutations were identified: a known ENG mutation (c.360+1G > A; p.Gly74_Tyr120del); a novel ENG mutation (c.1-127C > T); and a novel ACVRL1 mutation (c.252_253insC; p.Val85fsX168). We further validated that the 5'-UTR ENG mutation prevents translation of ENG from the biological translation initiation site of the mutant allele, and leads to degradation of the mutant transcripts.
Conclusions
This is the first experimental demonstration that a 5'-UTR mutation can prevent translation of ENG among HHT patients, and further supports the previous notion that haploinsufficiency is the primary mechanism of HHT1. Our data also underscore the importance of including exons encoding 5' UTR for HHT mutation screening.
Background
Hereditary hemorrhagic telangiectasia (HHT) is a rare genetic vascular disorder, inherited in an autosomal-dominant trait. With a regional variance, HHT affects about 1 in 5,000 to 8,000 people worldwide [1-3]. Major clinical diagnosis criteria include family history, recurrent epistaxis, mucocutaneous telangiectases, and arteriovenous malformations (AVMs) in internal organs such as the lung, brain, and liver [4,5]. Epistaxis is usually the earliest clinical symptom of HHT that appears at teens, while cutaneous and gastrointestinal telangiectases manifest at adulthood ages. However, both of these symptoms progressively worsen with age, and manifest in more than 90% of HHT patients in their 60's [1].
Five clinically indistinguishable types of HHT are defined by associated genetic loci. HHT1 (MIM #187300) and HHT2 (MIM #600376) that account for more than 80% of HHT are caused by heterozygous mutations in endoglin (ENG) [6-8] and activin receptor-like kinase 1 (ACVRL1; ALK1) [9-11] genes, respectively [12,13]. To date, 397 ENG and 332 ACVRL1 mutations and polymorphisms have been reported [14]. Two additional genetic loci on chromosomes 5 and 7 were identified for HHT3 (MIM #601011) and HHT4 (MIM #610655), respectively [15,16]. A group of juvenile polyposis patients harboring mutations in the SMAD4 gene display a combined syndrome with HHT (JP-HHT) (MIM #175050) [17]. Based on the fact that ENG, ALK1, and SMAD4 are receptors or intracellular mediator of transforming growth factor-β (TGF-β) family signals, it has been postulated that impaired signaling of TGF-β family member(s) would be an important pathological mechanism of HHT. Recent biochemical studies have suggested that BMP9 is the plausible physiological ligand of ALK1 [18-20].
Since clinical diagnosis of HHT has limitations due to the late onset, incomplete penetrance, and variability of disease manifestations, genetic diagnosis is valuable not only to confirm the clinical diagnosis but also to identify asymptomatic carriers among a HHT family. Early screening of asymptomatic patients for cerebral and pulmonary AVMs that are found in about 40% of HHT patients can prevent them from potential serious health risks such as stroke and brain abscess [1].
Despite dozens of HHT case reports for Korean HHT families, only one genetic study identifying genetic mutations has been reported [21]. Here, we report genetic analyses of three unrelated Korean HHT families diagnosed by clinical criteria. We found one known and one novel mutations in ENG, and one novel mutation in ACVRL1. With molecular analysis, we demonstrate that the novel ENG mutation found in the 5'-untranslated region (UTR) initiates aberrant translation that prevents translation of ENG from the biological translation initiation site of the mutant allele, and leads to nonsense-mediated mRNA decay.
Methods
Human subjects
Experimental procedures performed were reviewed and approved by the Ethics Committee, an institutional review board, at Lee Gil Ya Cancer and Diabetes Institute, Gachon University, Incheon, Korea. Patients were diagnosed with HHT when they possessed at least three of the four Curaçao established criteria: 1) an affected first degree family member; 2) recurrent nosebleeds; 3) multiple telangiectases along the mucocutaneous surface; and 4) AVMs in major organs [22]. HHT genetic analysis was then conducted after obtaining informed consent from patients and their family members. Clinical features of patients reported by clinicians are summarized in Table 1. Only obviously detected symptoms are listed. Since systematic screening was not employed, visceral AVMs and skin telangiectasia might have been missed from some patients.
Table 1.
Clinical features and mutations of three HHT families
| Family | Patient | Gender/Age | Clinical features | Mutations |
|---|---|---|---|---|
| I | I-1 | M/46 | Epistaxis | ENG c.360+1G > A, p.Gly74_Tyr120del |
| I-2 | M/43 | Epistaxis, PAVM, Epilepsy, Cerebral abscess |
ENG c.360+1G > A, p.Gly74_Tyr120del | |
| I-3 | F/41 | Epistaxis, PAVM | ENG c.360+1G > A, p.Gly74_Tyr120del | |
| I-4 | F/14 | Epistaxis, PAVM | ENG c.360+1G > A, p.Gly74_Tyr120del | |
| I-5 | F/12 | Epistaxis | ENG c.360+1G > A, p.Gly74_Tyr120del | |
| II | II-1 | F/82 | Epistaxis, Skin telangiectasia |
ENG c.1-127 C > T, new upstream translation start codon (TSC) |
| II-2 | M/65 | Epistaxis, CAVM | ENG c.1-127 C > T, new upstream TSC | |
| II-3 | M/53 | Epistaxis | ENG c.1-127 C > T, new upstream TSC | |
| II-4 | F/50 | Epistaxis | ENG c.1-127 C > T, new upstream TSC | |
| II-5 | M/13 | Epistaxis, CAVM, dead | n/d | |
| II-6 | M/18 | Epistaxis, PAVM, Seizure, Embolic cerebral infarction |
ENG c.1-127 C > T, new upstream TSC | |
| III | III-1 | M/55 | Epistaxis | ACVRL1 c.252_253insC, p.Val85fsX168 |
| III-2 | M/50 | Epistaxis | ACVRL1 c.252_253insC, p.Val85fsX168 | |
| III-3 | F/40 | Epistaxis | ACVRL1 c.252_253insC, p.Val85fsX168 |
n/d: not determined
Genomic DNA isolation
Blood samples were collected in vacuum blood collection tubes containing EDTA. Genomic DNA was isolated from 200-μl blood samples using the Exgene Blood SV mini kit (GeneAll, South Korea) according to the manufacturer's protocol.
PCR amplification and sequence analysis of ENG and ACVRL1 exons
All exons of ENG and ACVRL1 genes, including non-coding regions and flanking intronic sequences, were amplified by genomic polymerase chain reaction (PCR) using PrimeSTAR HS DNA Polymerase (TAKARA, Japan) and the appropriate primer sets (Additional file 1; Table S1). PCR products were separated in a 1-2% agarose gel and purified using QIAquick Gel Extraction Kit (QIAGEN, Germany) or Expin PCR SV kit (GeneAll, Korea). These purified PCR products were then directly sequenced. Accession numbers of the reference sequences used in this study are [GenBank:NM_000118.2] (cDNA) and [GenBank:NC_009551.1] (genomic DNA) for ENG, and [GenBank:NM_000020.2] (cDNA) and [GenBank:NC_009545.1] (genomic DNA) for ACVRL1.
Plasmid construction and transient in vitro transfection
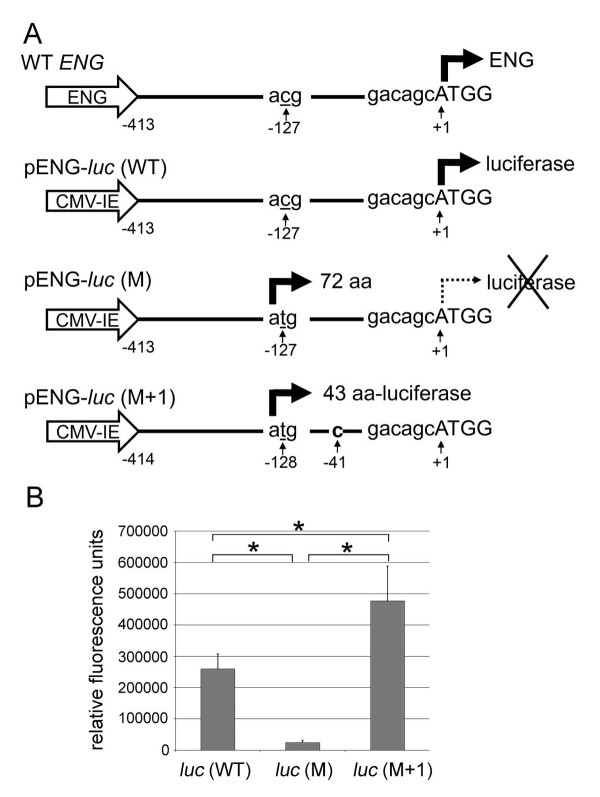
ENG 5'-UTRs (position -413 to -1) of wild-type (WT) and mutant (M) alleles from genomic DNA of HHT patients in Family 2 (Figures 1) were amplified via PCR and subcloned into T&A Cloning Vectors (Real Biotech Corporation, Taiwan), yielding pTA-ENG-WT and pTA-ENG-M. The ENG c.1-127 C > T mutation created a putative translation start codon in the 5'-UTR of pTA-ENG-M. WT and mutant 5'-UTRs were amplified with hENF-5UTR-F1 (gctagcctctacccggttggcaggcggcct) and hENF-5UTR-R1 (ccatgGtgtccacgtgggggcctgtgcg) from WT and mutant ENG alleles, respectively. For cloning purpose, an NheI site (underlined) was introduced at the 5'-end of the forward primer and the 'G' nucleotide at position -2 in the 5'-UTR was replaced with 'C', which produced an NcoI site (underlined) at the 5'-end of the reverse primer. The luciferase gene in pGL4.14 [luc2/hygro] (Promega, USA) was subcloned into the NcoI and XbaI sites in pTA-ENG-WT and pTA-ENG-M plasmids, generating pTA-ENG-WT-luc and pTA-ENG-M-luc, respectively. The eGFP gene in pEGFP-N1 (Clontech, USA) was replaced with the ENG 5'-UTR-luciferase fragments of pTA-ENG-WT-luc and pTA-ENG-M-luc. The substituted 'C' nucleotide at position -2 to produce the NcoI site reverted to the original 'G' nucleotide by site directed mutagenesis (COSMO GENETECH, Korea), generating pENG-luc(WT) and pENG-luc(M), respectively (Figure 2A). A plasmid with an artificial 5'-UTR, which was produced by insertion of a 'C' nucleotide between -41 and -40 of the mutant 5'-UTR in pENG-luc(M), was generated and designated pENG-luc(M+1) (Figure 2A). The inserted 'C' nucleotide between -41 and -40 resulted in a frameshift and produced a luciferase fusion protein translated from the aberrant translation start codon (Figure 2A).
Figure 1.
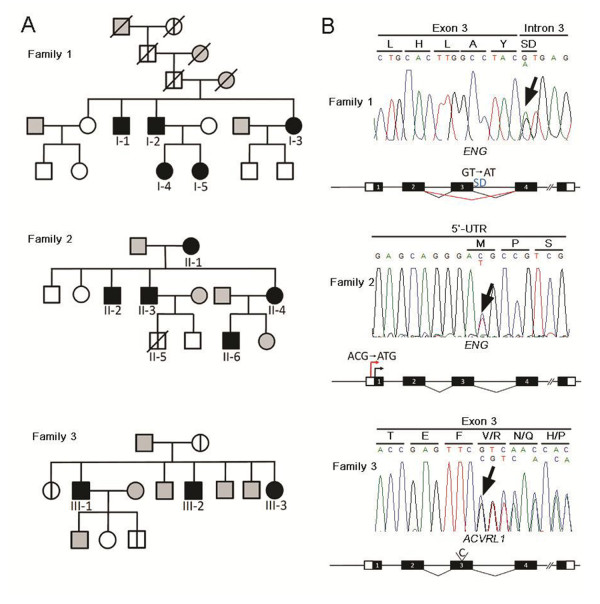
Pedigree and genetic analysis of three HHT families. (A) Pedigree of families with genetic mutations and/or symptoms of HHT. Pedigree symbols: filled symbol, affected individual; open symbol, unaffected individual; divided symbol, affected individual by hearsay; gray symbol, no phenotypical and genetic data available. A slashed symbol indicates that the individual is deceased. Genetic analysis was performed on all individuals represented by filled and open symbols. Genetic mutations and HHT-associated symptoms of numbered patients are summarized in Table 1. (B) Genetic studies of three representative family members with HHT. Family 1, ENG c.360+1G > A (p.Gly74_Tyr120del); Family 2, ENG c.1-127 C > T (aberrant translation); Family 3, ACVRL1 c.252_253insC (p.Val85fsX168). The amino acid translation is shown above each codon. The amino acid sequence in Family 2 is the predicted sequence translated from the aberrant translation start codon. Arrows indicate mutation sites. SD, splicing donor sequence.
Figure 2.
Functionality of the aberrant translation start codon. (A) Schematic representation of the pENG-luc constructs. Wild-type (WT) ENG promoter produces ENG mRNA including the 413-nt-5'-UTR. pENG-luc(WT) was designed to code for luciferase mRNA containing the same ENG 5'-UTR under the control of the immediate early promoter of CMV (CMV-IE). In pENG-luc(M), the 'C' at position -127 in pENG-luc(WT) was replaced with 'T' (underlined), which generated the aberrant translation start codon. The substitution was found in the mutant allele of HHT patients in Family 2. The aberrant translation start codon putatively produces a 72-amino acid-protein out of frame with the luciferase coding sequence. In pENG-luc(M+1), a 'C' nucleotide (bold) was inserted between -41 and -40 of pENG-luc(M), thus producing an aberrant translation start codon that was in frame with the luciferase coding sequence. Nucleotides are numbered with c.1 corresponding to the 'A' of the ATG translation start codon in the reference sequence [GenBank:NM_000118.2]. (B) Bar graph illustrating the results of luciferase assays. HepG2 cells were transfected with pENG-luc plasmids [pENG-luc(WT), pENG-luc(M), or pENG-luc(M+1)] plus the β-galactosidase plasmid. Luciferase activity was normalized to β-galactosidase activity. The luciferase activity of pENG-luc(M) was significantly reduced compared to that of pENG-luc(WT). The activity was restored to levels even higher than WT levels in pENG-luc(M+1), which produced an in-frame luciferase fusion protein (44 aa-luciferase). Separate transfections were performed for each of three separate experiments (n = 9). Data represent mean ± SD. *p < .0001, as determined by Student's t test.
The plasmids were transfected into HepG2, a human hepatocellular liver carcinoma cell line, with Magnetamin (LPS Solution, South Korea) according to manufacturer's instructions. Cells were harvested 48 h after transfection. The luciferase activity was measured with the Enhanced Luciferase Assay Kit (BD Biosciences, USA) and normalized with β-galactosidase activity using β-galactosidase Enzyme Assay System (Promega, USA).
Allele-specific expression analysis
Total RNAs were isolated from blood using RNAiso Plus (TAKARA, Japan). Reverse transcription was performed with Prime RT-Premix (2×) (Genet Bio, Korea) using ENG-specific R3 primer (5'-cctggagagtcagctccagctgtg-3'). The PCR primers, F2 (5'-ctgctgtcactgccatccattgga-3') and R2 (5'-agacctggctagtggtatatgtca-3'), were used for amplification of the ENG cDNA. Genomic DNA was amplified using F1 (5'-ccatccttcggacagcaactccag-3') and R1 (5'-ccaccctgggtccctggacaccta-3'). Amplified PCR products were digested with BtsCI, which recognizes GGATG (i.e. mutant specific) sequence, and analyzed on an 8% polyacrylamide gel to determine whether the PCR product possessed ENG c.1-127 C > T mutation. The PCR products were also subjected to direct sequencing.
Real-time RT-PCR
Total RNAs were isolated from blood samples of unaffected and affected individuals using RNAiso Plus (TAKARA, Japan) and first-strand cDNA was synthesized by PrimeScript RT Master Mix Kit (TAKARA, Japan) with oligo(dT) primer. Real-time PCR was carried out using SYBR Premix DimerEraser Kit (TAKARA, Japan) and CFX384 Real-Time system (Bio-Rad, USA). A human Cyclophilin primer set (Forward, 5'-tgccatcgccaaggagtag-3'; Reverse, 5'-tgcacagacggtcactcaaa-3') was used to normalize the amount of total cDNA in each sample. Predesigned PCR primer for amplifying the Exon 3 and 4 region of the ENG gene (PrimerBank ID 168693646b1) [23-25] was used to determine the amount of ENG cDNA in each sample. Each sample was run in triplicate. After 40 cycles of PCR reaction, the relative amount of ENG transcripts was determined using the ΔΔCt method.
Results
Clinical features and mutations
Clinical features and mutations of patients are summarized in Table 1. Pedigree of HHT families is represented in Figure 1A.
Family 1
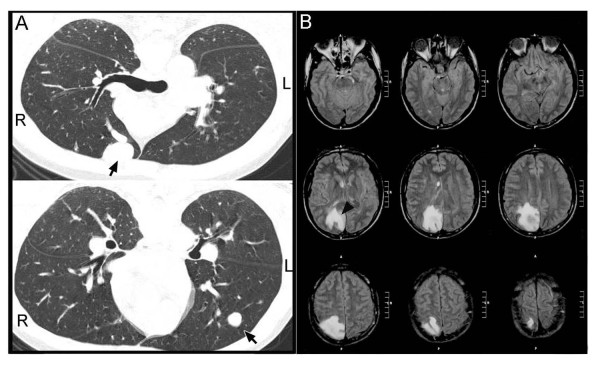
The proband is a 43-year-old man (I-2), diagnosed as HHT with recurrent epistaxis, pulmonary AVM (PAVM) (Figure 3A), and family history of epistaxis and/or PAVM. He had also suffered for epilepsy and cerebral abscess (Figure 3B) when he was 26 and 40 years old, respectively. These brain symptoms were most likely due to PAVMs that were recently treated with coil embolization. His two daughters (I-4 and I-5), brother (I-1), and sister (I-3), have experienced frequent nosebleeds and/or PAVM. This trait can be traced to great-grandmother of the proband.
Figure 3.
Clinical features of an HHT patient (I-2). PAVM (arrows) and cerebral abscess (arrow head) are detected by chest computed tomography (CT) (A) and brain CT (B), respectively.
Family 2
The proband is an 18-year-old man (II-6). He was diagnosed as HHT with recurrent epistaxis, PAVM, and family history of recurrent epistaxis [26]. He had seizure and embolic cerebral infarction [26]. These neurological complications might be associated with PAVM. Stroke, cerebral abscess, seizure, transient ischemic attacks, and migraine headaches were observed in 9%, 8%, 37%, and 43% PAVM patients, respectively [27]. A history of epistaxis was found in his relatives on the mother's side (patient II-1, II-2, II-3, II-4) [26]. His grandmother (II-1) has telangiectasis lesions on the lips and skin, and his uncle (II-2) had cerebral AVM (CAVM), and his cousin (II-5) died of CAVM.
Family 3
The proband is a 55-year-old man (III-1) who was suspected to be a HHT patient due to recurrent epistaxis and familial history. His brother (III-2) and sister (III-3) suffer from epistaxis. No history of visceral AVM was reported in the family.
Genetic analysis
PCR amplicons of whole exons and their flanking regions of ENG and ACVRL1 from genomic DNA of HHT patients and their family members (Figure 1, Table 1) were directly sequenced.
Family 1
We identified a heterozygous mutation in the splicing donor sequence in intron 3 of ENG (c.360+1G > A) from the proband sample. This mutation was previously reported that it causes an in-frame deletion of exon 3 (47 amino acids; p.Gly74_Tyr120del) and that the mutant protein fails to localize at the plasma membrane but stays in the cytosol [28]. Genetic analysis from symptomatic and asymptomatic family members identified the same mutation from family members with epistaxis and/or PAVM. No mutation was found in two children of the affected proband's sister (I-3).
Family 2
No mutation was found in the coding region and splicing junctions of either of ENG and ACVRL1 in the proband (II-6), but a substitution mutation in the 5'-UTR of ENG c.1-127 C > T. Interestingly, the same mutation was found in his mother (II-4, obligate carrier) and only in her symptomatic family members (II-1, -2, -3). The suspected mutation (-128ACG-126 with -128ATG-126) could generate a putative translation start codon in the 5'-UTR. If the putative translation start codon is functional, the ENG mutant allele would prevent the translation initiation from the translation start codon of normal ENG protein and produce an 84-amino acid-protein out of frame with the ENG-coding sequence instead. We confirmed with molecular analyses (described below) that this is indeed the case.
Family 3
We identified a novel 1-base pair (bp) insertional mutation in the ACVRL1 locus from the proband (III-1), and the same mutation was found in his affected sibling (III-2, -3). The 'C' nucleotide insertion in exon3 (c.252_253insC) would result in premature termination of translation (p.Val85fsX168) and production of truncated protein of size 167 amino acids instead of 503 amino acids full-length protein.
Functional tests of aberrant translation start codon found in Family 2
To investigate the extent to which the putative translation start codon created by mutation in the 5'-UTR of ENG identified in Family 2 inhibits normal ENG translation, we generated luciferase reporter constructs. These reporter constructs contain 5'-UTR regions (-413 to -1) of wild-type (WT) or of the mutant (M; c.1-127 C > T) ENG gene including the biological translation initiation sequence that was fused in frame with luciferase cDNA (Figure 2A). If the putative translation start codon by c.1-127 C > T mutation was functional and dominant over the endogenous start codon, the translated product from the putative start codon would be out of frame with the luciferase coding sequence, and thus no luciferase activity would be detected. As shown in Figure 2B, the luciferase activity from the cells transfected with pENG-luc(M) was dramatically lower than that with pENG-luc(WT), suggesting that the putative translation initiation sequence functions as the dominant translation start codon and prevents translation of functional luciferase. This notion was further supported by the results from the assay using the pENG-luc(M+1) reporter, in which single nucleotide was inserted between positions -41 and -40 of the pENG-luc(M). This single nucleotide insertion makes the translation from the putative translation start codon of pENG-luc(M+1) reporter in frame with the luciferase coding sequence. The luciferase activity from the cells transfected with the pENG-luc(M+1) reporter was even higher than that with the pENG-luc(WT) reporter, indicating that the putative translation start codon was able to produce a functional luciferase fusion protein. These results demonstrate that the aberrant translation start codon in the mutant ENG allele of Family 2 is functional and inhibits normal translation of the endogenous ENG protein.
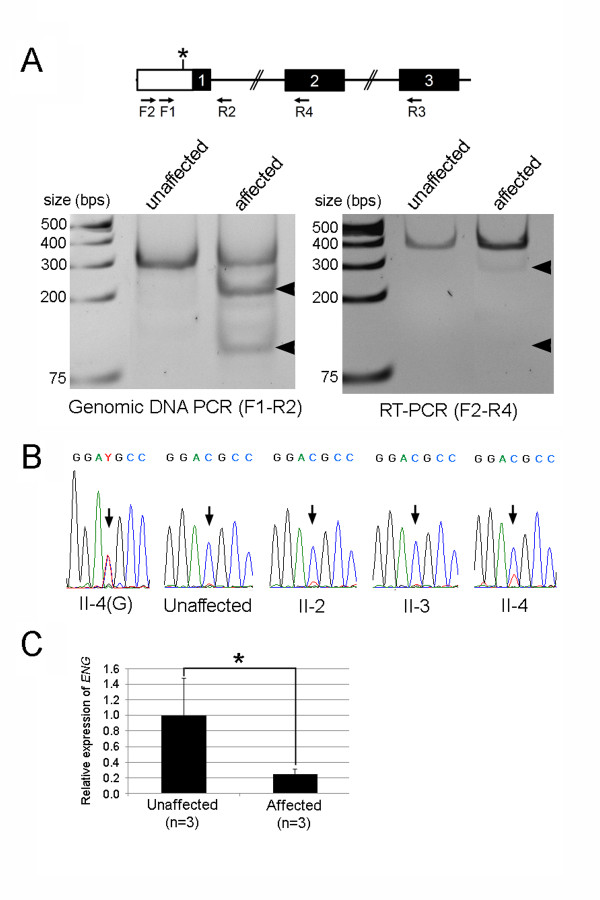
The aberrant translation start codon in the mutant allele creates a new open reading frame (ORF), which is 252 bp in length and has a termination codon in exon2. The mutant transcript could be a target of the nonsense-mediated mRNA decay [29,30], a surveillance mechanism, which degrades transcripts with nonsense mutations for preventing the production of erroneous proteins. To test this, we performed a restriction fragment length polymorphism (RFLP) analysis and direct sequencing of genomic DNA- and RT-PCR products using templates isolated from blood samples of unaffected and affected family members. The ENG c.1-127 C > T mutation introduces the BtsCI restriction site (GGATG) on the PCR product amplified from the mutant allele. As anticipated, BtsCI digestion on PCR products from genomic DNA templates demonstrated the presence of PCR amplicon of the mutant allele only from affected individuals (Figure 4A). On the other hand, the BtsCI-sensitive PCR amplicon was barely detectable from RT-PCR templates of affected individuals (Figure 4A), suggesting a much lower level of transcripts from the mutant allele compared with those from WT allele. Direct sequencing of RT-PCR products also confirmed the diminished mutant transcripts. Direct sequencing of PCR amplicons from genomic template of an affected family member (patient II-4) showed similar height of 'C' and 'T' peaks amplified from WT and mutant alleles, respectively (Figure 4B). However, the mutant 'T' peak was hardly detected in direct sequencing of RT-PCR products of three patients (patient II-2, -3, -4). To further confirm nonsense-mediated mRNA decay of ENG transcripts from the mutant allele, we quantified the amount of ENG mRNA using a real-time RT-PCR on RNAs isolated from unaffected and affected individuals (Figure 4C). The levels of ENG transcripts in the affected members (II-2, II-3, II-4) appear to be significantly lower than those in unaffected controls. Taken together, these results suggest that the substitution mutation (ENG c.1-127 C > T) in the 5'-UTR of ENG creates an aberrant translation start site which leads to frameshift and nonsense-mediated mRNA decay of the mutant transcript.
Figure 4.
Allele-specific expression analyses of the ENG c.1-127 C > T mutant allele. (A) Restriction fragment length polymorphism analysis. (Left panel) F1 and R1 primers were used for amplification of the ENG genomic DNA. BtsCI, which recognizes the mutant allele, digested of 313 bp PCR product from the mutant allele into 216 bp and 97 bp fragments (arrowheads). The mutant allele-specific BtsCI-digested PCR products were detected in affected family member (Patient II-3) but not in an unaffected family member. (Right panel) F2 and R2 primers were used to amplify ENG transcripts from ENG cDNA generated by reverse transcriptase using ENG-specific R3 primer. BtsCI digestion of RT-PCR product amplified from mutant allele is supposed to yield 284 bp and 97 bp fragments if the mutant transcripts are stably present. Unlike genomic PCR products, BtsCI-digested PCR fragments from RT-PCR products of affected individuals were barely detectable (arrowheads). (B) Direct sequencing of the PCR products amplified from genomic DNA of a patient II-4 and cDNAs of an unaffected family member and three patients (II-2, II-3, II-4). The ENG c.1-127 positions were indicated by arrows. (C) Real-time RT-PCR analysis of ENG transcripts. Total RNAs isolated from the blood of three unaffected persons and three patients (II-2, II-3, II-4) were used for first-strand cDNA synthesis. Mean values of ENG ΔΔCt ratios and standard deviations are shown as filled box and bar above each box, respectively. The relative amount of ENG transcripts in each sample was normalized with the amount of Cyclophilin in the sample. * p < 0.01, compared with the unaffected samples, as determined by Student's t test.
Discussion
We identified two ENG and one ACVRL1 mutations in genetic analyses of three unrelated Korean HHT families. Here, the ENG c.1-127 C > T mutation in Family 2 and the ACVRL1 c.252_253insC mutation in Family 3 are reported for the first time (Figure 1). Through in vitro luciferase reporter assay, we demonstrated that the C > T substitution (c.1-127 C > T) in the 5'-UTR of ENG is the disease-causing mutation in Family 2 (Figure 1, 2). Mutations in the 5'-UTR are very rare in HHT patients. Bossler et al. reported the c.1-10 C > T mutation in the ENG 5'-UTR of a patient with HHT [31], which is the only known 5'-UTR mutation among reported ACVRL1 and ENG mutations, except large deletion mutations that include the 5'-UTRs [14]. Although the patient with the ENG c.1-10 C > T mutation had recurrent epistaxis, PAVM, and a suggestive family history, no experimental data testing the functionality of the mutant allele was reported.
Generally, eukaryotic ribosomes bind to 5'-end cap of mRNA and scan along the mRNA in the 5' to 3' direction until the first translation start codon is found, thus initiating protein synthesis [32]. This 'position effect' is evident in cases in which a mutation creates an ATG codon upstream of the endogenous translation start codon [32]. The new ATG initiates translation and suppresses translation from the endogenous ATG. The optimal sequence surrounding the translation start codon is 5'-gcc(a/g)ccATGg-3' in mammals [32]. Within the context, the purine (A or G) at position -3 is the most important nucleotide. Additionally, the 'G' at position +4 is also highly conserved, especially in the absence of 'A' at position -3. According to mutagenesis experiments, the sequence contexts generating the most robust translation are 5'-annATGn-3' or 5'-gnnATGg [32]. When the context lacks both the purine at position -3 and the 'G' at position +4, most ribosomes skip the ATG and continue scanning the sequence downstream. In the case of a sequence in which only one of these nucleotides is conserved, initiation of translation from the suboptimal ATG depends on several factors including downstream secondary structure and the remaining nucleotides surrounding the ATG codon [32]. The endogenous and mutant translation start codons in the mutant ENG allele are of the following sequence contexts: 5'-gacagcATGg-3' (endogenous) and 5'-gcagggATGc-3' (mutant). The sequences surrounding the endogenous translation start codon contains a purine (A) at position -3 and 'G' at position +4. On the other hand, the mutant translation start context has a purine (G) at position -3, but 'C' instead of 'G' at position +4. Our data from the functional assays reported here demonstrate that the suboptimal mutant translation start context is functionally active and dominate over the translation from the endogenous translation start context (Figure 2). Furthermore, we present that the mutation results in mRNA degradation (Figure 4), which might be caused by nonsense-mediated mRNA decay [29,30]. A critical determinant of nonsense-mediated mRNA decay is the distance between the translation start codon and premature termination codon. Generally, premature termination codons located at least 50-55 nt upstream of the last exon-exon junction leads to strong nonsense-mediated mRNA decay responses. The ENG c.1-127 C > T mutation creates a new open reading frame (ORF), which is 252 bp in length. The distance between the termination codon of the new ORF and the last exon-exon junction is more than 1.5 kb, which satisfies the 50-55 nt boundary rule.
Conclusions
Three unrelated Korean HHT families were analyzed clinically and genetically. We found HHT-causing ENG or ACVRL1 mutations in these families. Consistent with HHT clinical reports worldwide, the mutation carriers exhibit diverse clinical symptoms even among family members, and epistaxis is the most common symptom among HHT1 and HHT2 patients. We presented experimental demonstration for the first time in the HHT field that a 5'-UTR mutation creates alternative translation initiation sequence, suppresses normal translation of ENG from its endogenous translation start codon, and leads to degradation of the mutant transcripts. In addition to other known cases [4,33,34], this finding provides further supporting evidence that haploinsufficiency is predominant mechanism of HHT1. Our data also underscore the importance of including exons encoding 5' UTR for HHT mutation screening.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YJL and SPO were the principal investigators and take primary responsibility for the paper. S-TK, HDL, and KYL recruited the patients and performed clinical diagnosis and treatment. M-JK, JS, and J-BL performed the mutational analysis. YJL, SPO, and M-JK co-ordinated the research. YJL and SPO wrote the paper. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
A title listing the PCR primers for ACVRL1 and ENG genomic PCR. PCR primer sequences that were used for amplifying exons and their flanking intronic sequences for sequencing ACVRL1 and ENG genes.
Contributor Information
Mi-Jung Kim, Email: kimmj@gachon.ac.kr.
Seon-Tae Kim, Email: rhinokim2002@hanmail.net.
Hyoung-Doo Lee, Email: hdlee@pusan.ac.kr.
Kyu-Yong Lee, Email: kylee@hanyang.ac.kr.
Jiyoung Seo, Email: jyseo@gachon.ac.kr.
Jae-Bom Lee, Email: ljbom@hotmail.com.
Young-Jae Lee, Email: leeyj@gachon.ac.kr.
Suk P Oh, Email: ohp@ufl.edu.
Acknowledgements and Funding
The authors would like to thank all patients and families participated in this study. This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST) (NRF-2008-331-E00041) and by a research grant of the Life Insurance Philanthropy Foundation (1-16).
References
- Shovlin CL. Hereditary haemorrhagic telangiectasia: pathophysiology, diagnosis and treatment. Blood Rev. 2010;24:203–219. doi: 10.1016/j.blre.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Kjeldsen AD, Vase P, Green A. Hereditary haemorrhagic telangiectasia: a population-based study of prevalence and mortality in Danish patients. J Intern Med. 1999;245:31–39. doi: 10.1046/j.1365-2796.1999.00398.x. [DOI] [PubMed] [Google Scholar]
- Dakeishi M, Shioya T, Wada Y, Shindo T, Otaka K, Manabe M, Nozaki J, Inoue S, Koizumi A. Genetic epidemiology of hereditary hemorrhagic telangiectasia in a local community in the northern part of Japan. Hum Mutat. 2002;19:140–148. doi: 10.1002/humu.10026. [DOI] [PubMed] [Google Scholar]
- Abdalla SA, Letarte M. Hereditary haemorrhagic telangiectasia: current views on genetics and mechanisms of disease. J Med Genet. 2006;43:97–110. doi: 10.1136/jmg.2005.030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenato GM, Guanti G. Hereditary Haemorrhagic Telangiectasia (HHT): genetic and molecular aspects. Curr Pharm Des. 2006;12:1173–1193. doi: 10.2174/138161206776361291. [DOI] [PubMed] [Google Scholar]
- McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, Helmbold EA, Markel DS, McKinnon WC, Murrell J. et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- McDonald MT, Papenberg KA, Ghosh S, Glatfelter AA, Biesecker BB, Helmbold EA, Markel DS, Zolotor A, McKinnon WC, Vanderstoep JL. et al. A disease locus for hereditary haemorrhagic telangiectasia maps to chromosome 9q33-34. Nat Genet. 1994;6:197–204. doi: 10.1038/ng0294-197. [DOI] [PubMed] [Google Scholar]
- Shovlin CL, Hughes JM, Tuddenham EG, Temperley I, Perembelon YF, Scott J, Seidman CE, Seidman JG. A gene for hereditary haemorrhagic telangiectasia maps to chromosome 9q3. Nat Genet. 1994;6:205–209. doi: 10.1038/ng0294-205. [DOI] [PubMed] [Google Scholar]
- Vincent P, Plauchu H, Hazan J, Faure S, Weissenbach J, Godet J. A third locus for hereditary haemorrhagic telangiectasia maps to chromosome 12q. Hum Mol Genet. 1995;4:945–949. doi: 10.1093/hmg/4.5.945. [DOI] [PubMed] [Google Scholar]
- Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, Yoon SJ, Stenzel TT, Speer M, Pericak-Vance MA, Diamond A. et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- Johnson DW, Berg JN, Gallione CJ, McAllister KA, Warner JP, Helmbold EA, Markel DS, Jackson CE, Porteous ME, Marchuk DA. A second locus for hereditary hemorrhagic telangiectasia maps to chromosome 12. Genome Res. 1995;5:21–28. doi: 10.1101/gr.5.1.21. [DOI] [PubMed] [Google Scholar]
- Lesca G, Burnichon N, Raux G, Tosi M, Pinson S, Marion MJ, Babin E, Gilbert-Dussardier B, Riviere S, Goizet C. et al. Distribution of ENG and ACVRL1 (ALK1) mutations in French HHT patients. Human mutation. 2006;27(6):598. doi: 10.1002/humu.9421. [DOI] [PubMed] [Google Scholar]
- Govani FS, Shovlin CL. Hereditary haemorrhagic telangiectasia: a clinical and scientific review. European journal of human genetics: EJHG. 2009;17(7):860–871. doi: 10.1038/ejhg.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HHT Mutation Database. http://www.hhtmutation.org/index.php Database Last Updated: 09/19/2009.
- Cole SG, Begbie ME, Wallace GM, Shovlin CL. A new locus for hereditary haemorrhagic telangiectasia (HHT3) maps to chromosome 5. Journal of medical genetics. 2005;42(7):577–582. doi: 10.1136/jmg.2004.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrak-Toydemir P, McDonald J, Akarsu N, Toydemir RM, Calderon F, Tuncali T, Tang W, Miller F, Mao R. A fourth locus for hereditary hemorrhagic telangiectasia maps to chromosome 7. American journal of medical genetics. 2006;140(20):2155–2162. doi: 10.1002/ajmg.a.31450. [DOI] [PubMed] [Google Scholar]
- Gallione CJ, Repetto GM, Legius E, Rustgi AK, Schelley SL, Tejpar S, Mitchell G, Drouin E, Westermann CJ, Marchuk DA. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363(9412):852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- David L, Mallet C, Keramidas M, Lamande N, Gasc JM, Dupuis-Girod S, Plauchu H, Feige JJ, Bailly S. Bone morphogenetic protein-9 is a circulating vascular quiescence factor. Circulation research. 2008;102(8):914–922. doi: 10.1161/CIRCRESAHA.107.165530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard N, Bidart M, Mallet C, Lesca G, Giraud S, Prudent R, Feige JJ, Bailly S. Functional analysis of the BMP9 response of ALK1 mutants from HHT2 patients: a diagnostic tool for novel ACVRL1 mutations. Blood. 2010;116(9):1604–1612. doi: 10.1182/blood-2010-03-276881. [DOI] [PubMed] [Google Scholar]
- David L, Mallet C, Mazerbourg S, Feige JJ, Bailly S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood. 2007;109(5):1953–1961. doi: 10.1182/blood-2006-07-034124. [DOI] [PubMed] [Google Scholar]
- Lee ST, Kim JA, Jang SY, Kim DK, Do YS, Suh GY, Kim JW, Ki CS. Clinical features and mutations in the ENG, ACVRL1, and SMAD4 genes in Korean patients with hereditary hemorrhagic telangiectasia. Journal of Korean medical science. 2009;24(1):69–76. doi: 10.3346/jkms.2009.24.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shovlin CL, Guttmacher AE, Buscarini E, Faughnan ME, Hyland RH, Westermann CJ, Kjeldsen AD, Plauchu H. Diagnostic criteria for hereditary hemorrhagic telangiectasia (Rendu-Osler-Weber syndrome) Am J Med Genet. 2000;91(1):66–67. doi: 10.1002/(SICI)1096-8628(20000306)91:1<66::AID-AJMG12>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic acids research. 2010. pp. D792–799. [DOI] [PMC free article] [PubMed]
- Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic acids research. 2003;31(24):e154.26. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K-W, Chang W-Y, Kim H-Y, Koh S-H, Han G-S, Lee Y-J, Lee K-Y. A Case of Hereditary Hemorrhagic Telangiectasia Presented with Seizure. Korean J Stroke. 2007;9(1):62–65. [Google Scholar]
- Dinkel HP, Triller J. Pulmonary arteriovenous malformations: embolotherapy with superselective coaxial catheter placement and filling of venous sac with Guglielmi detachable coils. Radiology. 2002;223(3):709–714. doi: 10.1148/radiol.2233010953. [DOI] [PubMed] [Google Scholar]
- Pece N, Vera S, Cymerman U, White RI Jr, Wrana JL, Letarte M. Mutant endoglin in hereditary hemorrhagic telangiectasia type 1 is transiently expressed intracellularly and is not a dominant negative. The Journal of clinical investigation. 1997;100(10):2568–2579. doi: 10.1172/JCI119800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96(3):307–310. doi: 10.1016/S0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. The Biochemical journal. 2010;430(3):365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- Bossler AD, Richards J, George C, Godmilow L, Ganguly A. Novel mutations in ENG and ACVRL1 identified in a series of 200 individuals undergoing clinical genetic testing for hereditary hemorrhagic telangiectasia (HHT): correlation of genotype with phenotype. Human mutation. 2006;27(7):667–675. doi: 10.1002/humu.20342. [DOI] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299(1-2):1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadick H, Riedel F, Naim R, Goessler U, Hormann K, Hafner M, Lux A. Patients with hereditary hemorrhagic telangiectasia have increased plasma levels of vascular endothelial growth factor and transforming growth factor-beta1 as well as high ALK1 tissue expression. Haematologica. 2005;90(6):818–828. [PubMed] [Google Scholar]
- Paquet ME, Pece-Barbara N, Vera S, Cymerman U, Karabegovic A, Shovlin C, Letarte M. Analysis of several endoglin mutants reveals no endogenous mature or secreted protein capable of interfering with normal endoglin function. Human molecular genetics. 2001;10(13):1347–1357. doi: 10.1093/hmg/10.13.1347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A title listing the PCR primers for ACVRL1 and ENG genomic PCR. PCR primer sequences that were used for amplifying exons and their flanking intronic sequences for sequencing ACVRL1 and ENG genes.