Abstract
Background
A substantial amount of data has been accumulated supporting the important role of genomic islands (GEIs) - including pathogenicity islands (PAIs) - in bacterial genome plasticity and the evolution of bacterial pathogens. Their instability and the high level sequence similarity of different (partial) islands suggest an exchange of PAIs between strains of the same or even different bacterial species by horizontal gene transfer (HGT). Transfer events of archetypal large genomic islands of enterobacteria which often lack genes required for mobilisation or transfer have been rarely investigated so far.
Results
To study mobilisation of such large genomic regions in prototypic uropathogenic E. coli (UPEC) strain 536, PAI II536 was supplemented with the mobRP4 region, an origin of replication (oriVR6K), an origin of transfer (oriTRP4) and a chloramphenicol resistance selection marker. In the presence of helper plasmid RP4, conjugative transfer of the 107-kb PAI II536 construct occured from strain 536 into an E. coli K-12 recipient. In transconjugants, PAI II536 existed either as a cytoplasmic circular intermediate (CI) or integrated site-specifically into the recipient's chromosome at the leuX tRNA gene. This locus is the chromosomal integration site of PAI II536 in UPEC strain 536. From the E. coli K-12 recipient, the chromosomal PAI II536 construct as well as the CIs could be successfully remobilised and inserted into leuX in a PAI II536 deletion mutant of E. coli 536.
Conclusions
Our results corroborate that mobilisation and conjugal transfer may contribute to evolution of bacterial pathogens through horizontal transfer of large chromosomal regions such as PAIs. Stabilisation of these mobile genetic elements in the bacterial chromosome result from selective loss of mobilisation and transfer functions of genomic islands.
Background
In the early 1980s large unstable chromosomal regions carrying virulence-associated genes were identified in uropathogenic E. coli [1]. Later, such large unstable chromosomal regions were designated pathogenicity islands (PAIs) [2-4]. A constantly increasing number of similar genetic elements detected in many pathogenic and non-pathogenic microorganisms led to the definition of a family of related genetic elements, termed genomic islands (GEIs), whose members share characteristic features [5-7]. Although PAIs, a subgroup of GEIs, are in several cases superficially similar, they structurally differ with respect to the encoded virulence factors, the size and the presence of different mobile and accessory elements. Due to the presence of mobility genes (integrases, transposases, IS elements) or the occurrence of recombination processes or point mutations, PAIs constantly undergo structural changes [4,8-12]. Upon acquisition and chromosomal insertion, islands together with additional large regions of flanking chromosomal sequence context can be transferred by conjugation and homologous recombination and thus contribute to genome plasticity and the simultaneous transfer of multiple traits [13].
Nevertheless, PAIs are in many cases not stably integrated into the E. coli host chromosome and may be lost upon deletion. This process can be studied by island probing [10,14-16]. The influence of different environmental conditions on the stability of five PAIs of UPEC strain 536 has already been investigated before [17] indicating that PAI I536, PAI II536, PAI III536, and PAI V536 delete with frequencies between 10-5 and 10-6, while loss of PAI IV536 could not be detected. In UPEC strain 536, PAI deletion is catalyzed by a P4-like bacteriophage integrase which is encoded on the respective island [18]. Similar deletion frequencies (10-5 - 10-6) were also reported for PAIs of REPEC strain 84/110-1 and S. flexneri 2a [12,19]. Higher deletion frequencies (10-3 - 10-4) have, however, been observed for O-islands 43 and 48 in enterohemorrhagic E. coli (EHEC) O157:H7 isolates [14]. Circular intermediate (CI) formation in the cytoplasm of UPEC strain 536 was demonstrated for PAI II536 and PAI III536. Since none of these two islands apparently contain an origin of replication, it has been hypothesized that CIs are lost upon cell division unless they reintegrate into the chromosome. Furthermore, horizontal gene transfer (HGT) of such circularized PAIs may occur with the help of bacteriophages or conjugative plasmids [17].
A close functional association between PAIs and bacteriophages was reported for several bacterial pathogens. In V. cholerae, the entire 39.5-kb Vibrio Pathogenicity Island (VPI) can be transfered by the general transducing phage CP-T1 [20]. The "high pathogenicity island (HPI)" of Yersinia pseudotuberculosis has been shown to be transfered by a bacteriophage [21]. The so-called Staphylococcus aureus pathogenicity islands (SaPIs) can excise and replicate upon induction by other resident S. aureus bacteriophages and then packed into phage-like particles which can be transferred to recipient cells [22-24]. SaPI transfer by transduction can even occur between representatives of different species. The intra- and interspecies transfer was demonstrated for the SaPI-2 element which could be transferred into a variety of different recipients [22,25,26]. The identification of self-replicating plasmid-like states of the excised SaPI element, however, is also reminiscent of plasmid-like ancestors [22]. Bacteriophage-mediated transfer is limited by the amount of DNA that can be packed into the phage capsid, but in some cases it can expand beyond 100 kb [27,28]. As multiple island-like genomic regions in other bacteria exhibit features of degenerate prophages as well, there may be the possibility to mobilize these islands by other phages.
The discovery of integrative conjugative elements (ICEs) and related genetic entities suggests another mechanism of PAI transfer [29-32]. With the help of excisionases and integrases PAIs and related integrative mobilisable elements are able to site-specifically delete from or integrate into the chromosome. After deletion they are able to replicate and can also be transmitted into a new host by their own conjugative machinery. A variant of the "high pathogenicity island" (HPI) has been described in E. coli strain ECOR31 to contain a 35-kb sequence with striking homology to conjugative plasmids [33]. The identification of this ICE-EC1 carrying a functional transfer determinant suggests that conjugative transfer may have played a role in the spread of the HPI, and possibly also in the transmission of other PAIs. The spread of the non-selftransmissible but mobilisable antibiotic resistance gene cluster of the Salmonella genomic island 1 (SGI1) also supports the existence of a conjugal transfer mechanism for PAIs as well as interstrain PAI transfer observed in Pseudomonas aeruginosa, Enterococcus faecalis and Streptococcus thermophilus [34-36]. Type IV secretion systems (T4SSs) have been shown to mediate the horizontal transfer of such DNA elements in a broad range of bacteria [32,37-40]. Alternatively, (co-)mobilisation of circular intermediates of islands and related genetic elements has been described [23,41-44].
To study whether archetypal PAIs of E. coli which usually lack traits that enable their distribution such as origins of replication and tra genes could be generally (co-)mobilised by a helper plasmid, we investigated the transferability of PAI II536, the largest PAI (102.2 kb) of UPEC strain 536, into an E. coli K-12 recipient and back into a PAI II536-negative mutant of strain 536.
Results
Transfer of the entire PAI II536 from UPEC strain 536 into E. coli K-12
Altogether, 31 mating experiments were carried out at 20°C and 37°C. Plating of conjugation batches with E. coli strains 536-19/1mob (donor) and SY327λpir (recipient) resulted in high numbers of chloramphenicol (Cm) and nalidixic acid (Nal)-resistant colonies and 899 resulting haemolytic clones were further investigated. Screening of clones that grew on the Cm-Nal selective blood agar medium with the E. coli K-12- and K15 capsule-specific PCRs, however, revealed that only 27.6% (248 clones) of them were true E. coli K-12 transconjugants, whereas the rest proved to be spontaneous nalidixic acid resistant mutants of strain 536. These clones were further analysed with four PAI II536-specific PCRs (Figure 1B) to determine whether the complete PAI II536 had been transferred. 93.1% (231 clones) of the 248 transconjugants acquired the complete island and 6.9% (17 clones) of the haemolytic transconjugant clones have only been partially transferred to the recipient strain.
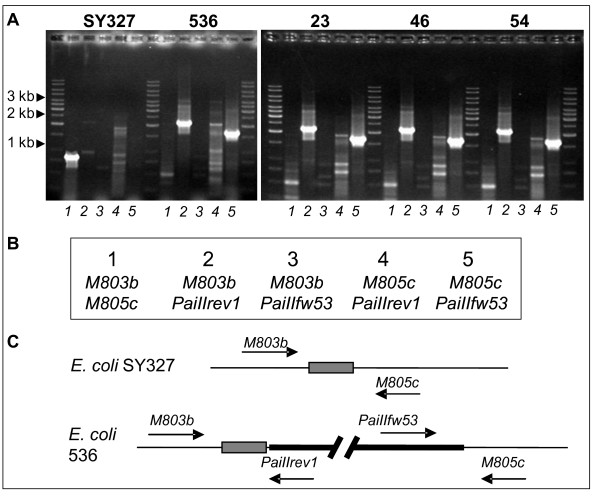
Figure 1.
Confirmation of the chromosomal insertion of the mobilised PAI II536 in recipient strain SY327. leuX and PAI II536- specific PCRs were carried out (A) with laboratory K-12 strain SY327λpir, wild type strain 536, and the transconjugant clones 23, 46, 54. For this purpose, four test primers (M803b, M805c, PaiIIrev1, PaiIIfw53) were used in different combinations indicated in (B). The orientation of the primers relative to leuX (grey box) in a K-12 strain and in the wild type strain 536 is depicted in the lower part of the figure (C).
The mating temperature slightly affected the proportions of the different types of PAI transfer. At 20°C, 81.5% (n = 88) of the transconjugants carried the chromosomally inserted PAI II536 construct, 14.8% (n = 16) had circular intermediates, and 3.7% (n = 4) resulted from partial PAI II536 transfer. Upon mating at 37°C, 70.0% (n = 98) of PAI II536 were chromosomally inserted, 20.7% (n = 29) were circular intermediates, and 9.3% (n = 13) were only partially transferred. The differences observed between the different types of transconjugants obtained at 20°C and 37°C were not significant.
Transfer frequencies were between 1 × 10-7 and 6.66 × 10-9 (data not shown), depending on the mating temperature (20°C or 37°C) as well as on the ratio of donor and recipient cells (3:1 or 9:1). The mean transfer frequency at both temperatures was always higher with a donor: recipient ratio of 9:1 relative to a 3:1 ratio. The differences observed were, however, not significant (Table 1).
Table 1.
Mobilisation and remobilisation of PAI II536
| Transfer rate of PAI II536 | ||
|---|---|---|
| 20°C | 37°C | |
| Mobilisation rate from E. coli 536 to E. coli SY327 | ||
| Donor-recipient ratio 3:1 | 3.47 × 10-08 ± 4.85 × 10-09 | 3.65 × 10-08 ± 5.46 × 10-09 |
| Donor-recipient ratio 9:1 | 4.93 × 10-08 ± 1.14 × 10-08 | 4.31 × 10-08 ± 6.11 × 10-09 |
| Remobilisation rate from E. coli SY327 to E. coli 536-21 | ||
| Donor with integrated PAI II536 | 1.41 × 10-07 ± 1.25 × 10-07 | 8.00 × 10-08 ± 7.47 × 10-08 |
| Donor with CI of PAI II536 | 4.32 × 10-05 ± 3.65 × 10-05 | 3.75 × 10-05 ± 3.18 × 10-05 |
31 and 10 independent conjugation experiments were performed for the mobilisation and remobilisation experiment, respectively. Plasmid RP4 was used as a helper plasmid for mobilisation of the excised PAI II536 construct from E. coli 536 into recipient E. coli SY327.
PAI II536 integrates site-specifically into the E. coli K-12 chromosome at the tRNA gene leuX
Upon conjugation, the transferred circularised form of the PAI II536 derivative can integrate into the recipient's chromosome. Additionally, the recipient strain SY327λpir also enables episomal replication of the transferred CI. Analysis by PCR of the transconjugants carrying the complete PAI II536 derivative allowed to distinguish between chromosomally inserted and episomal circular forms of the PAI II536 construct. Episomal CIs could not be detected in the clones with the chromosomally inserted PAI II536 derivative. As exemplarily shown for clones 23, 46, and 54, the orientation of the site-specifically integrated PAI II536 within the chromosome was determined by using combinations of the four primer pairs indicated in Figure 1. In these three clones as well as in donor strain 536, PCR screening products could only be obtained using primer pairs 2 and 5, which amplify the ends of PAI II536 with the adjacent core genome context. Primer pair 1 amplifies the empty leuX locus in the core genome context and gave only a PCR product in the recipient strain SY327. Accordingly, PAI II536 has been inserted into the leuX gene of the E. coli SY327 chromosome in the identical orientation as in the donor chromosome (Figure 1).
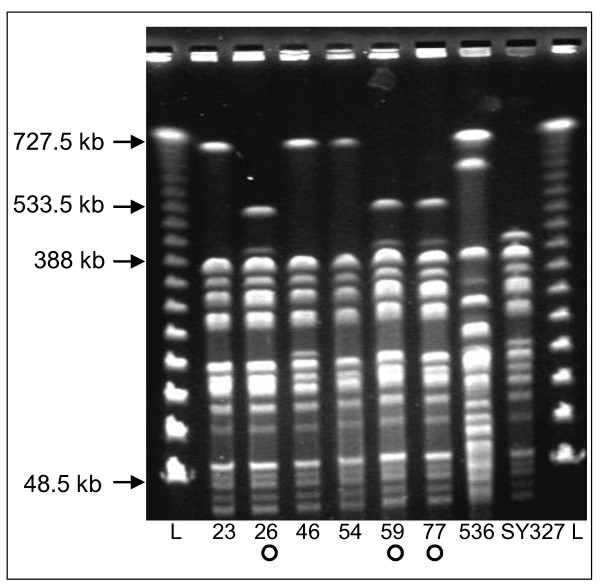
Genomic restriction patterns of representative transconjugants, carrying either the chromosomally inserted PAI II536 derivative or its episomal CI, were compared to each other and to those of the donor and recipient strain by PFGE in order to assess their genomic homogeneity (Figure 2). Generally, the restriction patterns of the transconjugants were very similar to that of recipient strain SY327λpir. The PFGE patterns of the selected transconjugants which carried the transferred PAI II536 in their chromosome exhibited only minor differences among each other. Similarly, the restriction patterns of the clones containing the stable episomal CI of PAI II536 were identical. Both groups of transconjugants could be clearly distinguished upon the presence of a ~400-kb and a ~530-kb restriction fragment in those recipient clones with a stable cytoplasmic PAI II536 CI which were absent from recipients in which chromosomal integration of the island occurred. Instead, a restriction fragment of about 700 kb was visible in the latter clones (Figure 2). This larger restriction fragment may comprise the 530-kb restriction fragment after chromosomal insertion of the transferred PAI II536 (107-kb) construct. These data demonstrate that PAI II536 can be mobilized upon excision from the chromosome by helper plasmids into suitable recipient strains. Upon transfer, the majority of CIs integrates site-specifically into the recipient's chromosome at the leuX locus or remains as an episomal CI.
Figure 2.
Analysis of the genomic restriction pattern of different recipient clones upon transfer of PAI II536 by PFGE. Genomic DNA of three representatives of the transconjugants carried either a chromosomally inserted PAI II536 or an episomal circular intermediate (CI) were digested with SfiI. Donor strain 536 and recipient strain SY327λpir are controls. Recipients 26, 59, and 77 (marked with 'o') carried a PAI II536-specific CI, whereas in strains 23, 46, and 54 PAI II536 has been chromosomally inserted at the leuX tRNA locus. L, Lambda Ladder PFGE marker, (New England Biolabs).
Remobilisation of the transferred PAI II536 into E. coli strain 536-21
Since two types of transconjugants resulted from the PAI II536 mobilisation, two types of remobilisation experiments were performed: K-12 strains harbouring either the CI or the chromosomally inserted PAI II536 were used as donors. Since the recipient strain 536-21 does not express the π-protein, only chromosomal integration of PAI II536 into the leuX gene was observed in all transconjugants. There was a marked difference in the conjugation efficiency between the remobilisation of the circular and the integrated forms. In those cases where strain SY327-77 carrying an episomal CI of PAI II536 was used as donor, average PAI transfer was about 100- to 1000-fold more efficient with transfer rates of 3.75 × 10-5 at 37°C and 4.32 × 10-5 at 20°C, respectively. However, if SY327-54 served as a donor, where PAI II536 was integrated into the chromosome, the average efficiency of transfer was 8 × 10-8 and 1.4 × 10-7, at 37°C and 20°C, respectively (Table 1). These results support that the mobilised PAI and the RP4 plasmid include all the factors required for excision of the chromsomally inserted PAI as well as for its efficient transfer.
Discussion
Horizontal gene transfer (HGT) plays an important role during prokaryotic evolution. Exchange and accumulation of a variety of fitness or virulence factors frequently carried on mobile genetic elements contributes to evolution of different pathogens and pathotypes from non- or less pathogenic variants [8,45]. One perfect environment for this evolutionary process is the mammalian gut with its large bacterial density which offers the possibility of close cell-to-cell contacts between closely or even remotely related bacteria. In this way, members of the gut flora, such as E. coli, may also increase their pathogenic potential and may evolve from commensals into e.g. extraintestinal pathogens. E. coli may, nevertheless, also exist outside of the gut, e.g. in the environment having the possibility to exchange genetic information with other bacteria. High bacterial cell densities could be observed, e.g. in bacterial biofilms, an important bacterial lifestyle in the environment. The PAI II536 transfer at 20°C indicates that E. coli can exchange PAIs not only upon growth at human body temperature but also at a temperature which is closer to the ambient temperature in the environment.
For the transfer of PAIs, different mechanisms have been postulated. For example, the presence of phage-related sequences on most PAIs suggests a key role of bacteriophages in HGT, and, indeed, a transfer by transducing phages has been reported for VPI and SaPI1 [20,26,27]. Flanking direct repeat sequences (DRs) and an active bacteriophage integrase play also an important role in the excision process of E. coli 536-specific PAIs [18], which is essential for a subsequent transfer. Alternatively, PAIs can be transfered by conjugation. The HPI of E. coli strain ECOR31 with its flanking DRs, an integrase gene and the right border region (RB-HPIECOR31) encoding a functional mating pair formation system and a DNA-processing region, fulfills all structural criteria of integrative and conjugative elements, ICE [29,31,33]. Although neither conserved repABC genes, other indications of a plasmid replicon, nor mobilisation have been detected, this HPI variant supports the hypothesis that PAI transfer can also occur by conjugal transfer [33]. Furthermore, high partial similarity between different polyketide biosynthesis determinants located on islands such as the HPI and the colibactin island of extraintestinal pathogenic E. coli, ICEs and different enterobacterial plasmids have been previously described. The presence of these polyketide determinants in different enterobacterial species and their (co-)localisation on different mobile genetic elements further support the idea that different chromosomal and episomal elements can recombine and thus due to HGT promote bacterial genome plasticity [46]. Additionally, self-transmissible conjugative elements can mobilize other genomic DNA regions in cis or in trans. The conjugative plasmid RP4, for example, can mediate transfer of mobilizable plasmids which code for an origin of transfer (oriT), a relaxase and nicking accessory proteins for interaction with oriT. A conjugative element then provides the mating pair formation functions for transfer [47].
Large-scale DNA transfer followed by homologous recombination can also be involved in the distribution of chromosomally inserted pathogenicity islands. Different HPI-transfer events have been detected in E. coli, in which not only the HPI itself but also flanking regions of the genomic backbone have been transfered. Schubert and colleagues demonstrated that the conjugative F plasmid can transfer and insert the HPI into the recipient chromosome by homologous recombination of flanking DNA regions. Upon chromosomal integration of an F plasmid, the recipient genome acquires an oriT and thereby becomes mobilisable. Resulting so-called "high frequency of recombination" (Hfr) strains can transfer large parts of their chromosomes at high frequency [13].
PAI deletion has been described for UPEC strain 536 and other pathogenic bacteria [10,14,17,48-50] as well as the occurrence of circular intermediates upon PAI excision of [12,23,26,30,33,35,36,50] suggesting that the latter could be formed during conjugal or phage-mediated transfer. Using a conjugative helper plasmid, transfer of a CI was also verified for the 43-kb Salmonella genomic island 1 (SGI1) [30]. In addition, the 35-kb HPI of Yersinia enterocolitica could be mobilised [51] when a modified RP4 plasmid was used as a shuttle vector during the transfer experiments. Several cases of plasmid mobilisation as a major mechanism for horizontal gene transfer of PAIs have been described [42-44].
With the PAI II536 construct used in this study, we were able to transfer this ~107-kb DNA region in the presence of the unmodified RP4 plasmid and thereby demonstrated that PAI II536 is mobilisable, but not self-transmissible. To increase the stability of the large PAI II536-specific CI and thus the transfer frequency, we also integrated an origin of replication into this PAI. In this respect, our model construct is artificial, but exhibits similar features of some ICEs including the HPIECOR31. In the latter case, the origin of replication seems to be inactivated by insertion of an IS630 homologue [33]. This may explain why HPIECOR31 is not transferable although CI formation of this island was shown in the same study. Whereas plasmids replicate autonomously, ICEs are generally thought to be incapable of autonomous replication. Instead, their replication depends on that of host chromosome [52]. Some ICE and ICE-like elements, however, have been reported to be capable of autonomous replication [53-57]. In the light F plasmid-mediated mobilization of the HPI [13], it would, nevertheless, also be interesting to analyse in the future if a PAI II536 construct, which is not a self-replicating entity, but only carries an oriT, could be mobilized upon provision of the appropriate conjugative machinery in trans on a plasmid.
The primary aim of our study was to demonstrate the transferability of a large archetypal island of UPEC strain 536 as this PAI can be excised site-specifically from the chromosome by its cognate integrase. On the other hand, we also tested conditions which may affect the transfer of an excised circular PAI intermediate. The frequency of PAI transfer in the mobilisation experiments was low (between 10-8 and 10-9). We postulate that the efficiency of PAI II536 transfer depends on several factors including the growth temperature, integrase activity, the size, and the chromosomal or episomal state of the PAI. In spite of the large size of PAI II536, complete transfer occurred at a high rate. 93.1% of the transconjugants received the complete 107-kb PAI II536 construct. The activity of the PAI-encoded integrase can contribute to the transfer efficiency by affecting the PAI excision as well as the integration frequency. The remobilisation efficiency was three log scales higher with a stable episomal CI compared to an integrated PAI, indicating that a more active integrase may increase the chance of transfer by frequent induction of PAI-excision from the chromosome (Table 1). PAI II536 transfer rates at 20°C and 37°C were not significantly different. Besides the gut, E. coli also faces the environment as a natural habitat since the bacteria are excreted each day in considerable amounts. As a part of naturally occurring biofilms in sewage or drinking water systems, they are exposed to stimuli described above, i.e. low temperature and high density of cells, what might explain their ability to efficiently exchange genetic elements also under these conditions.
In accordance with previously published results [18], the mobilisation and remobilisation experiments corroborated that the P4-like integrase of PAI II536 is highly specific. In both strain backgrounds, SY327λpir and 536-21, the PAI II536 was found only to be inserted into the leuX locus thereby restoring the complete tRNA gene in the latter strain. This result demonstrated that leuX is the preferred chromosomal integration site of PAI II536. Site-specific chromosomal integration of PAIs has already been described before. However, if multiple isoacceptor tRNA genes exist, chromosomal insertion may occur at all the available isoacceptor tRNA loci. The HPI of Y. pestis is usually associated with the asnT tRNA locus, but in Y. pseudotuberculosis the HPI can insert into any of the three chromosomal asn tRNA loci [58]. The same phenomenon has been observed as well, e.g. with LEE PAIs [12] and the PAPI-1 island of P. aeruginosa [36].
The lack of genes required for mobilisation and/or transfer on the archetypal PAIs of UPEC strains such as E. coli 536 has been considered to reflect an advanced stage of "homing" of these islands, i.e. an ongoing process of stabilisation of such chromosomal regions resulting from the selective inactivation and loss of corresponding genes [5,32]. Consequently, horizontal transfer of such islands, although they can be efficiently excised from the chromosome, could not be detected so far and the mechanism of acquisition remains speculative. This study further supports the important role of mobilisation and conjugation for transfer and dissemination of genomic islands and indicates that loss of mobilisation and transfer genes promotes stabilisation of horizontally acquired genetic elements in the recipient genome.
Conclusions
We provide evidence that a 107-kb chromosomal PAI derivative of UPEC can be mobilised into other E. coli recipient strains. This transfer was dependent on the presence of a helper plasmid and accessory transfer genes. The new host with the mobilisable PAI II536 could also serve as donor passing on this PAI to other recipients. These results underline that in a suitable genetic background dissemination of large genomic regions such as PAIs by conjugal transfer contributes to genome plasticity of E. coli and the evolution of bacterial pathogens. Stabilisation of beneficial genetic information localised on mobile genetic elements can be achieved by selective loss of transfer or mobilisation functions encoded by these elements.
Methods
Bacterial strains and growth conditions
The complete list of the strains and plasmids used in this study is shown in Table 2. Analysis of the complete genome sequence of E. coli strain 536 (O6:K15:H31) revealed the presence of six large typical PAIs [59]. For the mobilisation experiments strain 536-19/1mob was used as donor, and the laboratory strain SY327λpir [60] served as recipient. Two different donor strains were used for re-mobilisation of PAI II536. In strain SY327-77, the mobilised PAI II536 existed extrachromosomally as a circular intermediate. In strain SY327-23, the transferred island was chromosomally inserted at the leuX tRNA gene. Strain 536-21, a PAI I536- and PAI II536 deletion mutant was used as recipient for remobilisation.
Table 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| 536 wt | UPEC wild type strain, SmR | [69] |
| 536-21 | 536, ΔPAI I536 ΔPAI II536, SmR | [2] |
| 536-19/1mob | Donor strain in the mobilisation experiments, pirλatt, mobGP704 inserted in PAI II536, pRP4, SmR, ApR, CmR, TcR, KmR | This study |
| SY327λpir | F-, araD, Δ(lac pro), argE(Am), recA56, RifR, gyrA λpir | [60] |
| SM10λpir | thi1, thr1, leuB6, supE44, tonA21, lacY1, recA::RP4-2-Tc::Mu λpir KmR | [60] |
| SY327-23 | Mobilised PAI II536 is integrated into leuX | This study |
| SY327-77 | Mobilised PAI II536 is present as a CI | This study |
| Plasmids | ||
| pGEM®T-Easy | bla, T/A cloning vector | Promega |
| pGP704 | bla, oriR6K, mobRP4 | [60] |
| pSG704 | cat, oriR6K, mobRP4 | This study |
| pCVD442Tc | bla, tet, oriR6K, mobRP4, sacB | This study |
| pLDR8 neo, int expression vector, Ts | neo, int expression vector, Ts | [62] |
| pLDR9 | bla neo, cloning vector to integrate DNA into attB | [62] |
| pPAI II-CI | bla, positive control for detection of PAI II536-specific CIs | [17] |
Bacteria were grown in Luria broth (LB) or on LB agar at 20°C or 37°C. In the mobilisation experiments, selection of transconjugants was performed on blood agar plates, whereas lactose containing M9 minimal agar plates were used for the remobilisation experiments. If required, antibiotics were used in the following concentrations: ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), kanamycin (100 μg/ml), streptomycin (10 μg/ml), tetracycline (5 μg/ml) and nalidixic acid (4 μg/ml).
Oligonucleotides
The list of oligonucleotides used in this study is compiled in Table 3. Oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany) or Sigma-ARK (Steinheim, Germany).
Table 3.
Primers used in this study
| Designation | Sequence (5` - 3`) | Comment and reference |
|---|---|---|
| Primers for the mobilisation experiments | ||
| Cm_fw_PstI | CTGCAGGCTTGTCAGGGGGGCGGAG | cat cassette in pGP704 |
| Cm_rev_PstI | CTGCAGACCCTGCCCTGAACCGACGA | " |
| pir_fw_SacI | TTTGAGCTCCCGTCAAGTCAGCGTAATGCTC | Introduction of a stop codon into pir |
| pir_revStop_ EcoRI | TTTGAATTCTCAAGCTTCATCATCTTTATCGCCAGA | " |
| paiII_1XhoI | TTTCTCGAGGGGAAGCACGATATGCAGCC | Labelling of PAI II536 by homologous recombination with pSG704 |
| paiII_1SacI | TTGAGCTCGATATTTTTTGGCTGCTTCAGCTTTACG | " |
| paiII_2SacI | TTTGAGCTCGGACAAGAACACAAAATCACTCTACTGA | " |
| paiII_2XhoI | TTTCTCGAGCCCGGCTTCATCGACAATGA | " |
| ATT1 | GAGGTACCAGCGCGGTTTGATC | Confirmation of pir integration into the λ attachment site (E. coli) |
| ATT2 | CAGATGGCGCAATGCCATCTGG | " |
| 17 kD up | CCCGGCTGAACAGACGATT | Screening PCRs for PAI II536 |
| 17 kD in | GCAGCGGAGAGTCATTGTC | " |
| hlyDup | CGCGATAATCCGCTACATC | " |
| hlyDin | GGGTATGGCTGTCACTGCA | " |
| hec_down1 | CACACTGAAAGGCCGCAGC | " |
| hec_down2 | GTACAGCGTGCCGTTCTGC | " |
| dsdXin | GCTGTATCCCGACATCAGC | " |
| dsdAup | GCCACATCATTCTCCCGTA | " |
| ORFAin | CATCCCCTTCACTACAGGA | " |
| Na-Anti_pdo | CAGCGATGCTGGACCGTAT | " |
| PaiII_1fw | CAACCGTAGAAATACGTGCCG | Overlapping PCRs of PAI II536 |
| PaiII_1rev | CCAGTATGAGGCAAACCCTAAAG | " |
| PaiII_2fw | GACAATAGCTGTCCATACGGTG | " |
| PaiII_2rev | CAATGCTGGCCATATCCATCAG | " |
| PaiII_3fw | GACTCACTACTGGGAAGGTC | " |
| PaiII_3rev | GATGGTCATGTGCAGGGAGG | " |
| PaiII_4fw | GTGATGTCACTGGCGGTAATAATC | " |
| PaiII_4rev | GGCGATGACCATGATACGGTA | " |
| PaiII_5fw | CGGAATACTGAACTGCGGATAA | " |
| PaiII_5rev | GCTGGATGCCAATCAATGATCG | " |
| PaiII_6fw | GCATCCATCTCCGTTCACAG | " |
| PaiII_6rev | GCATGGCTGGTTGTTCCTAAAC | " |
| PaiII_7fw | CCTCCTTTGGACTGAAGTTCA | " |
| PaiII_7rev | GCACAGGCGCTCTTTTATTGTTTG | " |
| PaiII_8fw | GGACACATGGCAGGGTCTG | " |
| PaiII_8rev | GGAACACGTCTTCTTGTTGACA | " |
| PaiII_9fw | CAGCGATTTGTCACCACCTG | " |
| PaiII_9rev | GTACTTGACGCGGCGGACA | " |
| PaiII_10fw | GCTTCTGAAAAACGGGTGAAGTC | " |
| PaiII_10rev | CATGGCGCATCATGAAATCATCA | " |
| PaiII_11fw | CTCTCGCGTATATTCAGCAAAAAC | " |
| PaiII_11rev | CGTCACATCGGATAACATTCGG | " |
| PaiII_12fw | CTGATATCTCTTTCAGACTTCAGAAC | " |
| PaiII_12rev | GTCACCTCAAGAACGTCTAACC | " |
| PaiII_13fw | CTGGCACCTATGGATCAGGT | " |
| PaiII_13rev | GTTCAGCAACTGAAGGTTCATACT | " |
| PaiII_14fw | GATGACCATCAGTGTTTCCGCT | " |
| PaiII_14rev | CGGGGATTTAAGTATTGGTCAGTT | " |
| PaiII_15fw | GCAACAATCTGACCTGCAAGCAT | " |
| PaiII_15rev | GGATGATGAGCTTCAGGTTCAG | " |
| PaiII_16fw | TGCCGTACAGCTTGTCATTACC | " |
| PaiII_16rev | CTGGGTACTGCACTTTCCTCA | " |
| PaiII_17fw | CTGACATTGCCACCAGATTTTTGT | " |
| PaiII_17rev | GGTGTAATGCGCTAACCTGTTTC | " |
| PaiII_18fw | CTACAAATGTTCAATATGGTGGGTATATC | " |
| PaiII_18rev | CGCTGTTGCCACTGGATTAATG | " |
| PaiII_19fw | GCCATCCACTACATATCATGCC | " |
| PaiII_19rev | CGACGGGTTTCTATGCTGAG | " |
| PaiII_20fw | CCTCAACTGGAGCAATTTTCTGTC | " |
| PaiII_20rev | GGACTTGGATCACTGAAGCTTTAC | " |
| PaiII_21fw | CCACAAGCTGTTGATTTTGGTACG | " |
| PaiII_21rev | GCGATAGTGGGCAATTTGCTATTG | " |
| PaiII_22fw | CGTAAACGTCCTCCAGAATTTATATC | " |
| PaiII_22rev | GGACGATGGCGATATGTCTG | " |
| PaiII_23fw | GTTCTTCATTTCTACGTTGCTTTGTC | " |
| PaiII_23rev | CCTACCAGAGATAACCCATGG | " |
| PaiII_24fw | CGGTTTCTCCTGAACATAACTTTG | " |
| PaiII_24rev | GGTGAAGTCCGTAACCAGAATG | " |
| PaiII_25fw | GCCTGTTTTTGCTGCTGTTCAC | " |
| PaiII_25rev | GCAGGACTAAAGTTGCAGAGC | " |
| PaiII_26fw | CGCTTTCGCCCCGATTTCTA | " |
| PaiII_26rev | ATGACCGTCGTACTGTGGAC | " |
| PaiII_27fw | GTCAGCCCGCTTTTCTTCTG | " |
| PaiII_27rev | GCTCCGTCGTATACCGATGA | " |
| PaiII_28fw | CGGTCAAGAAAATACGATGAGCC | " |
| PaiII_28rev | GAACGACAGCAAAATCCTCTCC | " |
| PaiII_29fw | CAGCACCTGCGCCGTCA | " |
| PaiII_29rev | GCGATGCCACGGTGAAAACC | " |
| PaiII_30fw | CCGGTATTACTGAATGTCCCG | " |
| PaiII_30rev | GAACATGAAGACAGCACTGACC | " |
| PaiII_31fw | CCTGAAGCAGAACATCATCCAG | " |
| PaiII_31rev | CGGCTGATATCCTGAGACTG | " |
| PaiII_32fw | CTCGCTTCCACGGACGTTG | " |
| PaiII_32rev | GTTGGCAGTGCTGAAAGCAG | " |
| PaiII_33fw | GGTCAGAATGTCCTCAGTGAG | " |
| PaiII_33rev | CAATGAAATTGACAGGAGAGATACC | " |
| PaiII_34fw | CCACTGGCATGATTTTTACCCTG | " |
| PaiII_34rev | CGGGCACATTCCTGCTGTC | " |
| PaiII_35fw | GGATACTGGGCGATGAGCG | " |
| PaiII_35rev | GAGCACGGTGAGCGAAATAG | " |
| PaiII_36fw | CCGCATTAGGTGACTTTACACG | " |
| PaiII_36rev | GACGCCGTACTGACCGATG | " |
| PaiII_37fw | CCAGGTTTGTTATCGAGGTAAGG | " |
| PaiII_37rev | GGCGCTATCGACTACGTCC | " |
| PaiII_38fw | GGCAGTATATCGATTCGGCGA | " |
| PaiII_38rev | GCTTCCCAGCCTGTCACTTC | " |
| PaiII_39fw | GGGCATCTTCAAAGTCAAAGCC | " |
| PaiII_39rev | CGCCCGTCTGTTTTTCAGTTTC | " |
| PaiII_40fw | GGGGCATCAAGGTCGCTATTT | " |
| PaiII_40rev | CAGAACCGCAGCCAGCCAT | " |
| PaiII_41fw | GCTGCGATGCGGATCCAC | " |
| PaiII_41rev | GGTTACCGCAATGGTGAAAGG | " |
| PaiII_42fw | GCTTTTACTGCGCCGACATCA | " |
| PaiII_42rev | CGTTGCACGCGGCTATCTG | " |
| PaiII_43fw | CGATGGATACATTCGGGTTTAGC | " |
| PaiII_43rev | GCAACAGCGACATCATCCTG | " |
| PaiII_44fw | CTCTCTCTTCAGCCAGTCATC | " |
| PaiII_44rev | GCCAAAATCTGATCCCCAGC | " |
| PaiII_45fw | GCAACTACGCCATTGGTTTGTC | " |
| PaiII_45rev | GAAAAACTGGCACGTCATCAACG | " |
| PaiII_46fw | GAAGGCTGCCATTCGGGTATA | " |
| PaiII_46rev | CTGTACTGACTCGTCAGCACT | " |
| PaiII_47fw | CTTGAGATTCAGCAAGGTGGC | " |
| PaiII_47rev | GGAATCCCCTAATGCTGGTG | " |
| PaiII_48fw | GTATAACGGGATGAAAGTGGGG | " |
| PaiII_48rev | GTTGAGAATGTCGGGAATGGTAC | " |
| PaiII_49fw | GGATGTGTATCAGACAAAGCAATG | " |
| PaiII_49rev | TTTCTGGCGAATTTCTTCAGGAAG | " |
| PaiII_50fw | CAGCCATTTTTCCCTCTCCG | " |
| PaiII_50rev | CCTGACCATCTTCCGTCATG | " |
| PaiII_51fw | CTGCTGTTCACTGTGGCATC | " |
| PaiII_51rev | GAGTGGCAACCAGTTGAGACT | " |
| PaiII_52fw | CGCATAATTCCACCACACCTTC | " |
| PaiII_52rev | GGCTGGTCGGTACGCAC | " |
| PaiII_53fw | GGCAGGCATTTCACTGTGTGA | " |
| PaiII_53rev | CGAAGGCCGGACTCGAACA | " |
| K12R (rev) | ATCCTGCGCACCAATCAACAA | E. coli K-12 specific [67] |
| K12L (fw) | TTCCCACGGACATGAAGACTACA | " |
| K12IS-L (fw) | CGCGATGGAAGATGCTCTGTA | " |
| orf4bico | GGAATGAATGCCACTCCATTATTGACAGAAATG | E. coli 536 (K15 capsule)- specific |
| orf5bico | GATCAAACGAGTCAGCTAAATAATCCCCAC | E. coli 536 (K15 capsule)- specific |
| M803b | GCCTGGAGTGTGACAAAGGTTAC | leuX flanking primers [17] |
| M805c | GATGTTCACCAAGGTGGGCGT | " |
| leu 2 | ACCAAGCGCTGCAAAAAGAT | Concatemer PAI II536 |
| Concat 1 | CCGGATTGGATCTATCGCGA | " |
| yjgB1 | ACTTTATCGGCACCCATCG | Downstream of leuX |
| yjgB2 | GCATGAGGTGATTGGGCG | " |
Preparation and manipulation of DNA
Plasmid DNA and chromosomal DNA were isolated according to standard protocols [61]. Recombinant DNA manipulations were carried out with enzymes supplied by GE Healthcare (Freiburg, Germany) or New England Biolabs (Frankfurt am Main, Germany) according to the manufacturer's instructions and standard procedures [61].
For cloning experiments, the Expand Long Template PCR System including a DNA polymerase with proof reading activity was used (Roche Diagnostics, Mannheim, Germany) and PCR screenings were performed with the REDTaq® ReadyMix™ (Sigma, Deisenhofen). PCR products were subcloned into the pGEM®T-Easy cloning vector (Promega, Mannheim, Germany).
Construction of a mobilisable PAI II536 construct
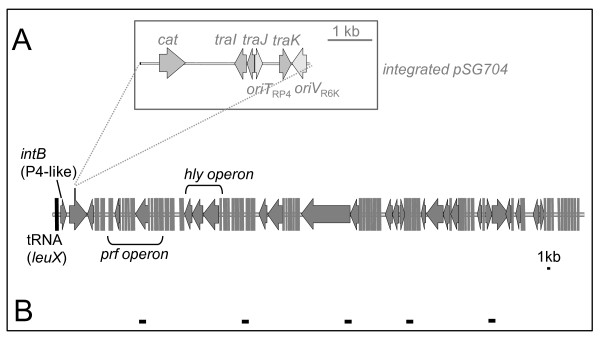
Introduction of a chloramphenicol resistance marker, mobilisation genes, an origin of replication, and an origin of transfer into PAI II536 was accomplished with the help of plasmid pSG704 which is a chloramphenicol-resistant derivative of the conjugative suicide vector pGP704 [60]. Prior to the insertion of the chloramphenicol acetyltransferase (cat) cassette into pGP704, the vector was digested with PstI. Thereby, a 700-bp fragment that encompasses two thirds of the ampicillin resistance gene bla was deleted and replaced by the cat cassette that was amplified from pACYC184 with flanking PstI sites. pSG704 resulted from ligation of two PCR products that correspond to non-coding sequences of PAI II536 located 2,500 bp downstream of leuX (amplified with the primer pairs paiII_1XhoI/paiII_1Sac and paiII_2Sac/paiII_2XhoI) into a SacI restriction site of this plasmid. Homologous recombination between these 4.4-kb pSG704-derived DNA and PAI II536 resulted stable integration of the cat cassette, the mobRP4 region with the traIJH genes, the oriTRP4, and the oriVR6K in PAI II536 (Figures 1A, 3, 4). This replication origin is only functional in the presence of the bacteriophage lambda π-protein.
Figure 3.
Genetic structure of PAI II536. For the transfer experiments, suicide vector pSG704 which carries the chloramphenicol acetyltransferase (cat) gene, an origin of replication and mobility genes (depicted in the enlarged insert) was stably integrated into a non-coding region of this island (A). Complete transfer of PAI II536 into the transconjugants was confirmed by detection of five regions of PAI II536 by PCR (B).
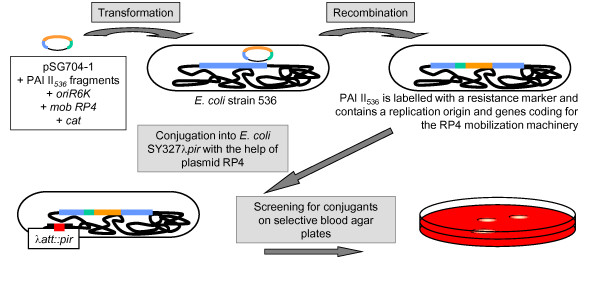
Figure 4.
Schematic presentation of the main steps of the PAI II536 mobilisation experiment.
Integration of the pir gene into the λ attachment site of uropathogenic E. coli strain 536
To stabilise the circular intermediate of PAI II536 after excision from the chromosome and thus enhance its transfer efficiency, we integrated the pir gene coding for the replication factor (π-protein) of the pSG704 oriV into the chromosomal λ attachment site of E. coli strain 536 (Figure 4). For this purpose, the pir gene was amplified from E. coli strain Sm10λpir with the primers pir_fw_SacI and pir_revStop_EcoRI. A resulting 950-bp PCR product comprising a truncated, but functional π-protein was subcloned into pLDR9 [62] using EcoRI and SacI. The resulting plasmid was used for pir integration into the λ attachment site as described before [62]. The correct pir integration was confirmed by PCR (primers ATT1 and ATT2). Expression of the active π-protein was confirmed by episomal propagation of a tetracycline-resistant derivative of the π-dependent suicide plasmid pCVD442 [63] in such strains.
Mobilisation of the labelled PAI II536 by the broad host range conjugative plasmid RP4
Plasmid RP4 was shown to be able to efficiently mobilise the IncQ plasmid RSF1010 which only encodes relaxosomal components [64]. After introduction of the mobRP4 region coding for the TraI, TraJ and TraK proteins, which form the relaxosome at oriT, and the oriVR6K into PAI II536, the RP4 plasmid was conjugated into the corresponding recombinant strain (Figure 4) since the mating pair formation (Mpf) system of a conjugative plasmid is also necessary for a successful PAI or CI transfer [65,66]. The resulting strain was designated E. coli 536-19/1mob. For further experiments, this clone was chosen as donor strain of the tagged PAI II536. The influence of the RP4 plasmid on PAI II536 instability was determined under different growth conditions. The deletion frequency of the island was not affected by the presence of RP4.
Conjugative transfer of PAI II536
Conjugation was carried out on LB agar plates under non-selective conditions. Donor and recipient strains were grown separately until late logarithmic growth phase and were then mixed with each other according to the following procedure. Donor and recipient strains were adjusted to a ratio of 3:1 or 9:1, were centrifuged and resuspended in LB medium to a final volume of 0.1 ml. This mixture was spotted on a dry agar plate and incubated at 20°C and 37°C, respectively. These temperatures were chosen to represent the environmental growth temperature or the human body temperature. The plates were incubated for two days. During the mobilisation experiments (donor: 536, SmR; recipient: SY327, NalR), selection for transconjugants was performed on blood agar plates containing chloramphenicol (20 μg/ml) and nalidixic acid (100 μg/ml). In the remobilisation experiments (donor: PAI II536 containing derivatives of E. coli SY327, NalR, CmR; recipient: 536-21, SmR) selection of clones with the remobilised PAI II536 was performed on M9 lactose medium containing streptomycin (10 μg/ml) and chloramphenicol (20 μg/ml). The frequency of transfer was calculated as follows: number of transconjugants/number of recipients.
Analysis of candidate transconjugants for PAI II536 transfer, deletion, and integration
A thorough analysis of the transconjugants obtained was necessary, because spontaneous nalidixic acid-resistant mutants of strain 536 could occur. Clones that appeared on Cm-Nal blood agar plates were analysed by a four-step PCR process. In the first step, clones were tested with two E. coli K-12 specific primer combinations (K12R/K12L or K12R/K12ISL [67]) and with the strain 536-specific primer combination (orf4bico/orf5bico [68]). The latter primer combination amplifies a 1.5-kb fragment that is specific for the region 2 of the K15 capsule locus. Clones that were positive with the K-12-specific primers and negative with the K15 capsule gene-specific primers, i.e. putative E. coli K-12 recipients, were additionally tested with PAI II536-specific primers in the second step. To confirm the presence of the transferred PAI II536, five primer pairs (17 kDup/17 kDin, hlyDup/hlyDin, hec_down1/hec_down2, dsdXin/dsdAup, ORFAin/Na-Anti_pdo) were used which amplify 800 to 1600-bp fragments of different regions of the PAI II536 (Figure 1B). Those clones that were positive in all five screening PCRs were subjected to a more detailed PCR analysis to verify transfer of the entire PAI II536 and to exclude possible internal deletions of the transferred PAI II536. For this purpose, 51 PCRs were designed to amplify overlapping sections of PAI II536 with an average size of 2 kb, thereby covering the entire PAI II536. In addition, transconjugants were tested for CI formation (PaiII_1rev/PaiII_53fw) and for site-specific integration of PAI II536 into the tRNA gene leuX (M803b/M805c). The latter two primer pairs also allowed the determination of the orientation of the integrated PAI (Figure 2).
Remobilization experiments were carried out with two PAI II536-positive clones of E. coli K-12 as donors that derived from the mobilisation experiments and a derivative of the wild type UPEC strain 536 as recipient. Donor and recipient strains were mixed in a 3 : 1 ratio and incubated at 20°C and 37°C, respectively. Experiments were divided into two sets according to the state of the mobilised PAI. In the first set, the donor strain SY327-77 harbored PAI II536 in the circular form. In the second set, clone SY327-23, harboring the chromosomally integrated PAI II536, served as donor strain. In both cases, strain 536-21, a non-hemolytic derivative of strain 536, which lacks the two islands encoding functional α-hemolysin determinants (PAI I536 and PAI II536) [2], served as the recipient. In the remobilisation experiments, the same PCR-based verification process as described above was carried out with exconjugants that grew on the Cm-lactose-M9 minimal agar plates. In addition, pulsed-field gel electrophoresis (PFGE) analysis of the randomly picked transconjugants was carried out. Genomic DNA for PFGE analysis was prepared and cleaved with NotI or SfiI as described before [2]. Gels were run for 21-24 h with pulse times of 0.5-50 s.
Phenotypic characterisation of transconjugants
PAI II536 comprises a α-hemolysin gene cluster. This determinant was used as a phenotypic marker in this study to verify the presence of PAI II536 in transconjugants after the mobilisation and remobilisation experiments. Therefore, transconjugants were screened post-experimentally on blood agar plates to analyse the hemolytic activity. UPEC strain 536 served as a positive control, while strains SY327 and 536-21 served as negative controls.
Statistical analysis
Statistical analysis of the conjugation rate was performed by the Mann-Whitney U test. The ratio/distribution of integrated, cointegrated and partial transconjugant clones at 20°C and 37°C was compared by the chi-square test. The difference was considered significant if p < 0.05.
Authors' contributions
GS and UD designed research together with BH, LE and JH. GS constructed the mobilisable PAI II536 variant and performed the mobilisation and transconjugation experiments assisted by VS. BM and BH provided bacterial strains and constructs and supported the construction of the mobilisable PAI II536 variant, suitable recipient strains as well as mobilisation experiments. GS and UD wrote the manuscript assisted by BM, LE and JH. All authors have read and approved the final manuscript.
Contributor Information
György Schneider, Email: gyorgy.schneider@aok.pte.hu.
Ulrich Dobrindt, Email: dobrindt@uni-muenster.de.
Barbara Middendorf, Email: barbara.middendorf@ukmuenster.de.
Bianca Hochhut, Email: bianca.hochhut@mail.uni-wuerzburg.de.
Valéria Szijártó, Email: vali.d@hotmail.com.
Levente Emődy, Email: levente.emody@aok.pte.hu.
Jörg Hacker, Email: joerg.hacker@leopoldina.org.
Acknowledgements and Funding
This work was carried out within the European Virtual Institute for Functional Genomics of Bacterial Pathogens (CEE LSHB-CT-2005-512061) and the ERA-NET PathoGenoMics I consortium "Deciphering the intersection of commensal and extraintestinal pathogenic E. coli" (Federal Ministry of Education and Research (BMBF) grant no. 0313937A) and the Hungarian Research Foundation (OTKA 62092 and 78915). UD was also supported by the German Research Foundation (DO 789/4-1). The excellent technical assistance of K. Lotzl (Pécs) and B. Plaschke (Würzburg) is appreciated. We acknowledge support by the Open Access Publication Fund of the University of Münster.
References
- Hacker J, Knapp S, Goebel W. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J Bacteriol. 1983;154(3):1145–1152. doi: 10.1128/jb.154.3.1145-1152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschäpe H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62(2):606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8(11):1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17(1):14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2(5):414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23(6):1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2(5):376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Dobrindt U, Hacker J, Hasnain SE. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat Rev Microbiol. 2008;6(5):387–394. doi: 10.1038/nrmicro1889. [DOI] [PubMed] [Google Scholar]
- Dobrindt U. (Patho-)Genomics of Escherichia coli. Int J Med Microbiol. 2005;295(6-7):357–371. doi: 10.1016/j.ijmm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65(11):4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumer L, Jores J, Kirsch P, Cavignac Y, Zehmke K, Wieler LH. Dissemination of pheU- and pheV-located genomic islands among enteropathogenic (EPEC) and enterohemorrhagic (EHEC) E. coli and their possible role in the horizontal transfer of the locus of enterocyte effacement (LEE) Int J Med Microbiol. 2003;292(7-8):463–475. doi: 10.1078/1438-4221-00229. [DOI] [PubMed] [Google Scholar]
- Tauschek M, Strugnell RA, Robins-Browne RM. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol Microbiol. 2002;44(6):1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, Weinert K, Tenaillon O, Matic I, Denamur E. Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog. 2009;5(1):e1000257. doi: 10.1371/journal.ppat.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Middendorf B, Tarr PI, Zhang W, Prager R, Aldick T, Dobrindt U, Karch H, Mellmann A. Chromosomal instability in enterohaemorrhagic Escherichia coli O157:H7: impact on adherence, tellurite resistance and colony phenotype. Mol Microbiol. 2011;79(4):1024–1044. doi: 10.1111/j.1365-2958.2010.07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorf B, Blum-Oehler G, Dobrindt U, Mühldorfer I, Salge S, Hacker J. The pathogenicity islands (PAIs) of the uropathogenic Escherichia coli strain 536: island probing of PAI II536. J Infect Dis. 2001;183(Suppl 1):S17–20. doi: 10.1086/318843. [DOI] [PubMed] [Google Scholar]
- Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun. 1998;66(9):4011–4017. doi: 10.1128/iai.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol. 2004;186(10):3086–3096. doi: 10.1128/JB.186.10.3086-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol. 2006;61(3):584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J Bacteriol. 2001;183(19):5535–5543. doi: 10.1128/JB.183.19.5535-5543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea YA, Boyd EF. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol Lett. 2002;214(2):153–157. doi: 10.1111/j.1574-6968.2002.tb11339.x. [DOI] [PubMed] [Google Scholar]
- Lesic B, Bach S, Ghigo JM, Dobrindt U, Hacker J, Carniel E. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol Microbiol. 2004;52(5):1337–1348. doi: 10.1111/j.1365-2958.2004.04073.x. [DOI] [PubMed] [Google Scholar]
- Maiques E, Ubeda C, Tormo MA, Ferrer MD, Lasa I, Novick RP, Penades JR. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. J Bacteriol. 2007;189(15):5608–5616. doi: 10.1128/JB.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Barry P, Penades JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc Natl Acad Sci USA. 2007;104(36):14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, Penades JR. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol. 2003;49(1):193–210. doi: 10.1046/j.1365-2958.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323(5910):139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29(2):527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- Boyd EF, Davis BM, Hochhut B. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 2001;9(3):137–144. doi: 10.1016/S0966-842X(01)01960-6. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46(3):601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- Doublet B, Boyd D, Mulvey MR, Cloeckaert A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol. 2005;55(6):1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32(1):99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33(2):376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Dufke S, Sorsa J, Heesemann J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol Microbiol. 2004;51(3):837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guedon G. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol. 2009;191(8):2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn PS, Baghdayan AS, Dolan GT, Shankar N. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Mol Microbiol. 2007;63(2):530–544. doi: 10.1111/j.1365-2958.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103(52):19830–19835. doi: 10.1073/pnas.0606810104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MQ, Chen J, Lory S. The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J Bacteriol. 2010;192(13):3249–3258. doi: 10.1128/JB.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AA. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J Bacteriol. 2004;186(18):6077–6092. doi: 10.1128/JB.186.18.6077-6092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger S, Trummler CS, Fauster A, Lang S, Sprenger H, Gorkiewicz G, Zechner EL. Interbacterial macromolecular transfer by the Campylobacter fetus subsp. venerealis type IV secretion system. J Bacteriol. 2011;193(3):744–758. doi: 10.1128/JB.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche D, Flechard M, Lallier N, Reperant M, Bree A, Pascal G, Schouler C, Germon P. ICEEc2, a new integrative and conjugative element belonging to the pKLC102/PAGI-2 family, identified in Escherichia coli strain BEN374. J Bacteriol. 2010;192(19):5026–5036. doi: 10.1128/JB.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord A, Ceccarelli D, Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol. 2010;78(3):576–588. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- Laverde Gomez JA, van Schaik W, Freitas AR, Coque TM, Weaver KE, Francia MV, Witte W, Werner G. A multiresistance megaplasmid pLG1 bearing a hylEfm genomic island in hospital Enterococcus faecium isolates. Int J Med Microbiol. 2011;301(2):165–175. doi: 10.1016/j.ijmm.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Manson JM, Hancock LE, Gilmore MS. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci USA. 2010;107(27):12269–12274. doi: 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikosa M, Sochacka-Pietal M, Baj J, Bartosik D. Identification of a transposable genomic island of Paracoccus pantotrophus DSM 11072 by its transposition to a novel entrapment vector pMMB2. Microbiology. 2006;152(Pt 4):1063–1073. doi: 10.1099/mic.0.28603-0. [DOI] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W. et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77(11):4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254(4):400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- Bach S, Buchrieser C, Prentice M, Guiyoule A, Msadek T, Carniel E. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect Immun. 1999;67(10):5091–5099. doi: 10.1128/iai.67.10.5091-5099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Alokam S, Kothapalli S, Porwollik S, Proctor E, Choy C, McClelland M, Liu SL, Sanderson KE. Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J Bacteriol. 2004;186(10):3214–3223. doi: 10.1128/JB.186.10.3214-3223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanna C, Wang J, Zhang D, Xu Z, Ali A, Hou YM, Karaolis DK. The Vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J Bacteriol. 2003;185(23):6893–6901. doi: 10.1128/JB.185.23.6893-6901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonenka U, Nölting C, Heesemann J, Rakin A. Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid. Mol Microbiol. 2005;57(3):727–734. doi: 10.1111/j.1365-2958.2005.04722.x. [DOI] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155(5):376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang GR, Shoemaker NB, Salyers AA. Production of two proteins encoded by the Bacteroides mobilizable transposon NBU1 correlates with time-dependent accumulation of the excised NBU1 circular form. J Bacteriol. 2001;183(21):6335–6343. doi: 10.1128/JB.183.21.6335-6343.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol. 2006;62(3):723–734. doi: 10.1111/j.1365-2958.2006.05396.x. [DOI] [PubMed] [Google Scholar]
- te Poele EM, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek. 2008;94(1):127–143. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Babic A, Grossman AD. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 2010;75(2):268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Loper JE, Paulsen IT, Thomashow LS. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol. 2009;9:8. doi: 10.1186/1471-2180-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30(5):965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Ölschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emődy L. et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. 2006;103(34):12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Diederich L, Rasmussen LJ, Messer W. New cloning vectors for integration in the lambda attachment site attB of the Escherichia coli chromosome. Plasmid. 1992;28(1):14–24. doi: 10.1016/0147-619X(92)90032-6. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59(12):4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Lurz R, Grahn AM, Bamford DH, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177(16):4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste JP, Pansegrau W, Ziegelin G, Kröger M, Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci USA. 1989;86(6):1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- Kuhnert P, Nicolet J, Frey J. Rapid and accurate identification of Escherichia coli K-12 strains. Appl Environ Microbiol. 1995;61(11):4135–4139. doi: 10.1128/aem.61.11.4135-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Dobrindt U, Brüggemann H, Nagy G, Janke B, Blum-Oehler G, Buchrieser C, Gottschalk G, Emődy L, Hacker J. The pathogenicity island-associated K15 capsule determinant exhibits a novel genetic structure and correlates with virulence in uropathogenic Escherichia coli strain 536. Infect Immun. 2004;72(10):5993–6001. doi: 10.1128/IAI.72.10.5993-6001.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger H, Hacker J, Juarez A, Hughes C, Goebel W. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J Bacteriol. 1982;152(3):1241–1247. doi: 10.1128/jb.152.3.1241-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]