Abstract
It is widely accepted that at least 10% of all mutations causing human inherited disease disrupt splice-site consensus sequences. In contrast to splice-site mutations, the role of auxiliary cis-acting elements such as exonic splicing enhancers (ESE) and exonic splicing silencers (ESS) in human inherited disease is still poorly understood. Here we use a top-down approach to determine rates of loss or gain of known human exonic splicing regulatory (ESR) sequences associated with either disease-causing mutations or putatively neutral single nucleotide polymorphisms (SNPs). We observe significant enrichment toward loss of ESEs and gain of ESSs among inherited disease-causing variants relative to neutral polymorphisms, indicating that exon skipping may play a prominent role in aberrant gene regulation. Both computational and biochemical approaches underscore the relevance of exonic splicing enhancer loss and silencer gain in inherited disease. Additionally, we provide direct evidence that both SRp20 (SRSF3) and possibly PTB (PTBP1) are involved in the function of a splicing silencer that is created de novo by a total of 83 different inherited disease mutations in 67 different disease genes. Taken together, we find that ∼25% (7154/27,681) of known mis-sense and nonsense disease-causing mutations alter functional splicing signals within exons, suggesting a much more widespread role for aberrant mRNA processing in causing human inherited disease than has hitherto been appreciated.
The sequences of mammalian exons perform at least two overlapping roles in gene expression. First, exons are encoded with the primary sequence determinants of proteins. This information is decoded by the ribosome and translated into functional polypeptides. Secondly, it has been understood for some time that exonic sequences also contribute to pre-mRNA splicing through both sequence and local structure (Watakabe et al. 1993; Wang and Cooper 2007; Warf and Berglund 2010). These latter observations are not surprising given the organization of mammalian genes, which typically contain small exons (∼140 bp) flanked by thousands of base pairs of intronic DNA sequence. The large size of many mammalian genes and the apparent degeneracy of mammalian splice sites marking the 5′ and 3′ termini of introns are also suggestive of a requirement for auxiliary cis-acting elements in facilitating exon recognition (Keren et al. 2010).
Exonic sequences contain a staggering array of cis-acting elements that direct the activation or repression of splicing (Liu et al. 1998; Fairbrother et al. 2002; Cartegni et al. 2003; Wang et al. 2004; Yeo et al. 2004; Shi et al. 2005; Goren et al. 2006). Typically, these functional elements are classified as either exonic splicing enhancers (ESE) or exonic splicing silencers (ESS) based on their ability to stimulate or inhibit splicing, respectively. ESE and ESS elements, acting in concert with their cognate trans-acting RNA-binding proteins, represent important components in a splicing code that specifies how, where, and when mRNAs are assembled from their precursors (Barash et al. 2010). Two of the major players in establishing exon identity are the serine- and arginine-rich proteins (SR proteins) and the heterogeneous nuclear ribonucleoproteins (hnRNPs) (for review, see Wang and Burge 2008). SR proteins promote the initial stages of spliceosome assembly by binding to ESEs and recruiting basal splicing factors to adjacent splice sites or by antagonizing the effects of ESS elements (Kohtz et al. 1994; Graveley et al. 2001; Zhu et al. 2001). In contrast, hnRNPs mediate the repressive effects of silencers and can alter recruitment of the core splicing machinery (Wang et al. 2006; Yu et al. 2008). The interactions between silencers, enhancers, and their cognate binding proteins play a critical role in the fidelity and regulation of pre-mRNA splicing (Eperon et al. 2000; Zhu et al. 2001).
At least 10% of all mutations identified as causing human inherited disease are known to alter consensus 5′- or 3′-splice sites, thereby inducing aberrant pre-mRNA splicing (Krawczak et al. 2007). Nonetheless, the role(s) played by pre-mRNA splicing in human genetic disease remain enigmatic (Cooper et al. 2009). Although the mechanistic consequences of mutations on splice sites are fairly easy to interpret, evaluating precisely how inherited disease-causing mutations influence the loss or gain of ESE/ESS motifs is much more challenging (Cartegni and Krainer 2002; Pagani and Baralle 2004). This is due in part to the considerable functional overlap between protein-coding sequences and the cis-acting elements involved in splicing regulation. Hence, many mis-sense and nonsense mutations that alter pre-mRNA splicing may be incorrectly assumed to have an impact solely on protein structure–function relationships as a consequence of amino acid substitution or protein truncation, rather than on splicing changes per se (Liu et al. 2001; Pagani and Baralle 2004). It is also possible that the impact of a disease allele may be due to the combination of an aberrant splicing event and the presence of a normal-length mutation-bearing transcript. Such multifunctional sites within coding regions have recently been identified by the intragenic mapping of common genetic variants known as single nucleotide polymorphisms (SNPs) (Majewski and Ott 2002; Fairbrother et al. 2004; Goren et al. 2008). As a consequence of purifying selection, SNPs appear somewhat depleted and synonymous codon bias restricted (GAA vs. GAG), revealing a silhouette of the “splicing code” that appears position-restricted relative to the edges of exons (Majewski and Ott 2002; Fairbrother et al. 2004; Chamary et al. 2006). Here we have investigated the relationship between coding sequence mutations and splicing regulation using a novel combination of bioinformatic and biochemical techniques.
Results
Disease-causing mutations overlap with the splicing code
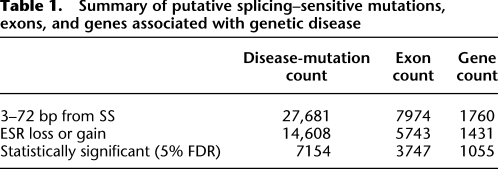
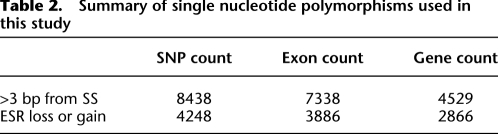
We extracted 27,681 exonic disease-causing (mis-sense and nonsense) (see Table 1) mutations from the Human Gene Mutation Database (HGMD; http://www.hgmd.org), a proprietary, hand-curated database requiring one or more pieces of causal evidence for inclusion (e.g., absence from normal controls, cosegregation of lesion and phenotype through pedigree, independent occurrence in an unrelated patient, etc.) (Stenson et al. 2008). For common genetic variants, we extracted 8601 exonic single nucleotide polymorphisms from the 1000 Genomes Project (http://www.1000genomes.org) (see Table 2; Durbin et al. 2010). These exonic SNPs were selected for neutrality by filtering average heterozygosity to 30%–50%, corresponding to a Hardy-Weinberg minor allele frequency of at least ∼0.18. In addition, we determined the ancestral allele (biallelic directionality) by comparison to the chimpanzee (Pan troglodytes) genome (Fairbrother et al. 2004; Karolchik et al. 2008).
Table 1.
Summary of putative splicing–sensitive mutations, exons, and genes associated with genetic disease
Table 2.
Summary of single nucleotide polymorphisms used in this study
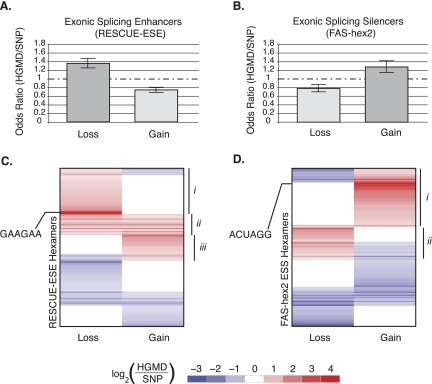
We used a set of 238 hexameric sequences corresponding to the RESCUE-ESE data set and 176 hexameric sequences corresponding to the FAS-hex2 ESS data set (Fairbrother et al. 2002; Wang et al. 2004). Each set of hexanucleotides was experimentally validated to enhance or silence splicing of an alternative exon in a minigene context. We used the directionality of the substitutions, based on ancestral > variant for SNPs and wild-type > disease for HGMD mutations to calculated odds ratios (OR), expressing the relative likelihoods that either disease-causing mutations or the putatively neutral polymorphisms are associated with the loss or gain of ESEs or ESSs (see Methods). Whereas disruption of ESEs was found to be strongly associated with the mutations from the HGMD data set by comparison with neutral SNPs, there was substantially less evidence for the gain of ESEs in the disease mutation data set (Fig. 1A). In contrast, a strong association was noted between disease-causing mutations and the creation of ESS motifs (Fig. 1B). Taken together, these data suggest that exon skipping may play a key role in human inherited disease not only via the loss of exonic splicing enhancers but also via the gain of exonic splicing silencers.
Figure 1.
Patterns of exonic splicing regulator loss or gain among pathological mutations (HGMD) as compared to putatively neutral SNPs. (A,B) Bar height corresponds to the odds ratio (OR) of HGMD/SNPs for the loss or gain of enhancers and silencers, respectively. Each error bar represents a two-tailed 95% confidence interval for the bar height (see Methods). Directionality was expressed in the form of the ancestral state > variant for the SNPs and healthy > disease for the HGMD mutations. (A) Hexamers corresponding to exonic splicing enhancers were obtained from the RESCUE-ESE database. Each hexamer was scored for the loss or gain (de novo creation) of an ESE by the inherited disease-causing mutations (relative to the wild-type allele) or putatively neutral SNPs (relative to the ancestral allele). (B) Hexamers corresponding to exonic splicing silencers were obtained from the FAS-hex2 database and scored for loss or gain as described in A. (C,D) Principal component analysis (PCA) of normalized ratios of HGMD versus SNP substitution for loss or gain of ESE and ESS hexamers, respectively. Each row corresponds to a single ESE or ESS hexamer, whereas each column represents loss or gain of the hexamer by a genomic variant. Any hexamers that were not significant at the 5% level were omitted from the heat map. Each box depicts the log ratio for the counts of HGMD/SNP causing loss or gain of a specific hexamer. A positive log ratio in red corresponds to a hexamer in a certain context (column) that is significantly enriched in inherited disease. Alternatively, a blue value represents a hexamer that is polymorphic across human populations. White boxes correspond to non-significant P-values given a false discovery rate (FDR) of 5%. (C) Hexamer clusters corresponding to ESE-loss (region i), ESE-loss and ESE-gain (region ii), and ESE-gain (region iii). Hexamer clusters corresponding to ESS-gain (region i) and ESS-loss (region ii). The loss/gain of SRSF1-like binding sites is indicated by GAAGAA in C, whereas the ACUAGG hexamer is indicated in D.
Disease-associated alterations of the splicing code
To determine if specific cis-acting elements are more susceptible to disease-causing mutations than others relative to a background level ascertained by reference to putatively neutral SNPs, we calculated the binomial enrichment P-value for loss or gain of individual hexamer sequences (see Methods). We visualized the distribution of genomic variants across individual hexamers from the ESE and ESS data sets using Principal Component Analysis for optimal leaf ordering (Rajaram and Oono 2010). Figure 1, C and D, depicts log2 ratios of HGMD mutations versus SNPs for loss or gain of individual ESE and ESS hexamers with a binomial P-value significant at a 5% false discovery rate (FDR). Of the 238 ESE hexamers considered in this analysis, 106 showed no significant difference for either loss or gain by inherited disease-causing mutations relative to SNPs given the 5% FDR. Similarly, 67 out of 176 ESS hexamers were not significantly different between the HGMD and SNP data sets. For both ESEs and ESSs, the heat maps clearly demonstrate that HGMD mutations are not uniformly distributed across all hexamers but, rather, are enriched in select subsets corresponding to losses or gains. For ESEs, we not only observe clusters of hexamers that are both exclusively either ablated or created by disease-causing mutations (Fig. 1C, regions i and iii) but also a small subset of hexamers that are subject to a significant degree of both loss and gain by disease-causing mutations (Fig. 1C, region ii). In contrast to the ESEs, a much larger number of hexamers are significantly enriched for disease-causing mutations that create ESSs rather than abolish ESSs (Fig. 1D, cf. regions i and ii). An expanded version of Figure 1, C and D, containing the hexamer sequences, is presented in Supplemental Figures 1 and 2. We also examined loss or gain of each hexamer sequence in several different contexts including their proximity to the nearest 5′ or 3′ splice sites and their presence within alternative or constitutive exons (see Methods). Supplemental Figures 3–6 show that, although the general observations described in Figure 1, C and D hold true, there is inconclusive evidence for a bias of hexamer loss or gain relative to either splice site or between constitutive or alternative exons.
ESEs ablated by disease-causing mutations share hallmarks of functional splicing enhancers
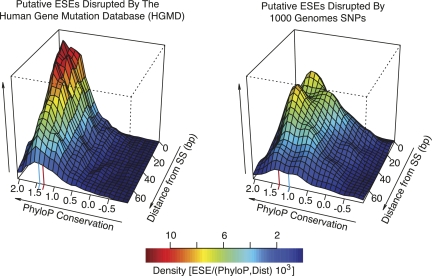
Since evolutionary conservation usually implies functionality (Boffelli et al. 2003; Margulies et al. 2003; Siepel et al. 2005), we opted to determine whether there was a difference in average evolutionary conservation between those ESE hexamers lost as a consequence of disease-associated mutation and those lost as a result of the introduction of a neutral SNP allele. Because ESEs are more abundant in the vicinity of splice sites and the activity of splicing enhancers decreases with increasing distance from splice sites (Graveley and Maniatis 1998; Yeo and Burge 2004; Parmley et al. 2006), we evaluated average phyloP scores across alignments of 46 placental mammals (Pollard et al. 2009) for HGMD- or SNP-disrupted ESE hexamers relative to their positions within exons. Figure 2 shows the bivariate density distributions of ESE hexamers lost by disease-causing mutations and neutral SNPs (Fig. 2, left and right panels, respectively). In both density plots, the axes displaying phyloP scores are marked with red lines corresponding to the statistical threshold for evolutionary conservation (phyloP score >1.3, α = 0.05) and blue lines corresponding to the median phyloP scores for ESE hexamers. The median phyloP score corresponding to the distribution of putative ESE hexamers ablated by disease-causing mutations is 1.42 (Fig. 2, left panel, blue line), easily exceeding the statistical threshold for evolutionary conservation. In contrast, ESE hexamers abolished by putatively neutral SNPs have much lower distributed average phyloP scores, such that the median (1.02) is well below the statistical threshold to reject the null hypothesis of neutrally evolving sequence. Although the median distance of ESEs from splice sites is not significantly different for those disrupted by SNPs or HGMD mutations, the distribution of phyloP scores for ESEs lost by disease-causing mutations shifts toward higher values approaching splice sites. In contrast, the distribution for ESEs disrupted by neutral SNPs is visibly shifted toward lower values near the edges of exons. Overall, the density plots in Figure 2 indicate that ESEs targeted by disease-causing mutations exhibit a bias not only toward higher conservation values, but also with respect to a location toward the edges of exons as compared to those ESEs targeted by neutral SNPs.
Figure 2.
Conservation of exonic splicing enhancers ablated by genomic variants. The two-dimensional density distributions (relative values given in color scale) of ESEs containing associated average phyloP (Pollard et al. 2009) scores and distances to the nearest splice site (3–72 bp). The density distributions for ESEs targeted for loss by inherited disease-causing (HGMD) mutations (left panel) or neutral SNPs (right panel). In each panel the red line designates a phyloP score corresponding to a P-value of 0.05. The blue line designates the median phyloP score of each density distribution.
Disease-causing mutations often affect conserved regions of proteins (Kumar et al. 2009). To determine if the conservation levels observed for ESE hexamers ablated by disease-causing mutations could result as a byproduct of this bias, we sampled random hexamers that did not cause loss/gain of any ESR from HGMD mutation- or SNP-containing exons as well as another subset that encompassed HGMD mutations (Supplemental Fig. 7). As expected, both random hexamers sampled from HGMD exons and those containing HGMD mutations displayed lower distributed evolutionary conservation values than the ESEs lost by HGMD mutations (median average phyloP scores of 1.32 and 1.36 compared to 1.45, respectively; Welch test two-tailed P-value < 1.79 × 10−39 and 3.81 × 10−15). Furthermore, the phyloP scores for ESEs targeted by neutral SNPs were distributed lower overall than the set of random hexamers sampled from SNP-targeted exons (median average phyloP scores of 1.09 and 1.15, respectively; Welch test two-tailed P-value < 0.039).
Functional validation of a splicing silencer mutationally linked to 67 different disease genes
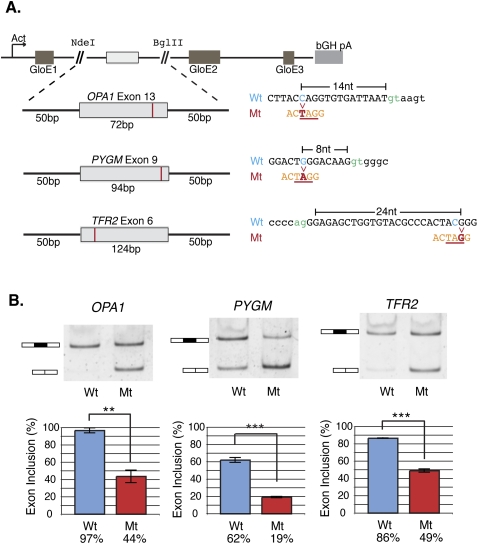
To test the hypothesis that those exonic splicing silencers that harbor a preponderance of disease alleles could represent functionally repressive elements, we opted to validate the activity of one of the most significant hexamers identified by our comparison of disease-causing and neutral polymorphisms within ESSs (designated in Fig. 1D, ACUAGG, binomial P-value < 2.2 × 10−16; Supplemental Table 4). This specific hexamer appears to have been created by a total of 83 different disease-causing mutations in 67 different genes. We searched this list of disease-causing mutations for sequences that were amenable for cloning into splicing reporter constructs and were present in exons of near average size and splice-site strength (Yeo and Burge 2004). Of the three different disease-causing mutations we selected—OPA1, PYGM, and TFR2—there was no a priori evidence for any effect on splicing from in vitro analysis (Bruno et al. 1999; Camaschella et al. 2000; Schimpf et al. 2008). However, aberrant splicing of OPA1, PYGM, and TFR2 is observed in patients carrying coding and non-coding mutations at other positions in these genes (Schimpf et al. 2006; Biasiotto et al. 2008; Nogales-Gadea et al. 2008).
We created matched pairs of beta-hemoglobin-based (HBB1) splicing reporter gene constructs containing the wild-type or mutant exon plus 50 bp of adjacent intron sequence (Rothrock et al. 2003). To investigate the effects of the ACUAGG silencer on splicing of the reporter genes, HeLa cells were transiently transfected with both wild-type or mutant constructs. Because inclusion of all three of the test exons is predicted to induce nonsense-mediated decay (NMD) by inducing an in-frame premature termination codon (PTC) (see Supplemental Fig. 8), we assayed splicing in the presence of the translation inhibitor emetine dihydrochloride, a potent inhibitor of NMD in vivo (Noensie and Dietz 2001). After RNA isolation and conversion to cDNA, each sample was tested for reporter RNA splicing efficiency. Inhibition of NMD was confirmed by assaying the splicing of the SRSF6 pre-mRNA, an endogenous PTC-containing gene known to undergo NMD (Lareau et al. 2007; Ni et al. 2007). The presence of the SRSF6 poison exon-containing mRNA shows that NMD was, indeed, inhibited in the emetine-positive samples (Supplemental Fig. 9). As shown in Figure 3B, introduction of the ACUAGG hexamer resulted in a remarkable degree of exon skipping in the OPA1, PYGM, and TFR2 reporters. Quantification of amplicons using an Agilent 2100 Bioanalyzer demonstrated that the ACUAGG silencer significantly decreased inclusion of the OPA1, PYGM, and TFR2 test exons from 97% to 44% (P-value < 2.39 × 10−3), 62%–19% (P-value < 6.83 × 10−4), and 86%–49% (P-value < 7.62 × 10−4), respectively (Fig. 3B). These data suggest that it is possible to predict splicing-relevant mutations based on the statistical enrichment of hexamers in disease-associated mutation data sets.
Figure 3.
Validation of mutations creating the enriched silencer ACUAGG using the beta-globin splicing reporter. (A) Splicing reporter constructs created from matched pairs of wild-type (Wt) or mutant (Mt) alleles that give rise to a gain of the ACUAGG silencer in constitutive exons in three different disease genes: OPA1, PYGM, and TFR2. GloE1, GloE2, and GloE3 designate exons 1–3 of beta-globin. The polyadenylation signal from the bovine growth hormone 1 gene is indicated by bGH pA. (Blue) Wild-type allele; (red) the mutant; (orange) the silencer sequence created by the mutation. (B) HeLa cells were transiently transfected in triplicate with both wild-type (Wt) and mutant (Mt) alleles. Twenty-four hours after transfection, cells were treated with emetine to inhibit NMD, RNA was harvested, and the splicing efficiency was determined by RT-PCR and visualized using 6% non-denaturing (29:1) polyacrylamide gel electrophoresis (PAGE). The graphs depict mean exon inclusion quantified using an Agilent 2100 Bioanalyzer with standard error bars (see Methods). Statistical hypothesis testing on means was executed using a Welch t-test for normal data with unequal sample size and variance using α-values of (*) 0.05, (**) 0.01, and (***) 0.001.
Identification of trans-acting splicing silencers
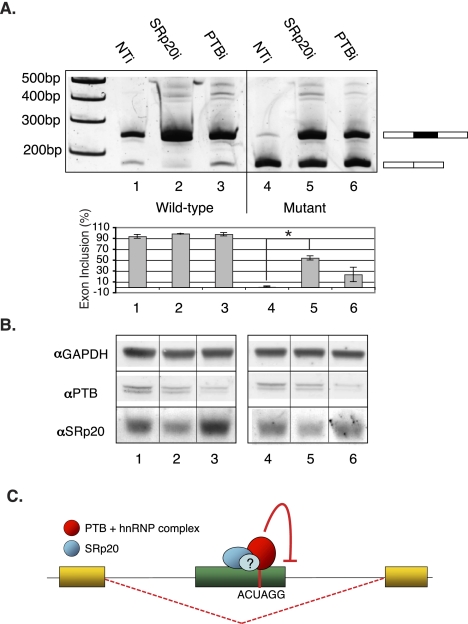
The data presented in Figure 4 suggest that the ACUAGG motif functions as a strong splicing silencer. Splicing silencers have been shown to interact with trans-acting factors such as hnRNPs and to alter the kinetics of the non-rate-limiting steps of spliceosome assembly when two 5′-splice sites are in competition (Zhu et al. 2001; Yu et al. 2008). Given that we did not observe activation of cryptic 5′-splice sites in the mutant OPA1 construct, we searched for potential trans-acting factors by RNA affinity chromatography using HeLa nuclear extracts. RNA-binding proteins captured by the wild-type and mutant RNA ligands at low and high stringency were identified by Multidimensional Protein Identification Technology (MudPIT) (Supplemental Fig. 10; Supplemental Tables 5–7). At both high and low concentrations of KCl, peptides corresponding to SRp20 (SRSF3), PTB (PTBP1), hnRNP D (HNRNPD), and hnRNP L (HNRNPL) were present on the mutant RNA ligand (Supplemental Fig. 10A,B; Supplemental Tables 5, 6), suggesting that these proteins may play a role in mediating the silencing activity of the ACUAGG hexamer.
Figure 4.
Identification of trans-acting factors implicated in skipping of the ACUAGG-containing OPA1 allele. (A) RT-PCR analysis of OPA1 splicing reporters from HeLa cells cotransfected with non-targeting siRNA (NTi), SRSF3 siRNA (SRp20i), PTBP1 siRNA (PTBi). Lanes 1–3 and 4–6, wild-type and mutant reporters, respectively. Statistical hypothesis testing on means was executed using a Welch t-test for normal data with unequal sample size and variance using α-values of (*) 0.05, (**) 0.01, and (***) 0.001. (B) Western blot showing relative depletion of SRp20 and PTB as compared to the GAPDH loading control. (C) Model for aberrant splicing by “ACUAGG” ESS. A point mutation creating the sequence ACUAGG results in recruitment of a silencer complex that may contain SRp20 and members of the hnRNP protein family, either directly or indirectly bound to the RNA sequence. The complex is involved in deterring inclusion of the mutant exon via mechanism(s) that still remain to be determined.
To test the role of SRp20 and PTB in splicing silencing, we cotransfected HeLa cells with either the wild-type or mutant OPA1 splicing reporter and siRNA targeting SRp20, PTB, or a non-targeting duplex. In cells transfected with non-targeting control siRNA, the mutant OPA1 reporter was inefficiently spliced relative to the wild-type reporter (Fig. 4A, cf. lanes 1 and 4). In contrast, depletion of SRp20 and, to a lesser extent, PTB partially rescued inclusion of the mutant OPA1 exon (Fig. 4A, cf. lane 4 with lanes 5 and 6). Quantification of the RT-PCR amplicons from duplicate experiments revealed that depletion of SRp20 and PTB restored inclusion of the mutant exon to ∼50% and ∼25% of the wild-type levels, respectively. Analysis of the exon inclusion ratios for the SRp20 and PTB depletion revealed that only knockdown of SRp20 resulted in statistically significant changes relative to the control (Fig. 4B). Depletion of both SRp20 and PTB was confirmed by fluorescent Western blot analysis of nuclear extracts prepared from transfected cells (Fig. 4B). Quantification of the Western blots revealed approximately twofold and 2.75-fold depletions of SRp20 and PTB, respectively, relative to cells transfected with non-targeting control duplex. Taken together, these data implicate SRp20 and PTB in both the recognition and function of the ACUAGG exonic splicing silencer motif. We did not test the role(s) of hnRNP D or hnRNP L in the function of this silencer motif.
Potential nonsense sequences are enriched in ESR hexamers
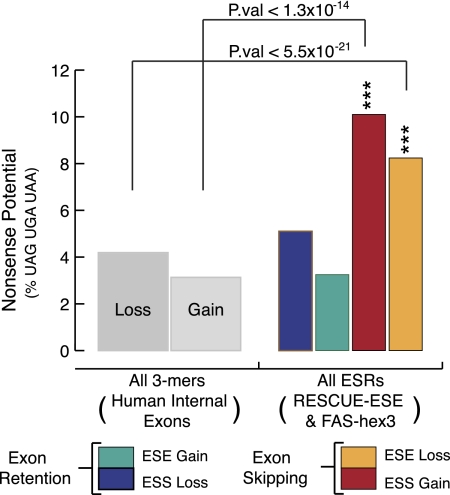
Nonsense-associated altered splicing (NAS) describes the phenomenon whereby exons encoding premature stop codons tend to be excluded from the mature RNA transcript during pre-mRNA splicing in the nucleus (Dietz et al. 1993; Li et al. 2002; Wachtel et al. 2004). Although the primary mechanism of NAS is still unknown, several different models have been proposed. These include a nuclear scanning model that invokes the action of a frame-sensitive mechanism in pre-mRNA splicing (Wang et al. 2002). Alternatively, NAS may be the direct result of ESE disruption as a means to abolish exon recognition (Shiga et al. 1997; Liu et al. 2001). In order for nonsense mutations to be specifically associated with the loss/gain of ESR sequences, there must be a sequence bias in the ESRs themselves. To investigate this hypothesis for ESR loss, we simulated mutations based on the transition/transversion rates observed for the 14,771 exonic HGMD mutations located near the edges of exons (Supplemental Fig. 11). For the ESR gains, we simply evaluated the proportion of nonsense 3-mers (UAG, UAA, UGA) compared to all of the 3-mers within the corresponding hexamers. As a control for the experiment, we used the same algorithm to evaluate the “loss” or “gain” of all 3-mers (excluding the first 3 bp) in a previously used set of 206,029 human internal exons (Fig. 5; Fairbrother et al. 2004). Using these data, we compared the nonsense potential of exon retention to exon skipping with respect to that of our control. For all ESR hexamers in our data sets, we observed at least an approximately twofold increase in nonsense potential, consistent with silencer gains (P-value < 5.50 × 10−21) and enhancer losses (P-value < 1.27 × 10−14) when compared to controls (χ2 goodness-of-fit test) (Fig. 5). As expected, minimal values of nonsense potential were observed for silencer loss (P-value < 0.19) and enhancer gain (P-value < 0.86), consistent with the nonsense potential seen for the respective “loss” and “gain” of all human exonic 3-mers (χ2 goodness-of-fit test) (Fig. 5). The lack of nonsense potential for enhancer gain is not surprising given the abundance of enhancers within exons (Fairbrother et al. 2002) and their being subject to protein-coding restrictions. These data therefore support a model that involves the disruption of enhancers and the creation of silencers to yield nonsense-associated altered splicing (NAS). Consistent with this postulate, it would appear as though enhancer loss and silencer gain are specifically associated with potential-nonsense codons through the sequence bias of the ESRs.
Figure 5.
An overview of the nonsense codon sequence bias in exonic splicing regulators. Bars correspond to the nonsense-coding potential of ESR loss or gain, the proportion (expressed as a percentage) of 3-mers matching UAG, UGA, or UAA out of total 3-mers. For ESR loss, this was calculated via simulated mutation based on HGMD transition/transversion probabilities (Supplemental Fig. 2). For all human internal exonic 3-mers, the nonsense-coding potential was calculated using the same algorithm as the ESRs, except using a set of all human internal exonic sequences instead of ESR hexamers. The frequencies were normalized, and the values for the data given for ESR loss or gain were analyzed statistically (P-values from χ2 goodness-of-fit test) using an α-value of (***) 0.001.
Discussion
The results presented here demonstrate that nearly ∼25% (7154/27,681) of exonic (i.e., mis-sense and nonsense) mutations that cause human inherited disease are likely to induce exon skipping either via the loss of evolutionarily conserved splicing enhancers or alternatively through the creation of potent splicing silencers (Table 2). Given that it has already been recognized that at least 10% of disease-causing mutations ablate 5′- or 3′-splice site consensus sequences (Krawczak et al. 2007), we conservatively estimate that approximately one-third of disease-causing mutations may induce aberrant splicing. Recently published work from another group using an independent strategy reached a similar estimate (22%) of splicing sensitive mis-sense/nonsense disease-causing mutations (Lim et al. 2011). Future studies that include mutations that affect intronic cis-elements may well increase this proportion. Overall, our results provide new insights into the underlying mechanisms that link mutation-induced aberrant splicing and human inherited disease. Understanding these mechanisms is a prerequisite for the optimization of treatment regimens as we enter the era of personalized medicine.
One surprising result from our study is that although genomic variants that create ESEs or abolish ESSs are more frequently associated with neutral SNPs (Fig. 1A,B), some individual ESE and ESS hexamers show a remarkable enrichment for disease-causing mutations when gained or lost, respectively. We believe that this class of mutations may induce aberrant splicing of adjacent exons as previously described for a polymorphism in the MST1R1 gene (Ghigna et al. 2005). We find it interesting that specific ESR hexamers, based on their HGMD/SNP log ratios, appear to be disproportionately represented by disease-causing mutations (Fig. 1C, region i; Fig. 1D, region i). Within each of these clusters there are individual hexamers that appear to be mutated very frequently in genetic disease, suggesting that specific trans-acting factors may be associated with several genetic disorders. For example, one of the sequences in the enhancer loss-enriched cluster displays a remarkable degree of similarity to the canonical binding site for the splicing factor SF2/ASF (SRSF1) (Fig. 1C, GAAGAA; Tacke and Manley 1995; Sanford et al. 2009). Indeed, the presence of an SF2/ASF consensus motif in this cluster supports previous evidence for the loss of ESEs as an important cause of human inherited disease (Sanford et al. 2009). Finally, there are a striking number of ESEs and ESSs that are relatively untouched by disease-causing mutations but appear to be more polymorphic in different human populations. These data suggest at least two non-mutually exclusive possibilities. The first is that those hexamers that are over-represented in the SNP data set may be redundant with the function of other hexamers and hence more prone to variation across human populations. Alternatively, the polymorphic ESRs identified here may be associated with allele-specific alternative splicing that confers a gain of fitness rather than a disease phenotype (Fraser and Xie 2009). Each of these hypotheses will require further testing.
As described above, we investigated the function of a specific ESS hexamer, ACUAGG (Fig. 2D), that has been created de novo by no fewer than 83 different single nucleotide substitutions (both mis-sense and nonsense) in 67 different genes as a cause of human inherited disease (see Supplemental Table 4). Reporter constructs derived from three different disease genes—OPA1, TFR2, and PYGM—all demonstrated that ACUAGG promotes skipping of test exons derived from the mutant alleles of each gene. We also prepared a reporter construct corresponding to an ACUAGG introduction near the 3′ss of exon 13 from MYH7. This mutation failed to induce appreciable skipping of the test exon (data not shown). For the case of ACUAGG insertion in OPA1, the effects of this ectopic silencer element on exon skipping appear to be mediated, at least in part, by SRp20 and possibly PTB. This is somewhat surprising since SR proteins are typically thought to promote exon inclusion by binding to splicing enhancers (Ram and Ast 2007). It is possible that interactions between SRp20, PTB, and other hnRNPs create an exon silencing complex that promotes exon skipping. A comprehensive analysis of PTB–RNA interactions identified many examples of alternative cassette exons that are skipped through the action of PTB-binding sites located near the 5′-splice site (Xue et al. 2009). Our data suggest that the ACUAGG hexamer is a potent splicing silencer that functions at both 5′- and 3′-splice sites.
The impact of premature termination (nonsense) codons on gene expression remains an important consideration in the elucidation of the pathogenic basis of disease-causing mutations. It is now well established that the NMD pathway plays a central role in preventing the accumulation and translation of nonsense-containing mRNA isoforms (Maquat 2004; McGlincy and Smith 2008). However, PTCs are also suggested to directly influence alternative splicing decisions (Wang et al. 2002; Wachtel et al. 2004). The most plausible model is that PTCs disrupt ESEs and induce exon skipping (Liu et al. 2001; Cartegni et al. 2002; Pagani et al. 2003; Zatkova et al. 2004). Our results presented in Figure 5 extend this model by suggesting that such a surveillance mechanism might evolve via the acquisition of ESR sequences to counteract PTC-containing exons associated with a greater likelihood of skipping. The apparent bias of ESR sequences toward potential nonsense codons would appear to be the most logical explanation for nonsense-associated altered splicing (Valentine and Heflich 1997). To test this postulate, we examined the very first observation of NAS where exon skipping was observed in the fibrillin (FBN1) gene due to a nonsense-causing T > G transversion 26 bp from a constitutive 3′-splice site (Dietz et al. 1993). Consistent with our model, the mutation appears to create a disease-enriched silencer CUUAGG (Supplemental Table 4, binomial P-value < 2.2 × 10−16), with the core of the motif containing the previously observed nonsense codon, UAG. We suspect that many NAS observations may be consistent with this model due to ESR sequence bias or else attributable to PCR amplification artifacts after NMD (Cartegni et al. 2002). It remains to be determined if such a mechanism might arise as an attempt to preserve the transcript at the expense of a single exon or as a hammer to ensure that NMD is successfully elicited by the PTC.
Methods
Data set preparations
Mutations from the Human Gene Mutation Database (HGMD; http://www.hgmd.org) and SNPs (30%–50% heterozygosity) from the 1000 Genomes Project (http://www.1000genomes.org) were extracted and mapped to hg19 internal exons as annotated by the UCSC Known Gene track (Karolchik et al. 2008). Intersecting alleles found in both the HGMD and SNP data sets were removed from the SNP data set. Biallelic SNPs whose ancestral allele could not be determined were also removed from the SNP data set. Alleles mapping to within the first 3 bp from a splice site were removed from the SNP data set due to possible splice-site consensus sequence overlap. HGMD mutations were divided into subsets corresponding to the nearest splice site (5′ or 3′) and according to whether the mutation mapped to constitutive or alternative exons using the UCSC Alt Events track. As a quality control measure, HGMD mutations mapping past half the average HGMD exon length (144 bp) from a splice site were also removed, leaving only mutations within 3 to 72 bp from the nearest splice site.
Odds ratios and binomial estimation
ESE loss was defined as an event involving a directional change from one allele to another that served to convert an ESE to neutral or an ESS. ESE gain was defined as an event involving a change from either neutral or ESS to an ESE. We counted the numbers of HGMD mutations or SNPs causing ESE loss/gain. An odds ratio (OR), was calculated given
where the event can be loss or gain of a given ESE hexamer and P(loss|data set) = 1 − P(gain|data set). Odds ratios are plotted as bars, with 95% confidence intervals (two-tailed) as error bars calculated using standard methods (Pagano and Gauvreau 2000). The same assumptions and calculations were used when considering loss or gain of ESS hexamers in neutral SNP and HGMD data sets.
To assess the significance of the enrichment of individual ESE hexamers in the HGMD data set as compared to the neutral SNP data set, we used the binomial distribution. For each hexamer i, the probability of P(HGMD[i] = k) is distributed such that HGMD[i] ∼ Bin(n, p[i]), where HGMD[i] is a random variable corresponding to the number of mutations causing loss of a particular ESE, p[i] is the neutral background probability for the particular hexamer to be targeted for loss, and n is the total number of HGMD mutations causing loss of any ESE. The neutral background probability (p[i]) for each ESE hexamer i is calculated as the number of times a neutral SNP causes loss of i plus a pseudocount normalized over the total number of SNPs causing loss to ESEs (SNP[i] + 0.5/∑i (SNP[i] + 0.5)). Based on large values for n and exceptionally low values for p[i], binomial P-values are approximated using the Poisson distribution such that λ[i] ∼ np[i]. We also applied this to ESE gain and both ESS loss and ESS gain, each with their own set of neutral background probabilities. For any mutation that alters multiple ESR hexamers, only the hexamer with the lowest binomial P-value is used in statistical tests. The significance of each P-value is determined for multiple hypotheses using a Benjamini-Hochberg false discovery rate (FDR) of 5% (Benjamini and Hochberg 1995).
Conservation of ESE loss hexamers
To assess the evolutionary conservation of lost ESE motifs, we calculated the average phyloP score from multiple orthologous alignments of 46 placental mammals (Karolchik et al. 2008; Pollard et al. 2009) for each ESE hexamer ablated by a directional allele. Typically, phyloP scores are used to determine the conservation of individual sequence alignment columns between species, given a null model of neutral evolution at single nucleotide resolution. These scores in the human genome range from values as low as −13.79 to 2.94 representing the −log10(P-value) to reject the null hypothesis. In this study, the average phyloP score is used to determine the relative conservation of hexamers rather than as a strict statistical test. Using each ESE phyloP score and its corresponding distance to the nearest exon–intron boundary, we performed two-dimensional (2D) Gaussian kernel density estimation and plotted the three-dimensional (3D) density using R. To compare the distribution of phyloP scores for ESEs disrupted by HGMD mutations to that due to chance alone, we randomly sampled 13,000 hexamers from both the HGMD- and SNP-targeted exons that did not match a hexamer in the ESE data set. We also sampled an equal size of random hexamers containing HGMD mutations that did not cause loss or gain of known ESRs. Statistical hypothesis testing on means was executed using a Welch t-test for normal data with unequal sample size and variance using α-values of (*) 0.05, (**) 0.01, and (***) 0.001. Given such large sample sizes, normality assumptions are approximately satisfied through the asymptotic relationship to the normal distribution provided by the central limit theorem. Additionally, we performed a non-parametric alternative, the Wilcoxon rank-sum test, which provided similarly significant P-values for each test shown.
Splicing reporter assay and RNAi
To assess the functional relevance of non-synonymous HGMD mutations to splicing, DNA inserts containing the entire exon plus 50 bp of flanking intron sequence for both the matched wild-type and mutant versions of selected mutations were created using Custom Gene Synthesis from IDT (http://www.idtdna.com) flanked by NdeI and BglII restriction sites. Test alleles were subcloned from the pSMART vector using NdeI and BglII restriction sites into the pSC14mw vector. All constructs were validated by sequencing. Splicing reporters were transiently transfected into HeLa and 293T cells in six-well plates using Lipofectamine 2000 (http://www.invitrogen.com) following the manufacturer's instructions. Cells were harvested 24 h post-transfection, and cytoplasmic RNA was subsequently isolated using Tri-Reagent LS (Sigma-Aldrich). RNA samples were converted to cDNA using GoScript (Promega). One hundred nanograms of cDNA was used as templates for PCR using Bulls Eye rTaq (Midwest Scientific). The sequences of the PCR primers used in this study are the following: Reporter Forward, CAAACAGACACCATGGTGCACC; Reporter Reverse, AACAGCATCAGGAGTGGACAGATC; SRSF6 Forward, TACGGCTTCGTGGAGTTCGAG; SRSF6 Reverse, TCTTGCCAACTGCACCGACTAG. Following PCR, the amplicons were purified using Purelink microcentrifuge columns (Invitrogen). Amplicons corresponding to alternative mRNA isoforms were separated with 6% (29:1) polyacrylamide gel electrophoresis and visualized using syberSAFE staining (Invitrogen). Bands corresponding to exon inclusion and exclusion were cut out and validated by DNA sequencing (data not shown). The linearity of the PCR reaction was confirmed by assaying splicing at increasing PCR cycles (Supplemental Fig. 12). For the experiments described above, quantification was performed following 29 cycles of PCR. Ratios corresponding to splicing efficiencies (% exon inclusion) were used to assay the effects of single nucleotide substitutions between samples rather than the absolute amount of each product. Molar ratios of mRNA isoforms were quantified using peak integration on a DNA1000 chip using an Agilent 2100 Bioanalyzer. To assay for activity of nonsense-mediated decay, we treated cells with 100 μg/mL emetine dihydrocholoride hydrate (Fluka) 10 h before harvesting.
For the RNA interference assay, HeLa cells were transiently cotransfected with both the construct and appropriate siRNA (NTi, SRp20i, or PTBi) using DharmaFECT Duo (http://www.thermoscientific.com) according to the manufacturer's instructions and harvested at 48 h post-transfection. Nuclear protein was resolved on Novex 10% bis tris polyacryamide gels and transferred to Immobilon FL (Millipore) using a Genie Blotter (Idea Scientific). Antibodies corresponding to PTB (mAb BB7), SRp20 (Sigma-Aldrich), and GAPDH (Calbiochem) were visualized with fluorescent-labeled secondary antibodies (GE) using the Fluoro-Chem Q system (Cell Bioscience). Following purification of cytoplasmic RNA using Tri-reagent LS, amplicons were generated using One-step RT-PCR (Invitrogen), and the following cycling program: 30 min at 55°C; 2 min at 94°C; and 30 cycles of 30 sec at 94°C, 30 sec at 59°C, and 60 sec at 72°C.
RNA affinity chromatography
RNA affinity chromatography was performed as previously described (Caputi and Zahler 2001) with the following modifications: For the OPA1 ligands, we selected a region 35 nt upstream of and 25 nt downstream from the 5′-splice site from exon 12 of the OPA1 gene. Both the wild-type and mutant alleles were sequenced using IDT Custom Gene Synthesis. RNA was transcribed in vitro using T7 RNA polymerase (Ambion) and gel-purified from 6% (19:1) polyacrylamide gels. Fifteen hundred picomoles of purified RNA was oxidized by metaperiodate treatment and coupled to adipic acid dihydrazide agarose beads (Sigma-Aldrich). 1.5 mg of HeLa nuclear extract was incubated with the beads coupled to wild-type or mutant RNA bait, washed, and eluted with increasing concentrations of KCl. One-half of the sample was resolved by 10% Novex Nupage gel electrophoresis and silver-stained (Silver SNAP; BD Bioscience). The remaining half was precipitated with 20% TCA and washed in acetone. The protein pellet was analyzed by Mud-PIT Mass spectrometry at the Vincent J. Coates Proteomic Laboratory at University of California, Berkeley. Differences in peptide coverage between the wild-type and mutant eluates were quantified using MASCOT (Perkins et al. 1999), and peptide spectra from each sample were compared using CONTRAST (Tabb et al. 2002). A complete table of all peptides identified in both eluates can be found in Supplemental Tables 5 and 6. Criteria settings for CONTRAST can be found in Supplemental Table 7.
Acknowledgments
We thank K. Lynch (University of Pennsylvania Medical School) for generously providing the beta-hemoglobin splicing reporter used in this study. We thank J. Caceres for comments on the manuscript and M. Ares, A. Zahler, Y. Liu, and S. Mooney for helpful discussions. MudPIT analysis was performed at the Victor Coates Proteomics Laboratory at UC Berkeley. This work was supported by the Ellison Medical Research Foundation New Scholar Award to J.R.S., grant R01GM085121 from the U.S. National Institutes of Health to J.R.S, and financial support from BIOBASE GmbH to D.N.C. and M.M.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.118638.110.
References
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Biasiotto G, Camaschella C, Forni GL, Polotti A, Zecchina G, Arosio P 2008. New TFR2 mutations in young Italian patients with hemochromatosis. Haematologica 93: 309–310 [DOI] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394 [DOI] [PubMed] [Google Scholar]
- Bruno C, Tamburino L, Kawashima N, Andreu AL, Shanske S, Hadjigeorgiou GM, Kawashima A, DiMauro S 1999. A nonsense mutation in the myophosphorylase gene in a Japanese family with McArdle's disease. Neuromuscul Disord 9: 34–37 [DOI] [PubMed] [Google Scholar]
- Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P 2000. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 25: 14–15 [DOI] [PubMed] [Google Scholar]
- Caputi M, Zahler AM 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J Biol Chem 276: 43850–43859 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31: 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet 7: 98–108 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ Jr, Pyeritz RE, Cutting GR 1993. The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259: 680–683 [DOI] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Caceres JF, Hayward DG, Krainer AR 2000. Selection of alternative 5′ splice sites: Role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol 20: 8303–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013 [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Holste D, Burge CB, Sharp PA 2004. Single nucleotide polymorphism–based validation of exonic splicing enhancers. PLoS Biol 2: e268 doi: 10.1371/journal.pbio.0020268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Xie X 2009. Common polymorphic transcript variation in human disease. Genome Res 19: 567–575 [DOI] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20: 881–890 [DOI] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G 2006. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol Cell 22: 769–781 [DOI] [PubMed] [Google Scholar]
- Goren A, Kim E, Amit M, Bochner R, Lev-Maor G, Ahituv N, Ast G 2008. Alternative approach to a heavy weight problem. Genome Res 18: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Maniatis T 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell 1: 765–771 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. 2008. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res 36: D773–D779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL 1994. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368: 119–124 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN 2007. Single base-pair substitutions in exon–intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat 28: 150–158 [DOI] [PubMed] [Google Scholar]
- Kumar S, Suleski MP, Markov GJ, Lawrence S, Marco A, Filipski AJ 2009. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res 19: 1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Li B, Wachtel C, Miriami E, Yahalom G, Friedlander G, Sharon G, Sperling R, Sperling J 2002. Stop codons affect 5′ splice site selection by surveillance of splicing. Proc Natl Acad Sci 99: 5277–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG 2011. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci 108: 11093–11098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev 12: 1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Cartegni L, Zhang MQ, Krainer AR 2001. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet 27: 55–58 [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res 12: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Margulies EH, Blanchette M, Haussler D, Green ED 2003. Identification and characterization of multi-species conserved sequences. Genome Res 13: 2507–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW 2008. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33: 385–393 [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noensie EN, Dietz HC 2001. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol 19: 434–439 [DOI] [PubMed] [Google Scholar]
- Nogales-Gadea G, Rubio JC, Fernandez-Cadenas I, Garcia-Consuegra I, Lucia A, Cabello A, Garcia-Arumi E, Arenas J, Andreu AL, Martin MA 2008. Expression of the muscle glycogen phosphorylase gene in patients with McArdle disease: The role of nonsense-mediated mRNA decay. Hum Mutat 29: 277–283 [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE 2004. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5: 389–396 [DOI] [PubMed] [Google Scholar]
- Pagani F, Buratti E, Stuani C, Baralle FE 2003. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J Biol Chem 278: 26580–26588 [DOI] [PubMed] [Google Scholar]
- Pagano M, Gauvreau K 2000. Principles of biostatistics 2nd ed Duxbury Press, Pacific Grove, CA [Google Scholar]
- Parmley JL, Chamary JV, Hurst LD 2006. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol Biol Evol 23: 301–309 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A 2009. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram S, Oono Y 2010. NeatMap—non-clustering heat map alternatives in R. BMC Bioinformatics 11: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Ast G 2007. SR proteins: a foot on the exon before the transition from intron to exon definition. Trends Genet 23: 5–7 [DOI] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW 2003. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell 12: 1317–1324 [DOI] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimpf S, Schaich S, Wissinger B 2006. Activation of cryptic splice sites is a frequent splicing defect mechanism caused by mutations in exon and intron sequences of the OPA1 gene. Hum Genet 118: 767–771 [DOI] [PubMed] [Google Scholar]
- Schimpf S, Fuhrmann N, Schaich S, Wissinger B 2008. Comprehensive cDNA study and quantitative transcript analysis of mutant OPA1 transcripts containing premature termination codons. Hum Mutat 29: 106–112 [DOI] [PubMed] [Google Scholar]
- Shi FD, Zhang JY, Liu D, Rearden A, Elliot M, Nachtsheim D, Daniels T, Casiano CA, Heeb MJ, Chan EK, et al. 2005. Preferential humoral immune response in prostate cancer to cellular proteins p90 and p62 in a panel of tumor-associated antigens. Prostate 63: 252–258 [DOI] [PubMed] [Google Scholar]
- Shiga N, Takeshima Y, Sakamoto H, Inoue K, Yokota Y, Yokoyama M, Matsuo M 1997. Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Invest 100: 2204–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball E, Howells K, Phillips A, Mort M, Cooper DN 2008. Human Gene Mutation Database: towards a comprehensive central mutation database. J Med Genet 45: 124–126 [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR III 2002. DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Manley JL 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J 14: 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine CR, Heflich RH 1997. The association of nonsense mutation with exon-skipping in hprt mRNA of Chinese hamster ovary cells results from an artifact of RT-PCR. RNA 3: 660–676 [PMC free article] [PubMed] [Google Scholar]
- Wachtel C, Li B, Sperling J, Sperling R 2004. Stop codon-mediated suppression of splicing is a novel nuclear scanning mechanism not affected by elements of protein synthesis and NMD. RNA 10: 1740–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA 2007. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chang YF, Hamilton JI, Wilkinson MF 2002. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol Cell 10: 951–957 [DOI] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845 [DOI] [PubMed] [Google Scholar]
- Wang Z, Xiao X, Van Nostrand E, Burge CB 2006. General and specific functions of exonic splicing silencers in splicing control. Mol Cell 23: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA 2010. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci 35: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y 1993. The role of exon sequences in splice site selection. Genes Dev 7: 407–418 [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB 2004. Variation in alternative splicing across human tissues. Genome Biol 5: R74 doi: 10.1186/gb-2004-5-10-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatkova A, Messiaen L, Vandenbroucke I, Wieser R, Fonatsch C, Krainer AR, Wimmer K 2004. Disruption of exonic splicing enhancer elements is the principal cause of exon skipping associated with seven nonsense or missense alleles of NF1. Hum Mutat 24: 491–501 [DOI] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8: 1351–1361 [DOI] [PubMed] [Google Scholar]