Abstract
X-chromosome inactivation (XCI) is a dosage compensation mechanism that silences the majority of genes on one X chromosome in each female cell. To characterize epigenetic changes that accompany this process, we measured DNA methylation levels in 45,X patients carrying a single active X chromosome (Xa), and in normal females, who carry one Xa and one inactive X (Xi). Methylated DNA was immunoprecipitated and hybridized to high-density oligonucleotide arrays covering the X chromosome, generating epigenetic profiles of active and inactive X chromosomes. We observed that XCI is accompanied by changes in DNA methylation specifically at CpG islands (CGIs). While the majority of CGIs show increased methylation levels on the Xi, XCI actually results in significant reductions in methylation at 7% of CGIs. Both intra- and inter-genic CGIs undergo epigenetic modification, with the biggest increase in methylation occurring at the promoters of genes silenced by XCI. In contrast, genes escaping XCI generally have low levels of promoter methylation, while genes that show inter-individual variation in silencing show intermediate increases in methylation. Thus, promoter methylation and susceptibility to XCI are correlated. We also observed a global correlation between CGI methylation and the evolutionary age of X-chromosome strata, and that genes escaping XCI show increased methylation within gene bodies. We used our epigenetic map to predict 26 novel genes escaping XCI, and searched for parent-of-origin-specific methylation differences, but found no evidence to support imprinting on the human X chromosome. Our study provides a detailed analysis of the epigenetic profile of active and inactive X chromosomes.
X-chromosome inactivation (XCI) is a mechanism of dosage compensation that equalizes the expression of sex-linked genes between 46,XY males and 46,XX females (Lyon 1961). This results in the silencing of the majority of genes on one of the two X chromosomes in each somatic cell of females (Carrel and Willard 2005) and a transition to a heterochromatic state. XCI is accompanied by several epigenetic modifications to the inactive X (Xi), such as an accumulation of variant histones and transcripts from the XIST gene (Clemson et al. 1996; Costanzi and Pehrson 1998), a delay in replication timing (Willard and Latt 1976), and relocalization to the nuclear periphery (Barr and Bertram 1949).
Several studies have shown that DNA methylation of the Xi plays an important role in the maintenance of its inactive state. While the active X (Xa) and Xi have very similar global levels of methylation (Bernadino et al. 1996), studies have shown that CpG islands (CGIs) have a tendency to be methylated on the Xi and unmethylated on the Xa (Tribioli et al. 1992; Hellman and Chess 2007). In contrast, the CGIs of genes escaping XCI often remain unmethylated on both the Xi and Xa (Weber et al. 2007). Mouse knockouts for the DNA methyltransferase enzyme Dnmt1 show defects in X inactivation (Panning and Jaenisch 1996). Furthermore, treatment of cells in vitro with the demethylating agent 5-azacytidine has been shown to cause the Xi to decondense (Haaf 1995), replicate earlier in the cell cycle (Jablonka et al. 1985), and in several studies using somatic cell hybrids has led to the reactivation of previously silenced genes (Hansen et al. 1996). However, despite its clear importance in the XCI process, to date only a single study has attempted to perform chromosome-wide studies of the distribution of DNA methylation on the X chromosome (Yasukochi et al. 2010).
DNA methylation also plays an important role in imprinting, a phenomenon in which the expression of a gene is dependent on its parent of origin. Because of their role in a variety of human phenotypes, there is considerable interest in identifying imprinted loci, and to date, about 70 human imprinted transcripts are known (http://www.geneimprint.com/). Although no imprinted genes have been identified on the human X chromosome, differences in both cognitive function and the frequency of autism between Turner syndrome patients with a maternally versus paternally derived X suggest the presence of X-linked imprinted gene(s) (Skuse et al. 1997), and in mouse a cluster of X-linked imprinted genes has been identified, although these apparently lack human homologs (Davies et al. 2005; Raefski and O'Neill 2005). Furthermore, in several non-primate mammals, the entire XCI process is subject to imprinting, with the paternal X chromosome being preferentially silenced in extra-embryonic tissues (Takagi and Sasaki 1975).
To characterize the epigenetic state of Xa and Xi chromosomes, we have used methylated DNA immunoprecipitation (MeDIP) followed by hybridization to high-density oligonucleotide arrays covering the entire X chromosome to profile DNA methylation patterns in both 46,XX females and Turner syndrome patients with a 45,X karyotype. As 45,X individuals carry a single Xa, while 46,XX individuals carry one Xa and one Xi, the difference in methylation between these two groups allows measurement of methylation changes that occur with XCI. As our study included multiple Turner syndrome patients with a single X chromosome of either maternal (XMAT) or paternal (XPAT) origin, we also used our epigenetic map to search for parent-of-origin-specific methylation differences on the X chromosome that might indicate imprinting. Our study provides an epigenetic profile of X inactivation giving novel insights into the phenomenon of dosage compensation.
Results
Characterization of methylation differences between active and inactive X chromosomes
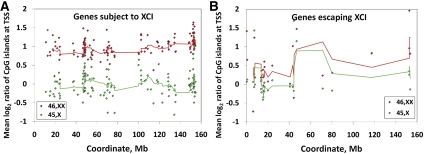
As 45,X Turner syndrome individuals carry a single Xa, while 46,XX females carry one Xa and one Xi, the difference in methylation between these two groups provides a measure of methylation specifically on the Xi. Initial visual comparison of methylation profiles revealed frequent large differences in methylation levels between 45,X and 46,XX individuals overlapping gene promoter regions and CGIs (Fig. 1; Supplemental Table 1). To gain a global view of methylation changes on the X chromosome with XCI, we generated composite plots of mean methylation levels within 271 RefSeq genes that are either subject to or escape XCI (Fig. 2), and 3060 CGIs (Fig. 3; Bock et al. 2007). We observed that for genes subject to XCI, a general increase in methylation on the Xi occurs at gene promoters. In comparison, genes escaping XCI show higher levels of methylation within gene bodies on both the Xa and Xi, but reduced promoter methylation only on the Xi. Additionally, for genes escaping XCI, a slight but consistent increase in gene body methylation is evident on the Xi. For CGIs, we observed that on average these show significantly higher methylation on the Xi, consistent with previous studies (Supplemental Table 2; Tribioli et al. 1992; Weber et al. 2007; Yasukochi et al. 2010).
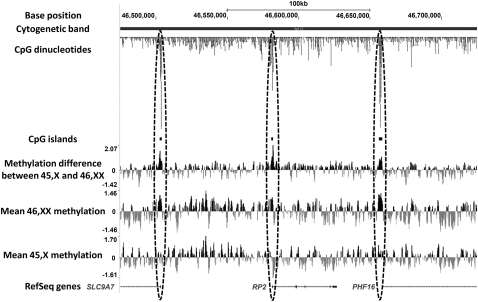
Figure 1.
Comparative methylation profiling of the X chromosome in 45,X and 46,XX individuals shows that X inactivation results in increased methylation of CpG islands at gene promoters. Regions containing high densities of CpG dinucleotides, corresponding with the transcription start sites of genes subject to X inactivation, show raised methylation levels on the Xi. The image shows the screenshot of a 250-kb region of Xp11.3 (chrX:46,475,000–46,725,000, hg18), with methylation data uploaded as custom tracks in the UCSC Genome Browser.
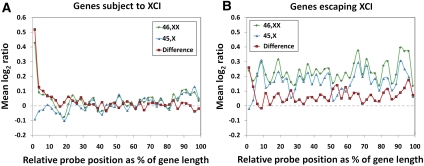
Figure 2.
Methylation patterns of genes subject to and escaping X inactivation on active and inactive X chromosomes. (A) For genes subject to XCI, methylation differences between 46,XX and 45,X individuals occur specifically at gene promoters, with 46,XX individuals showing increased methylation compared to 45,X cases. As 83% of RefSeq transcripts listed on the X chromosome have a CGI within 1 kb of their TSS and CGIs are heavily methylated on the Xi versus the Xa, this enrichment of methylation at gene promoters on the Xi is a correlate of CGI methylation. (B) In contrast, for genes escaping XCI, there is a generalized increase in methylation in the bodies of genes in both 45,X and 46,XX individuals compared to genes subject to XCI, with the biggest difference between 46,XX and 45,X occurring at the promoter region. Additionally, a slight but consistent increase in methylation in the bodies of genes escaping XCI is evident in 46,XX compared to 45,X individuals. Two hundred sixteen RefSeq genes listed on chrX scored as subject to XCI (score = 0) and 52 RefSeq genes scored as escaping XCI (score = 8 or 9) by Carrel and Willard (2005) were analyzed. Where multiple splice forms were listed for a single gene, we chose the maximal boundary of all isoforms to define the transcribed region. Probes were then assigned into one of 40 bins depending on their relative position along the gene body, defined by the maximal transcription start and end coordinates. Similar data were observed when considering only those genes of intermediate length (10–100 kb) (Supplemental Figs. 11, 12).
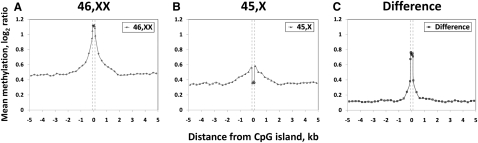
Figure 3.
Differential methylation between active and inactive X chromosomes occurs specifically at CpG islands. Each panel shows a composite plot of mean methylation levels within and flanking 3060 CGIs on the X chromosome in (A) 46,XX females; (B) 45,X Turner syndrome cases, corresponding to the Xa; and (C) the difference between these two groups, corresponding to the Xi. Three thousand sixty CGIs defined using epigenetic criteria (Bock et al. 2007) were analyzed. Those separated by <500 bp (n = 726) were merged into single regions, and the mean methylation level both within CGIs, and +5 kb and −5 kb was calculated. The apparent increase in methylation in flanking regions in 46,XX individuals is likely due to the clustering of CGIs on the X chromosome, which results in neighboring CGIs being sampled in the 5-kb flanking regions (10% of CGIs are separated by <1 kb, 36% by <5 kb). (Gray dashes) Borders of the CGIs. Mean methylation within CGIs was calculated by assigning probes into 10 windows proportional to the length of each CGI. Mean methylation in flanking regions was plotted in 250-bp windows. Underlying mean log2 values with variance and associated P-values are shown in Supplemental Table 2.
Next, we examined methylation levels at individual CGIs in relation to their physical position on the Xi. We observed wide variation in the change in CGI methylation with XCI. Although the majority of CGIs show increased methylation on the Xi (68% have higher methylation in 46,XX vs. 45,X cases, p < 0.01), we observed that 7% of CGIs showed significantly lower levels of methylation on the Xi (n = 159 had lower methylation in 46,XX compared to 45,X cases, p < 0.01), indicating that XCI results in reduced methylation at many loci (Supplemental Table 1). Many of these sites of reduced methylation cluster together, and their location is strongly correlated with the position of genes that escape XCI (Fig. 4; Supplemental Figs. 2, 3). Intersecting CGIs with 410 RefSeq genes of known X-inactivation status (Carrel and Willard 2005) yields correlation r = −0.81 between mean difference in CGI methylation between 45,X and 46,XX cases and XCI score, showing a strong trend that CGIs overlapping genes that escape XCI have lower methylation than the CGIs overlapping genes that are subject to XCI.
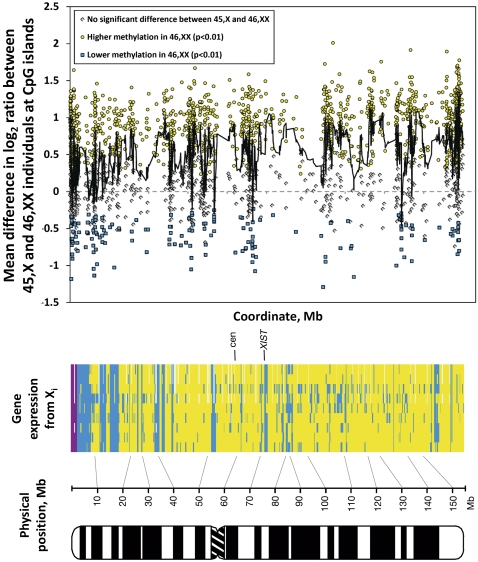
Figure 4.
X inactivation results in highly variable changes in methylation of CpG islands that correlate with the location of genes escaping X inactivation. While the majority of CGIs (61%) show increased methylation on the Xi (yellow circles), 7% of CGIs have significantly lower levels of methylation in 46,XX compared to 45,X individuals (blue squares; p < 0.01), contradicting the notion that XCI is always associated with increased methylation. The location of these sites of reduced methylation highly correlates with the physical position of genes known to escape XCI (r = −0.81). (Gray diamonds) The remaining 32% of CGIs showed no significant difference in methylation (p > 0.01). Each point represents the change in mean methylation between 45,X and 46,XX individuals at a CGI (Bock et al. 2007), with the black line showing the moving average of methylation at 10 CGIs. The heat map shows genes expressed from the Xi (blue), silent genes (yellow), pseudoautosomal genes (purple), and untested genes (white) in each of nine cell lines (adapted from Carrel and Willard 2005 and reprinted with permission from Nature Publishing Group © 2005).
As previous epigenetic studies of XCI have reported only increased methylation on the Xi, we sought to confirm this observation using an independent data set. We analyzed published data of methylation levels at individual CpG dinucleotides on the X chromosome measured in 600 brain samples produced by hybridization of bisulfite-converted DNA to Illumina bead arrays (Gibbs et al. 2010). Using this alternative technology, we observed a similar distribution of changes in methylation with XCI as with MeDIP, with 35% of the 1084 probes targeted to X-linked gene promoters showing lower methylation levels in females than males. Further analysis of these data indicated that the vast majority of gender differences in methylation result from probes mapping to the X chromosome, indicating XCI as the underlying mechanism, and that as observed by MeDIP, XCI is accompanied by both gains and losses of methylation at different sites (Supplemental Figs. 1, 2). We also validated our results using PCR and sequencing of bisulfite-converted DNA in other individuals with 45,X and 46,XX karyotypes that had not been tested by array. At sites indicated by MeDIP array analysis to have lower methylation on the inactive X, we observed that while 45,X individuals showed complete methylation, 46,XX females showed a mix of both methylated and unmethylated DNA, indicating either methylation specific to the Xa or a mix of methylation on both the Xa and Xi (Supplemental Fig. 4).
Based on the correlation between CGI methylation and the location of genes escaping XCI, we investigated the relationship between the X-inactivation status of a gene and its promoter methylation more closely. We analyzed CGIs located within 1 kb of the transcription start site (TSS) of genes consistently scored as either subject to or escaping XCI (score = 0 or 9) (Fig. 5; Supplemental Tables 1, 3; Carrel and Willard 2005). CGIs at promoters of genes subject to XCI show statistically significant increases in methylation on the Xi compared to the Xa. In contrast, the majority of CGIs at promoters of genes escaping XCI show methylation levels on the Xi similar to those on the Xa.
Figure 5.
Methylation status of CpG islands at gene promoters varies depending on gene inactivation status. (A) CGIs at promoters of genes subject to XCI show much higher methylation levels in 46,XX compared to 45,X individuals (mean 46,XX log2 methylation = 0.99, mean log2 methylation difference = 1.04, p = 1.3 × 10−23). (B) In contrast, the majority of CGIs at promoters of genes escaping XCI show much lower methylation levels in 46,XX individuals, similar to those seen in 45,X individuals (mean 46,XX log2 methylation = 0.54, mean log2 methylation difference = 0.34, p = 0.00013). Each point represents the mean log2 methylation of a CGI located within 1 kb of the TSS of a RefSeq gene scored by Carrel and Willard (2005) as being (A) silenced on the Xi (expressed in 0 of 9 hybrids containing an inactive X, n = 135), or (B) expressed from the Xi (expressed in ≥8 of 9 hybrids containing an inactive X, n = 29). Colored lines show the moving average of methylation at 10 and 3 CGIs for genes subject to and escaping XCI, respectively. Data for CGIs that lie outside of RefSeq gene promoters are shown in Supplemental Figure 14.
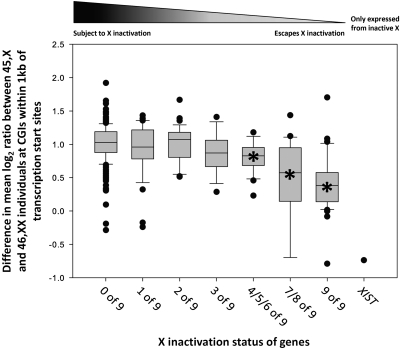
We also investigated genes that exhibit variable or unstable XCI, being expressed from the Xi in some individuals but silent in others. We observed an inverse correlation between the frequency with which a gene is expressed from the Xi and the methylation level of CGIs located near the TSS on the Xi (R2 = 0.18, p < 0.001) (Fig. 6). As we had measured methylation in multiple individuals, we investigated whether genes that show polymorphic XCI also undergo polymorphic methylation, but could find no evidence to support this hypothesis. Genes that exhibit variable XCI (scores 1–8) (Carrel and Willard 2005) did not show higher inter-individual variance in CGI methylation at their promoters than genes that are always active or inactive (score 0 or 9) (Supplemental Table 1).
Figure 6.
Difference in methylation levels between 46,XX and 45,X individuals at transcription start sites is inversely correlated with X-inactivation status. Boxplots show the mean methylation of CGIs located within 1 kb of the TSS of 363 RefSeq genes scored by Carrel and Willard (2005) for their expression status on the Xi (Supplemental Table 3). Genes are divided based on their expression status on the Xi, with a score from 0 to 9 corresponding to the number of somatic cell hybrids containing an inactive X chromosome that express that gene. Genes with score 0 are always subject to XCI, genes with score 9 always escape XCI, and genes with intermediate scores show polymorphic or unstable inactivation. The XIST gene is unique in being transcribed only from the Xi, and shows lower methylation in 46,XX versus 45,X individuals. Linear regression analysis of XCI score with methylation difference between 46,XX and 45,X yields R2 = 0.18, p < 0.001. ANOVA followed by Tukey analysis showed that genes with scores 4–9 showed statistically significant methylation differences compared to genes with score 0 (Bonferroni-adjusted p < 0.001, indicated by an asterisk within each box). Boxplots define lower and upper quartiles of each distribution, the internal band shows the median, while whiskers correspond to top and bottom deciles. Categories containing less than 15 genes were combined to yield sufficient sample sizes for meaningful comparison.
Finally, we observed a significant trend between CGI methylation and chromosomal position specifically on the Xi (linear regression, R2 = 0.102, p = 7.58 × 10−16 in 46,XX individuals; R2 = 0.001, p = 0.2 in 45,X individuals) (Supplemental Fig. 6). In general, CGIs on the Xi show smaller increases in methylation levels in the distal short arm compared to long arm, mirroring the known evolutionary ages of different portions of the human X chromosome (Lahn and Page 1999).
Characterization of sequence features in relation to methylation
We investigated the relationship between CpG density and methylation changes on the Xi and observed a positive correlation between CpG density and the difference in log2 methylation ratios between 46,XX and 45,X individuals (Supplemental Fig. 7). Given this potentially confounding relationship, we investigated whether this phenomenon might account for some of our prior observations. However, we observed no apparent relationship between the density of CpG dinucleotides within CpG islands at gene promoters or genes and their X-inactivation status, or between CpG density within CpG islands and physical position on the X chromosome (Supplemental Figs. 7, 8, 9). We therefore conclude that variations in promoter CpG density do not explain susceptibility of gene to XCI, and that the correlation we observed between methylation and evolutionary age of X chromatin does not result from variations in the CpG density of CpG islands. Investigation of repeat content of genes subject to and escaping XCI confirmed previous observations of reduced LINE content in genes escaping XCI relative to genes that are subject to XCI (Supplemental Fig. 11; Wang et al. 2006).
We hypothesized that regions susceptible to XCI might be enriched for sequence features associated with X inactivation, and we could therefore use our methylation map to search for motifs that might play a role in the XCI process. Although MEME analysis identified three high-frequency motifs 10–20 bp in size that showed potential enrichment in a set of 115 CGIs that showed the largest increase in methylation on the Xi (data not shown), these motifs were highly degenerate and of low complexity in nature. Subsequent analysis by FIMO suggested that these motifs were also enriched in a set of 115 CGIs from the opposite end of the distribution (those showing the largest decrease in methylation on the Xi) and in a set of 115 randomly chosen CGIs from the X chromosome, suggesting that any association of these motifs with XCI methylation was non-specific. Thus, we do not consider there to be any specific association of these motifs with XCI. DRIM analysis did not identify any motifs that were significantly correlated with CGI methylation.
Prediction of novel genes escaping X inactivation based on promoter methylation and validation by RNA-seq
We investigated the utility of our methylation map of the Xi and Xa to classify genes based on their X-inactivation status. For genes known to escape XCI (score 8 or 9), the mean difference in TSS CGI methylation between 45,X and 46,XX individuals is generally much lower than at genes subject to XCI, with 50% of genes escaping XCI showing a difference in log2 <0.39. Using this as a metric to score the genes tested by Carrel and Willard (2005), 14 of the 29 (48%) genes escaping XCI are below this threshold, while only three of 135 (2%) genes subject to XCI satisfy this criterion. Furthermore, in 46,XX individuals, the absolute methylation at the CGI of genes consistently subject to XCI (score = 0) is generally high, with the bottom fifth percentile of this distribution being log2 = 0.65. Using this as a metric to classify genes previously tested (Carrel and Willard 2005), 20 of 31 (65%) genes escaping XCI have methylation levels below this threshold, while only seven of 135 (5%) genes subject to XCI satisfy this criterion. Using a stringent combination of both criteria together (absolute methylation in 46,XX <0.65, difference in methylation between 46,XX and 45,X <0.44) identifies 12 of the 29 (41%) genes previously classed as escaping XCI, and only one of the 135 (0.7%) RefSeq genes scored as inactive. Thus, both the absolute and relative methylation levels of gene promoters in 45,X and 46,XX individuals strongly correlates with their expression from the Xi, and promoter CGI methylation is predictive of XCI status.
We hypothesized that we could therefore use our methylation map of the X chromosome to prospectively identify the majority of genes that escape XCI on the entire X chromosome based on their promoter methylation state. Classifying all known genes on the X chromosome by promoter methylation, as described above, allowed us to predict 31 novel genes (21 RefSeq genes and 10 additional transcripts listed as UCSC genes) that likely escape XCI with high confidence based on two criteria: (1) Their mean promoter CGI methylation in 46,XX individuals is <0.65; and (2) the difference in mean promoter methylation between 46,XX and 45,X is <0.39 (Supplemental Table 3).
We validated a subset of these predictions using transcribed SNPs in RNA-seq data from 46,XX lymphoblastoid cell lines that showed highly skewed X-chromosome inactivation. In a female in which every cell has silenced the same parental X chromosome, escape from XCI would manifest as biallelic expression of that gene, while monoallelic expression would indicate that the gene was subject to XCI. Of the 31 novel genes we predicted to escape XCI, eight contained a heterozygous SNP covered at sufficient read depth to be assayed in females with skewed XCI. Six of these eight genes showed biallelic expression in all individuals tested, thus confirming their predicted status as escaping XCI (DDX3X, EIF1AX, EIF2S3, TXLNG, RPS4X, HDHD1) (Supplemental Table 4). One further gene (UBA1) was biallelically expressed in one individual, but monoallelically expressed in a second individual, suggesting that it undergoes polymorphic XCI. Only one of the eight genes tested (MAGED1) showed monoallelic expression in all cases analyzed, suggesting that this gene is subject to XCI and that our prediction of escape from XCI was incorrect. Although we were only able to test a minority of the 31 predicted genes that escape XCI, largely due to the fact that many genes had low or absent expression in lymphoblasts combined with the limited availability of transcribed polymorphisms, these analyses demonstrate that our predictions of genes escaping XCI are largely correct.
Imprinting analysis of the X chromosome using patients with Turner syndrome
Previous phenotype studies have suggested the presence of one or more imprinted loci on the human X chromosome that influence cognitive function (Skuse et al. 1997). We reasoned that as most imprinted genes are accompanied by parent-of-origin-specific epigenetic marks, a search for differentially methylated regions between patients with 45,XMAT (n = 4) versus those with 45,XPAT (n = 3) might uncover imprinted loci on the X chromosome. Previously, we used an identical MeDIP and array hybridization protocol in patients with uniparental disomy of chromosome 15 to successfully detect novel DMRs associated with imprinted loci on chromosome 15 (Sharp et al. 2010). After applying the same analysis to compare methylation patterns in 45,XMAT versus 45,XPAT samples, none of the >1 million probes covering the X chromosome showed a difference that was even suggestive of parent-of-origin-specific X-chromosome methylation (all probes had FDR-adjusted p > 0.9999). Despite this, we further analyzed our data set in an attempt to identify weak signals of differential methylation that did not survive multiple testing correction. We searched for clusters of probes with nominal significance and identified 11 X-chromosome loci comprising three or more probes separated by <500 bp, each with an unadjusted P-value <0.01 (Supplemental Table 5). However, of these 11 loci, 10 showed characteristics that made them highly unlikely to be genuine DMRs and were excluded from further analysis: Three had a very low CpG density (<1 CpG per 250 bp), three were composed solely of common repetitive elements embedded in duplicated portions of the genome, and four showed differences in methylation intensity, but not direction, that resembled false-positive signals we had observed in previous studies of chromosome 15 (Sharp et al. 2010). We designed bisulfite PCR primers to amplify the one remaining locus and performed bisulfite sequencing in two 45,XMAT, two 45,XPAT samples, and two normal controls, but observed no methylation differences between maternally and paternally derived X chromosomes at this locus. Thus, we could find no evidence of significant parent-of-origin-specific methylation on the X chromosome.
Discussion
We have performed a detailed analysis of DNA methylation patterns on the active and inactive X chromosomes. Consistent with previous studies, we observe that XCI is accompanied by gains in methylation at the majority of CGIs of genes silenced on the Xi. However, our data show that methylation increases are not limited to gene promoter regions, and we demonstrate that the majority of intergenic CGIs also show increased methylation on the Xi, although to a lesser extent than that seen at gene promoters. Our study also shows that, in contrast to current thinking, XCI actually results in reduced methylation at a significant proportion of CGIs. These CGIs that show higher methylation on the Xa tend to occur outside of gene promoters. We also observed a relative increase in methylation within the transcribed region of genes escaping XCI compared to those that are subject to XCI, consistent with previous reports of both decreased methylation in the bodies of epigenetically silent genes (Hellman and Chess 2007) and increased methylation in expressed genes versus inactive genes (Li et al. 2010). However, as both 45,X and 46,XX individuals have an Xa and the MeDIP assay does not provide allelic methylation information, we are unable to distinguish if there is increased methylation specific to the active X. An alternative possibility that we cannot exclude is that the reduced methylation observed at some sites in 46,XX versus 45,X individuals represents a mosaic mix of methylation on both the Xa and Xi. However, in favor of the former hypothesis, allele-specific studies in a 46,XX female with completely skewed XCI showed monoallelic, rather than biallelic, methylation (Supplemental Fig. 5).
As we observed that CGI methylation directly correlates with the probability of a gene being subject to XCI, we used our methylation map to predict genes that escape XCI based on their absolute and relative promoter methylation on the Xi and Xa. By first calibrating suitable thresholds based on the inactivation status of 362 RefSeq genes that have a CGI within 1 kb of their TSS studied by Carrel and Willard (2005), we were able to identify a high-confidence set of 31 additional genes that likely also escape XCI. Multiple lines of evidence show that these predictions based on epigenetic profiling are largely accurate. Using RNA-seq data from 46,XX cell lines that showed completely skewed XCI, we were able to validate seven out of eight of these predicted genes as being expressed biallelically from both the Xi and Xa, confirming their escape from XCI. In addition, we note that this set of 31 predicted genes that escape XCI includes CD99, KDM5C, SMC1A, TBL1X, and KDM6A, all of which have already been shown to escape XCI by other studies (Dracopoli et al. 1985; Wu et al. 1994; Brown et al. 1995; de Conciliis et al. 1998; Greenfield et al. 1998). Of the other 26 genes, NLGN4X, PCDH11X, TBL1X, and TXLNG have functional Y-homologs, STS and KAL1 have pseudogene Y-homologs, and PLCXD1 is located in the Xp pseudo-autosomal region, all of which are features shared by many other genes that are known to escape XCI. In addition, 14 of these 31 genes were highlighted as potentially escaping XCI based on their consistently higher expression in females versus males (Johnston et al. 2008), and two were also previously identified as likely escaping XCI due to a lack of promoter methylation in females (Yasukochi et al. 2010). We conclude that the use of comparative methylation profiling provides a powerful method for the classification of genes escaping XCI that is complementary to methods based on expression profiling.
We also tested the hypothesis that variable XCI results from polymorphic promoter methylation but could find no evidence to support this. In 46,XX females, there was no significant difference in the inter-individual methylation variance at genes that were scored as showing variable inactivation XCI compared to those that are always active or always inactive (Carrel and Willard 2005). Thus, our results do not support the notion that variable XCI results from inter-individual epigenetic variation. Instead, our observations favor the alternative scenario that variability in XCI is caused by lower promoter methylation levels, likely resulting in less stable maintenance of XCI at these genes (Fig. 5; Lock et al. 1987; Keohane et al. 1998; Lingenfelter et al. 1998).
Our study also reveals a correlation between CGI methylation and chromosomal position on the Xi that corresponds with the evolutionary age of different X-chromosome strata (Supplemental Fig. 6). Previous studies have shown that the human X chromosome was formed from an ancestral X, corresponding to Xq, with multiple translocation events progressively adding additional autosomal chromatin onto Xp (Lahn and Page 1999). We observed a significant trend in which CGIs on the Xi show smaller increases in methylation levels in the younger evolutionary strata (distal Xp) compared to the ancestral portions (Xq). These data suggest that the chromatin regions recently added to the X chromosome are less heavily influenced by the XCI signal than the ancestral portions of the X, consistent with the higher density of genes escaping XCI in these regions.
Multiple lines of evidence suggest that our methodology provides accurate measurement of methylation on both the Xi and Xa. The patterns of CGI methylation we observed at numerous individual genes agree closely with those reported by previous studies that have used different methodologies, such as methylation-sensitive restriction enzyme digestion (Tribioli et al. 1992; Hellman and Chess 2007) or hybridization of bisulfite-converted DNA to bead arrays (Gibbs et al. 2010). Furthermore, based solely on measurements of CGI methylation, we are able to accurately predict and validate genes that are either known to or have been hypothesized to escape XCI. Our conclusions also largely mirror those from a previous study of gender differences in X-chromosome methylation using sequencing of HpaII and MspI restriction fragments (Yasukochi et al. 2010). As we observed, this study also identified a high correlation between low promoter methylation and escape from XCI, and that such genes tend to occur in evolutionarily younger strata.
We chose to study Turner syndrome patients with a 45,X karyotype as this provides a cleaner system that allows more accurate determination of methylation patterns on the active X chromosome. This is because the large areas of homology between the X and Y chromosomes would cause cross-hybridization artifacts on the microarray if we had studied 46,XY males, introducing significant noise to our data. It should be noted that >90% of 45,X concepti die in utero, and it is therefore possible that this might indicate additional genome abnormalities in Turner syndrome that could bias our results. However, as one of the common features of imprinted genes is the presence of differentially methylated regions (DMRs), our study design also enabled us to compare data from Turner syndrome patients with a maternally derived X against those with a paternally derived X as a method to search for imprinted loci on the X chromosome. While our previous use of this same methodology to study patients with UPD15 has successfully identified many novel DMRs on chromosome 15 (Sharp et al. 2010), our analysis of maternal and paternal X chromosomes failed to identify any sites of differential methylation, suggesting a lack of imprinting on the human X chromosome. We note, however, that the use of MeDIP and array hybridization has some limitations. Firstly, its resolution is limited, being only able to detect methylation changes at clusters of multiple CpGs. Thus, MeDIP is likely to miss subtle differences comprising only a few CpGs, or those occurring at high-copy sequences not represented on our microarray design. More specifically for the study of imprinting, some DMRs associated with imprinted genes are only differentially methylated in specific tissues, while other imprinted genes have been identified that apparently lack nearby DMRs. Given these caveats, while we cannot exclude the presence of cryptic DMRs or imprinting not detected by our methodology, our analysis was unable to find any evidence of imprinted genes on the human X chromosome. Finally, we also attempted to identify motifs that were enriched in sequences that underwent large changes in methylation on the Xi. However, although motifs were identified in our initial analysis, these sequences were highly redundant and of low complexity in nature, and further testing of these motifs yielded apparent enrichments in many different sequence sets, suggesting that these statistical associations do not represent genuine enrichments. On balance, we conclude that we were unable to identify any motifs that could be unambiguously linked to the XCI process.
In summary, our study provides a comprehensive profile of methylation patterns on human active and inactive X chromosomes. These data provide novel insights into the X-inactivation process and demonstrate the utility of epigenetic profiling as a method to study mechanisms of genome regulation.
Methods
Genomic DNA was extracted from peripheral blood samples from (1) four unrelated Turner syndrome patients with a single maternally derived X chromosome (45,XMAT), (2) three unrelated Turner syndrome patients with a single paternally derived X chromosome (45,XPAT), and (3) three unrelated control females with a normal 46,XX karyotype. The ages of each sample were unknown. For each individual with Turner syndrome, cytogenetic analysis of 30 stimulated T-lymphocyte nuclei showed the presence of a 45,X karyotype in all cells examined. Parental origin analysis of the X chromosome was determined using a panel of microsatellite markers mapping to the X chromosome in each Turner syndrome patient and their parents. Neither cytogenetic nor molecular analysis showed any signs of mosaicism in any of the Turner syndrome cases analyzed.
Methylated DNA was immunoprecipitated using monoclonal antibodies that recognize methylated cytosine (Weber et al. 2007). Briefly, 15 μg of DNA was sonicated to generate fragments 200–800 bp in size (Branson 450D Sonifier), incubated with 10 μg of anti 5-methyl cytidine (Diagenode), immunoprecipitated using Protein A Sepharose beads (Life Technologies), and purified by phenol:chloroform extraction. Immunoprecipitated and input DNA from each case were labeled by random priming using Cy5 and Cy3-conjugated random nonamers (TriLink BioTechnologies), and hybridized to tiling oligonucleotide arrays.
We used arrays composed of 2.1 million 50-mer to 75-mer oligonucleotides covering chromosomes 20, 21, 22, X, and Y at a median probe density of 1 per 100 bp (Roche NimbleGen, catalog #05543142001). Of these, 1,148,358 probes map to the 152-Mb sequenced portion of chromosome X. DNA labeling, array hybridizations, and washes were performed according to the manufacturer's recommendations, and slides were scanned using a G2565 scanner at 5-μm resolution (Agilent Technologies). Array images were analyzed using NimbleScan v2.5 software (Roche NimbleGen) with default parameters incorporating spatial correction, and the resulting files of probe log2 ratios were used for subsequent analysis. The log2 value is the Cy5:Cy3 fluorescence ratio (methylated DNA recovered by IP:total input DNA) for each probe, converted to a log2 scale, and represents a relative measure of the amount methylated DNA at each locus. We applied quantile normalization (Bolstad et al. 2003) to the raw data and filtered outlier probes to remove low-quality data points (Sharp et al. 2010). Technical replicates of the MeDIP and array hybridizations were performed for four of the seven individuals in this study, and correlation coefficients between these technical replicates were high (mean r = 0.93 after quantile normalization and outlier replacement). Subsequent data analyses were performed in Galaxy (http://main.g2.bx.psu.edu/). For all plots of differences between 45,X and 46,XX cases, we calculated the mean log2 methylation in multiple individuals with the same karyotype. However, in order to account for potential inter-individual variability in methylation patterns or noise resulting from probe variability, for all statistical analyses reported we used log2 values for individual probes in each individual. Data from each probe in each 45,X and 46,XX individual tested were then combined into two separate groups for the purposes of statistical comparison.
We used data sets of CGIs identified by epigenetic criteria (Bock et al. 2007) and downloaded additional information on RefSeq genes and UCSC genes from build36/hg18 of the UCSC Genome Browser (http://genome.ucsc.edu/). Data on the inactivation status of X-linked genes were taken from a previous study that scored expression for 624 loci in a panel of nine somatic cell hybrids containing a single inactive X chromosome (Carrel and Willard 2005). However, when mapped to hg18, we found that a significant number of these loci did not match well with current gene annotations. We therefore decided to be conservative in our approach and included only those loci tested by Carrel and Willard that we could map unambiguously to RefSeq annotations (n = 410). We conducted analysis of G+C nucleotide content, CpG dinucleotide content, and common repeat content of genes and CGIs to investigate sequence features that might influence our methylation results.
We used a custom analysis pipeline to detect regions of differential methylation between Turner syndrome individuals with X chromosomes of maternal and paternal origin (Sharp et al. 2010). Since invariant probes are less likely to contribute to the distinctions between subgroups, probes with a standard deviation across all samples <0.2 were removed from further analyses, and a moderated t-test (Smyth 2004) was conducted to identify probes that showed significantly different methylation between 45,XMAT and 45,XPAT samples using a 5% false discovery rate (FDR) correction (Benjamini and Hochberg 1995). All statistical analyses were performed using software from the Bioconductor project (Gentleman et al. 2004).
To assess X-chromosome methylation patterns assayed using a different technique, we analyzed a published data set of methylation at >27,000 CpG sites measured in 600 brain samples derived from 150 individuals by hybridization of bisulfite-converted DNA to Illumina bead arrays (Gibbs et al. 2010). Of these, 1084 probes mapped to the X chromosome. Samples were divided by gender, and analysis performed both for individual tissues, and by combining all four brain regions together, producing globally very similar results in each case.
For validation of array results and putative DMRs, we designed primers to amplify bisulfite-converted DNA using Methyl Primer Express v1.0 (Life Technologies). Two micrograms of genomic DNA from 45,X and 46,XX individuals was converted and purified using Epitect Bisulfite Kits (QIAGEN). Bisulfite-treated DNA was amplified using JumpStart REDTaq DNA Polymerase (Sigma-Aldrich), unincorporated primers and nucleotides were removed by incubation with Exonuclease I and Shrimp Alkaline Phosphatase (New England Biolabs), and the products were subjected to Sanger sequencing.
For validation of genes predicted to escape XCI, we performed analysis of published RNA-seq data from 30 female lymphoblastoid cell lines from the CEU HapMap population (Montgomery et al. 2010). SNP genotypes from the 1000 Genomes Project Pilot 1 were downloaded to annotate transcribed polymorphisms in X-linked RefSeq genes in each individual. Two hundred seventy-one RefSeq genes that had previously been shown to be consistently subject to XCI (Carrel and Willard 2005) were initially used to identify which of the 30 female cell lines showed completely skewed patterns of XCI. Female samples were scored as having skewed XCI based on the presence of monoallelic expression of at least three different highly expressed genes per individual that were known to be subject to XCI. Skewed XCI was defined by the observation in mRNA of only one allele of a transcribed SNP that had ≥20 overlapping reads. To allow some tolerance for sequencing errors, where ≥40 overlapping reads were present, ≥97.5% of reads were required to be derived from the same allele. By this criterion, six of the 30 females analyzed showed highly skewed patterns of XCI. Although this clonality rate of 20% is quite high, it agrees well with previous measurements in EBV-transformed lymphoblasts (Plagnol et al. 2008). For these six females that showed almost exclusive inactivation of one X chromosome, the allelic expression status of the 31 genes that had been predicted to escape XCI was measured. Biallelic expression of these genes would be indicative of escape from XCI, while monoallelic expression would indicate that the gene was subject to XCI. To enable testing of a useful number of these 31 genes in the six individuals while still maintaining statistical rigor, we chose a lower threshold corresponding to ≥10 reads overlapping a transcribed heterozygous SNP. At this threshold, eight of the 31 predicted genes could be tested. Under the null hypothesis that reads are sampled at random from either allele, this corresponds to p < 0.001 that a biallelically expressed gene would be incorrectly scored as showing monoallelic expression.
MEME (Bailey and Elkan1994) and DRIM (Eden et al. 2007) were used to search for sequence motifs enriched in CGIs that showed extreme gains or losses of methylation. For MEME analysis, all CGIs on the X chromosome were ranked based on their mean probe log2 ratio, and the top and bottom 5% tails of this distribution were tested for enriched motifs 5–20 bp in size. FIMO (http://meme.sdsc.edu/meme/fimo-intro.html) was then used to test the frequency and enrichment of the identified motifs in CGIs on the X chromosome. For DRIM analysis, motifs 6–8 bp were tested.
Data access
Array data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE22551.
Acknowledgments
We thank Professor Patricia Jacobs for supplying DNA samples from 45,X patients of known parental origin, and Dr. Mauro Delorenzi for useful discussions. The research leading to these results has received funding from the Fondation Jerome LeJeune, the European Commission Seventh Framework Program under grant agreement 219250 to A.J.S., the European Commission Sixth Framework Program under grant agreement LSH-2005-1.1.5-1 (anEUploidy), and a Swiss National Science Foundation grant to S.E.A.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.112680.110.
References
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36 AAAI Press, Menlo Park, CA [PubMed] [Google Scholar]
- Barr ML, Bertram EG 1949. A morphological distinction between neurones of male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163: 676–677 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bernardino J, Lamoliatte E, Lombard M, Niveleau A, Malfoy B, Dutrillaux B, Bourgeois CA 1996. DNA methylation of the X chromosomes of the human female: an in situ semi-quantitative analysis. Chromosoma 104: 528–535 [DOI] [PubMed] [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T 2007. CpG island mapping by epigenome prediction. PLoS Comput Biol 3: e110 doi: 10.1371/journal.pcbi.0030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Miller AP, Carrel L, Rupert JL, Davies KE, Willard HF 1995. The DXS423E gene in Xp11.21 escapes X chromosome inactivation. Hum Mol Genet 4: 251–255 [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404 [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB 1996. XIST RNA paints the inactive X-chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393: 599–601 [DOI] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, et al. 2005. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet 37: 625–629 [DOI] [PubMed] [Google Scholar]
- de Conciliis L, Marchitiello A, Wapenaar MC, Borsani G, Giglio S, Mariani M, Consalez GG, Zuffardi O, Franco B, Ballabio A, et al. 1998. Characterization of Cxorf5 (71-7A), a novel human cDNA mapping to Xp22 and encoding a protein containing coiled-coil alpha-helical domains. Genomics 51: 243–250 [DOI] [PubMed] [Google Scholar]
- Dracopoli NC, Rettig WJ, Albino AP, Esposito D, Archidiacono N, Rocchi M, Siniscalco M, Old LJ 1985. Genes controlling gp25/30 cell-surface molecules map to chromosomes X and Y and escape X-inactivation. Am J Hum Genet 37: 199–207 [PMC free article] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z 2007. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3: e39 doi: 10.1371/journal.pcbi.0030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, et al. 2010. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 6: e1000952 doi: 10.1371/journal.pgen.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, et al. 1998. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7: 737–742 [DOI] [PubMed] [Google Scholar]
- Haaf T 1995. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: Implications for methylation-associated cellular processes. Pharmacol Ther 65: 19–46 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Cranfield TK, Fjeld AD, Gartler SM 1996. Role of late replication timing in the silencing of X-linked genes. Hum Mol Genet 5: 1345–1353 [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A 2007. Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Jablonka E, Goitein R, Marcus M, Cedar H 1985. DNA hypomethylation causes an increase in DNase-I sensitivity and an advance in the time of replication of the entire inactive X chromosome. Chromosoma 93: 152–156 [DOI] [PubMed] [Google Scholar]
- Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT 2008. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet 4: e9 doi: 10.1371/journal.pgen.0040009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohane AM, Lavender JS, O'Neill LP, Turner BM 1998. Histone acetylation and X-inactivation. Dev Genet 22: 65–73 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, et al. 2010. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol 8: e1000533 doi: 10.1371/journal.pbio.1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, Chapman VM, Disteche CM 1998. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet 18: 212–213 [DOI] [PubMed] [Google Scholar]
- Lock LF, Takagi N, Martin GR 1987. Methylation of the Hprt gene on the inactive X occurs after X chromosome inactivation. Cell 48: 39–46 [DOI] [PubMed] [Google Scholar]
- Lyon MF 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning B, Jaenisch R 1996. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev 10: 1991–2002 [DOI] [PubMed] [Google Scholar]
- Plagnol V, Uz E, Wallace C, Stevens H, Clayton D, Ozcelik T, Todd JA 2008. Extreme clonality in lymphoblastoid cell lines with implications for allele specific expression analyses. PLoS ONE 3: e2966 doi: 10.1371/journal.pone.0002966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raefski AS, O'Neill MJ 2005. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet 37: 620–624 [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Migliavacca E, Dupre Y, Stathaki E, Sailani MR, Baumer A, Schinzel A, Mackay DJ, Robinson DO, Cobellis G, et al. 2010. Methylation profiling in individuals with uniparental disomy identifies novel differentially methylated regions on chromosome 15. Genome Res 20: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, James RS, Bishop DV, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA 1997. Evidence from Turner's syndrome of an imprinted X-linked locus affecting cognitive function. Nature 387: 705–708 [DOI] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642 [DOI] [PubMed] [Google Scholar]
- Tribioli C, Tamanini F, Patrosso C, Minalesi L, Villa A 1992. Methylation and sequence analysis around EagI sites: identification of 28 new CpG islands in Xq24-q28. Nucleic Acids Res 20: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Willard HF, Mukherjee S, Furey TS 2006. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol 2: e113 doi: 10.1371/journal.pcbi.0020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Willard HF, Latt SA 1976. Analysis of deoxyribonucleic acid replication in human X chromosomes by fluorescence microscopy. Am J Hum Genet 28: 213–227 [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ellison J, Salido E, Yen P, Mohandas T, Shapiro LJ 1994. Isolation and characterization of XE169, a novel human gene that escapes X-inactivation. Hum Mol Genet 3: 153–160 [DOI] [PubMed] [Google Scholar]
- Yasukochi Y, Maruyama O, Mahajan MC, Padden C, Euskirchen GM, Schulz V, Hirakawa H, Kuhara S, Pan XH, Newburger PE, et al. 2010. X chromosome-wide analyses of genomic DNA methylation states and gene expression in male and female neutrophils. Proc Natl Acad Sci 107: 3704–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]