Abstract
Emerging evidence suggests that poor glycemic control mediates post-translational modifications to the H3 histone tail. We are only beginning to understand the dynamic role of some of the diverse epigenetic changes mediated by hyperglycemia at single loci, yet elevated glucose levels are thought to regulate genome-wide changes, and this still remains poorly understood. In this article we describe genome-wide histone H3K9/K14 hyperacetylation and DNA methylation maps conferred by hyperglycemia in primary human vascular cells. Chromatin immunoprecipitation (ChIP) as well as CpG methylation (CpG) assays, followed by massive parallel sequencing (ChIP-seq and CpG-seq) identified unique hyperacetylation and CpG methylation signatures with proximal and distal patterns of regionalization associative with gene expression. Ingenuity knowledge-based pathway and gene ontology analyses indicate that hyperglycemia significantly affects human vascular chromatin with the transcriptional up-regulation of genes involved in metabolic and cardiovascular disease. We have generated the first installment of a reference collection of hyperglycemia-induced chromatin modifications using robust and reproducible platforms that allow parallel sequencing-by-synthesis of immunopurified content. We uncover that hyperglycemia-mediated induction of genes and pathways associated with endothelial dysfunction occur through modulation of acetylated H3K9/K14 inversely correlated with methyl-CpG content.
Large-scale prospective Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC), and more recently, the UKPDS trials (UK Prospective Diabetes Study (UKPDS) Group 1998; Writing Team for the Diabetes Control and Complications Trial 2002) and follow-up studies (Holman et al. 2008; Nathan et al. 2009) indicate that the establishment of tight glycemic control is critical for long-term control of diabetes. These studies signify the long-term beneficial effects of tight glycemic control in the prevention of macrovascular coronary diabetic complications, many years after the end of both original studies (DCCT and UKPDS) and after the HbA1c of the intervention group returned back to the level of the control group. These observations also emphasize that poor glycemic control—even when followed by intensive therapy—can mediate subinflammatory conditions and vascular complications. In summary, these studies support the central concept that the clinical consequences of early exposure to hyperglycemia can be far-reaching, and manifest as clinical signs in a delayed manner, a phenomenon that has been termed metabolic or hyperglycemic “memory” or the “legacy effect” (Chalmers and Cooper 2008). These intriguing observations suggest that the phenomenon of legacy could represent the ability of tight glycemic control to prevent cardiovascular complications rather than the ability of short-term glycemic spikes to cause them. The precise mechanisms that mediate hyperglycemic memory still remain controversial and not well understood. Thus, it is fundamental to elucidate the mechanism behind this effect, which could be common for both type 1 and type 2 diabetes.
Type 1 diabetes is associated with long-term vascular complications (Nathan et al. 2005) and the pathway mediating hyperglycemia-induced reactive oxygen species production is central to the activation of pro-inflammatory molecules (Pieper and Riaz ul 1997; Hofmann et al. 1998; Yerneni et al. 1999). Recent experimental data indicate that the sustained activation of inflammatory genes is associated with post-translational modifications to the histone H3 tail (El-Osta et al. 2008). Adding to the complexity of H3 tail variation is the type of modification such as acetylation or methylation. For example, in smooth muscle cells the expression of pro-inflammatory cytokines is associated with decreased gene-suppressive histone H3 lysine 9 (H3K9) methylation (Villeneuve et al. 2008), whereas gene-activating H3K4 methylation is demonstrable with increased gene expression and inversely correlated with H3K9 methylation in human vascular cells (Brasacchio et al. 2009). Experimental analyses of acetylated histones H3K9/K14 and methylated histones H3K4 indicate that transient hyperglycemia is associated with the activation of gene expression patterns (El-Osta et al. 2008). The introduction of genome-wide profiling of chromatin immunopurified H3K9m2 fragments using hybridization to microarrays was recently performed on human blood cells, providing proof-of-principle that histone methylation can be associated with the expression of pro-inflammatory genes (Miao et al. 2007). Whether the chromatin modifications exhibited by the lymphocyte population overlap the same changes in the endothelial cell that is relevant to type 1 diabetes and its complications remains unknown (Bell et al. 2010). What is emerging is a transcriptional network of remarkable complexity, and along with the general observation that chromatin contains a wealth of potential chemical variations—such as genomic methylation and histone post-translational modifications—that could be mediated by hyperglycemia. Defining the epigenetic identity of the vascular cell remains a formidable challenge (Ling and Groop 2009). Indeed, the identification of distinguishable histone modification patterns emphasizes the importance of precisely mapping genome-wide chromatin modifications to fully understand gene regulatory networks (Park 2009). The development of genome-wide approaches to investigate histone determinants and methyl-CpG content has led to significant advances in our understanding of chromatin structure and function (Schones and Zhao 2008). The application of a sensitive sequence-by-synthesis platform offers higher base-pair resolution that allows broader coverage than array-based methodologies with less signal-to-noise bias, and provides significant improvements over hybridization-based protocols (Fazzari and Greally 2010; Suzuki et al. 2010).
Current literature defines epigenetics as a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence (Berger et al. 2009). A fundamental question relevant to diabetes and its complications is whether subtle environmental cues such as transient hyperglycemia could confer gene-regulating changes. There is intense general interest in explaining long-lasting gene-regulating changes associated with diabetic complications (Katari et al. 2009; Vaquero and Reinberg 2009; Einstein et al. 2010; Thompson et al. 2010). However, the mechanism by which hyperglycemia regulates differential gene expression patterns and associated epigenomic changes in primary cell types is currently unknown. More specifically, the close correspondence of histone acetylation with that of genome-wide expression and CpG methylation states of the vascular endothelium, a primary site of dysfunction, remains unexplored. This is relevant because diabetes increases the risk of macrovascular and microvascular complications, and there is demonstrable evidence supporting the hypothesis that hyperglycemia can initiate the complex series of molecular events that result in the sustained expression of proinflammatory molecules (El-Osta et al. 2008; Brasacchio et al. 2009). To be able to study histone and genome modification in a model of hyperglycemic variability, we sought to carefully reproduce the extracellular conditions to maintain primary human aortic endothelial cells.
In this study we examined the immediate glucose-mediated transcriptional response and specifically determine histone and DNA methylation changes in primary aortic endothelial cells. We show that hyperglycemia induces changes in mRNA expression with the identification of genes involved in metabolic/cardiovascular disease, and endocrine disturbances that were associated with genome-wide changes in histone acetylation and CpG methylation. We focus on next-generation sequencing and, for the first time, illustrate the remarkable complexity of the vascular endothelial epigenome, which has not been previously examined. A close correspondence between gene expression and chromatinized acetylation was readily explained when the genomic methylation pattern was considered in response to hyperglycemia.
Results
Massive parallel sequencing reveals regionalized H3K9/K14 acetylation patterns in hyperglycemic primary human vascular cells
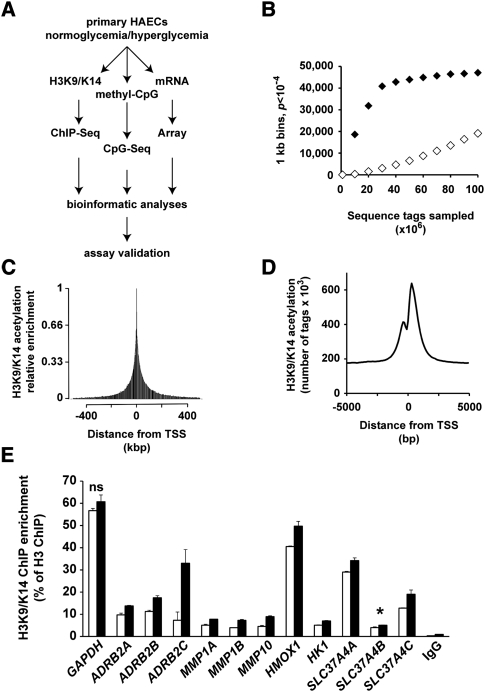
To characterize genome-wide changes in histone acetylation and CpG methylation we exposed human aortic endothelial cells (HAECs) in high-glucose (HG, 30 mM) or low-glucose (LG, 5.5 mM) conditions for 2 d, the overall strategy is illustrated in Figure 1A. Chromatin immunopurifications (ChIPs) were performed using an antibody recognizing acetylated histone H3 at lysine residues 9 and 14 (H3K9/K14). The immunopurification of soluble chromatin was tested for enrichment using qPCR at genomic regions that are consistent with the active and inactive chromatin states using GAPDH and MYOD1 as positive and negative markers, respectively (Supplemental Fig. 1A). Hyperacetylation of H3K9/K14 at GAPDH and MYOD1 genes was inversely correlated to the repressive H3K9m2 and H3K9m3 marks (Supplemental Fig. 1B). These experimental results indicate the validity in our approach, showing enrichment of H3K9/K14 acetylation on the active GAPDH gene. Next, we processed and analyzed hyperacetylated sequences of immunopurified chromatin by massive parallel sequencing using the Illumina Genome Analyzer II (GAIIx). Libraries generated from H3K9/K14 hyperacetylation ChIPs were analyzed on 16 lanes, generating >130 million 35-bp tags (Supplemental Table 1). For H3K9/K14 hyperacetylation after 50 million reads, we observed a plateau of 50,000 using the Bonferroni false-discovery rate correction (P < 10−4) (Fig. 1B).
Figure 1.
Study-power analysis and distribution of sequenced tags around the TSS of genes. (A) General schematic plans of the experiments and analyses performed in this study. (B) ChIP-seq-derived tags from independent experiments were pooled in 1-kb windows. For each 1-kb window, a Poisson test was applied to achieve P-value. (♦) H3K9/K14 acetylation LG ChIP-seq versus input; (◊) H3K9/K14 acetylation LG versus HG ChIP-seq. (C,D) The positions of all sequenced tags, pooled in 2.5-kbp and 100-bp windows, are expressed relative to the nearest genomic TSS determined on the human NCBI genome build 36.1. (C) Visualization of relative enrichment on a 1-Mbp window identifies that H3K9/K14 acetylation decreases exponentially on both sides of the TSS. (D) Visualization of relative enrichment on a 10,000-bp window centered on the TSS. (E) Validation by quantitative PCR of ChIP-seq differential tag enrichments. H3K9/K14 acetylation was monitored in 12 genomic regions from seven genes derived from independent experiments. Enrichments were normalized to H3. Specificity was assessed using IgG antibody. Differences in all analyzed results are statistically significant (P < 0.05, unpaired t-test) excluding GAPDH ([ns] not significant, P = 0.21). SLC37A4 amplicon B ([*] P = 0.057).
The acetylation of histone H3 is regionalized close to the transcriptional start site (TSS) and up to 5 kb downstream from actively transcribed genes (ENCODE Project Consortium 2007), which extends into the gene body and is associated with RNA polymerase II enrichment (Wang et al. 2008, 2009). Plotting the density of all generated sequence tags versus the entire set of RefSeq (as well as CAGE-generated) TSS positions in our experiments indicates that H3K9/K14 acetylation is predominantly regionalized (5 kbp upstream/downstream) in these genes (Fig. 1C). ChIP-seq acetylation events identified outside of the TSS correlated with alternative and/or multiple promoters, which would spread the tag enrichment around the single TSS we assigned to each RefSeq entry. Closer scrutiny of the sequence tags at transcription start sites show regional depletion of H3K9/K14 acetylation, closely resembling accessible nucleosome-free regions (Fig. 1D).
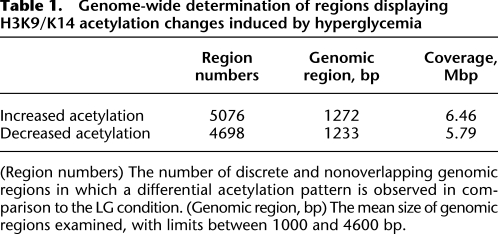
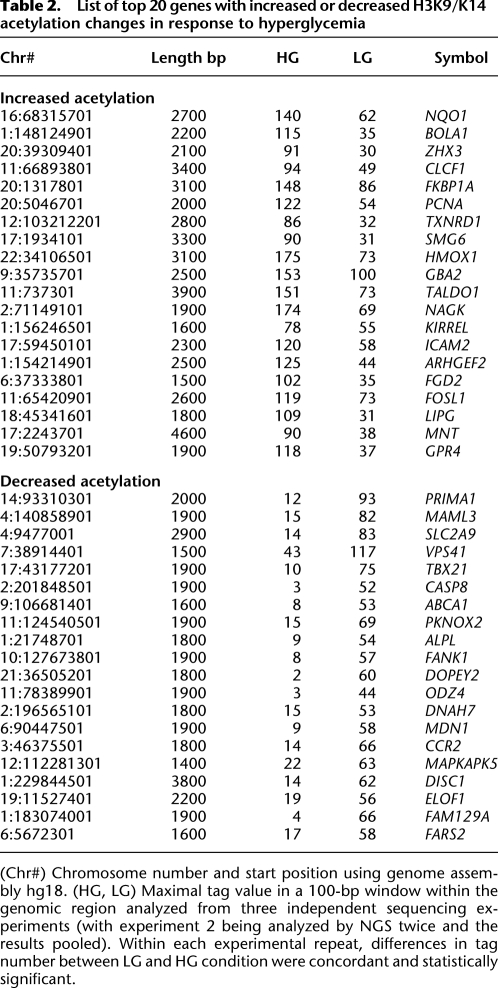
Sequence tags were combined from eight lanes of ChIP-seq from HAECs exposed to either LG and HG conditions, and H3K9/K14 enrichment was analyzed in 100-bp bins. ChIP-seq generated tags were aligned to the human genome reference hg18. We identified, in total, 41,867 regions in the genome with acetylation peaks when compared with the input data set. In response to hyperglycemia, 5076 regions showed significant hyperacetylation, while 4698 regions were classified as hypoacetylated (Table 1). Acetylation of gene sequences that did not achieve saturation suggests that deeper sequencing could reveal other histone H3K9/K14 acetylation sites (Fig. 1B, white diamonds). Western blotting of protein extracts prepared from cells exposed to high glucose was unremarkable for H3K9/K14 hyperacetylation and indicates the importance of identifying genome-wide changes to chromatin modification by massive parallel sequencing (Supplemental Fig. 1C). Table 2 classifies the highest ranked genomic regions with statistically significant acetylation changes conferred by hyperglycemia. We validated high-throughput sequencing by qPCR using the SLC37A4 gene (Supplemental Fig. 2A). Independent experiments validated increased histone acetylation on three different amplicons on this gene (Supplemental Fig. 2B). Quantification of the three aforementioned amplicons are consistent with H3K9 hyperacetylation patterns in human vascular cells (ENCODE Project Consortium 2007). Parallel qPCR analyses also confirmed hyperglycemia-induced hyperacetylation for the HMOX1 (Supplemental Fig. 2C) and ADRB2 genes (Supplemental Fig. 3B). In summary, qPCR validation of ChIP-seq regions and the close parallel to publicly available histone hyperacetylation profiles in human vascular cell types suggest that the ChIP-seq data and platform approach we have adopted provides reliable coverage of hyperglycemia-induced histone hyperacetylation in HAECs.
Table 1.
Genome-wide determination of regions displaying H3K9/K14 acetylation changes induced by hyperglycemia
Table 2.
List of top 20 genes with increased or decreased H3K9/K14 acetylation changes in response to hyperglycemia
To further qualify the technical specificity of our assays, we normalized against histone H3 as well as tested the specificity of immunopurifications with IgG antibodies (Fig. 1E). We confirm that hyperglycemia increased H3K9/K14 acetylation on the ADRB2, MMP1A, MMP1B, MMP10, and HK1 genes. The specificity of immunopurification was examined using IgG antibody (Fig. 1E) as well as no-antibody control (data not shown). These experimental results illustrate the close correspondence between qPCR and ChIP-seq enrichment for immunopurified H3K9/K14 chromatin.
Identifying genes associated with diabetes and vascular disease
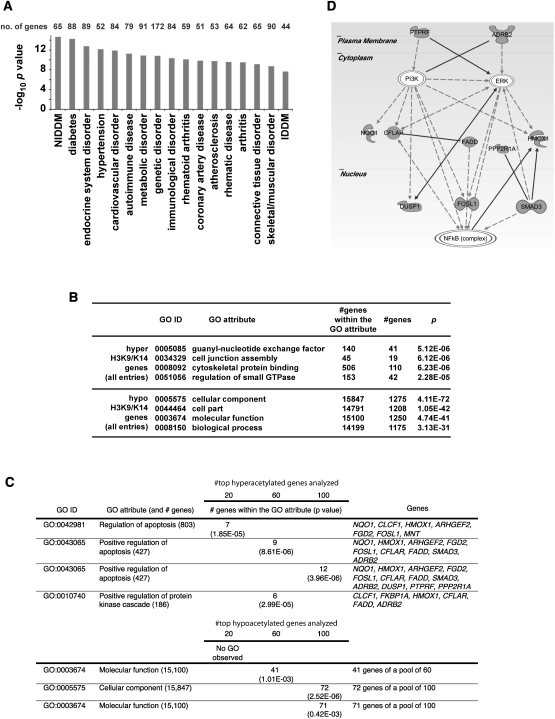
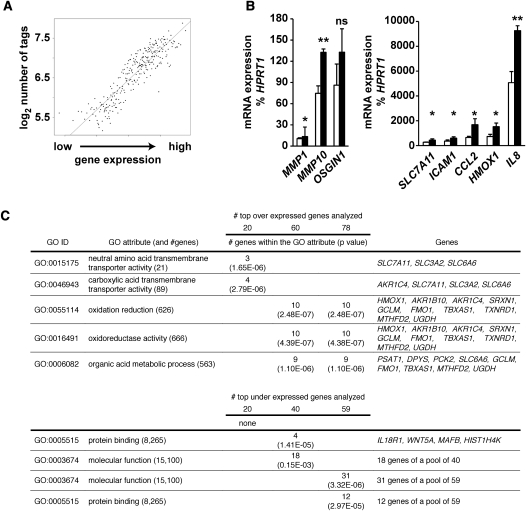
To determine the significance of histone acetylation patterns in aortic endothelial cells, we integrated ChIP-seq data with gene ontology resources. Ingenuity Pathway Analysis classified genes associated with differentially acetylated regions conferred by hyperglycemia and pathophysiology. Groups of genes were annotated to diabetes, coronary artery disease, and other cardiovascular complications (Fig. 2A; Supplemental Table 2). We examined the link between these clinical conditions associated with biological pathways by assigning gene-ontology (GO) annotations associated with hyperacetylation (Fig. 2B) and with the highest ranked genes categorized with hyper- or hypoacetylation events (Fig. 2C). GO annotations for hyperacetylated sequences identified apoptosis mediators and protein kinases, as well as ontologies related to intracellular junctions, cytoskeletal control, and GTPase-regulated signal transduction events. Notably, we identified gene sets that were associated with the regulation of apoptosis and the NF-κB pathway activation using IPA analysis (Fig. 2D). In contrast, gene ontology analysis of genes identified and associated with hyperglycemia-induced hypoacetylation was less informative, with no definitive conclusion to pathophysiology.
Figure 2.
Distribution of hyperglycemia-induced genes with differentially acetylated regions in primary human vascular cells. (A) Annotations for genes containing H3K9/K14 acetylation conferred by hyperglycemia were performed by Ingenuity System software analysis. Functional annotations are ranked according to their degree of significance with pathological condition. The number of genes contributing to the analysis is reported with 17 of the top 21 hits identified with functional or physiopathological links to hyperglycemic damage. The full list of genes is presented in Supplemental Table 2. (NIDDM) Non-insulin-dependent diabetes mellitus; (IDDM) insulin-dependent diabetes mellitus. (B) Gene ontologies (GOs) have been defined for H3K9/K14 hyperacetylation and hypoacetylation. For each GO association, the number of genes within a GO attribute and the number of observed genes is reported. (C) Hyperglycemia determined GOs have been ranked and defined for 20, 60, and 100 genes associated with genomic regions displaying the most significant increase or decrease in H3K9/K14 acetylation. (D) Representative Ingenuity Pathway Analysis map generated for the subset of hyperacetylated genes conferred by hyperglycemia representing the GO attribute “Positive regulation of apoptosis.”
Connectivity map analysis of hyperglycemia-induced histone hyperacetylation predicted that histone deacetylase inhibitors could increase H3K9/K14 patterns (Lamb et al. 2006). We used suberoylanilide hydroxamic acid (SAHA) to determine whether this HDAC inhibitor could also confer hyperacetylation patterns. Supplemental Figure 3 shows that HAECs exposed to 20 μM SAHA for 12 h increased ADRB2 gene expression, which is consistent with histone hyperacetylation by hyperglycemia. This close correspondence between glucose response and H3K9/K14 acetylation indicates that pharmacological inhibition by histone deacetylases may provide some benefit in preventing hyperglycemic damage and endothelial dysfunction.
Histone hyperacetylation reveals key transcriptional events associated with hyperglycemia
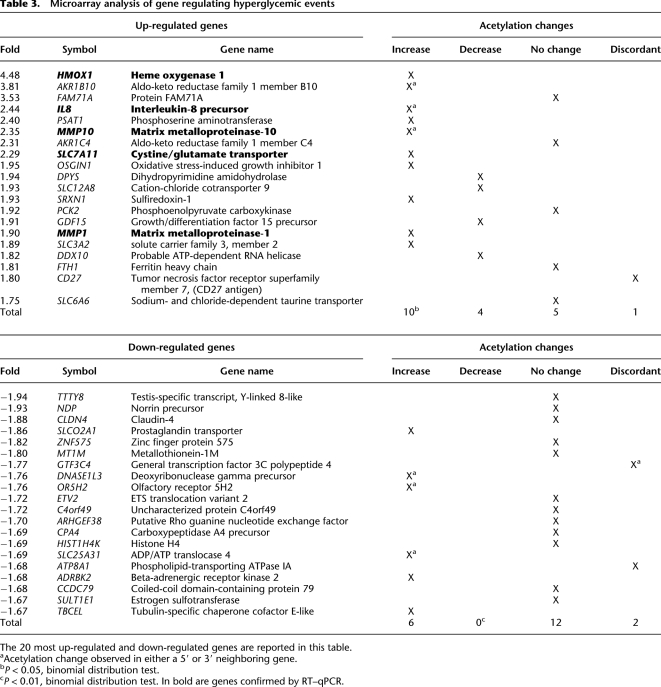
Since histone H3K9/K14 acetylation is known to associate with active genes (Heintzman et al. 2009), we examined mRNA expression from the same primary endothelial cells on Illumina human WG6 arrays. The experimental results show a tight association between mRNA abundance and histone acetylation (Fig. 3A). Analyses revealed that 137 annotated genes were differentially expressed (threshold fold change of 1.5), and of those, 78 genes were up-regulated and 59 were down-regulated in response to hyperglycemia (Table 3; Supplemental Table 3). The majority of up-regulated genes have a defined role in the physiopathology of diabetes, including diabetic complications. Gene-ontology analysis also identified a subset of hyperglycemic responsive genes involved with transmembrane amino acid transport and oxido-reductive cellular processes. Highly expressed genes included HMOX1, IL8, MMP10, SLC7A11, and MMP1 and were validated using RT–qPCR in independent experiments (Fig. 3B). Real-time–qPCR was also performed to monitor known glucose-responsive genes, including CCL2, also known as monocyte chemoattractant peptide 1 (MCP-1) and intracellular adhesion molecule-1 (ICAM1). We confirmed that hyperglycemia regulated CCL2 and MCP-1 as previously described (Piga et al. 2007). In contrast, no specific gene-ontology functions were identified within the subset of down-regulated genes (Fig. 3C).
Figure 3.
Relationship between mRNA expression and histone acetylation with gene ontologies in primary human vascular cells. (A) Genes were pooled according to expression in groups of 20 and the number of tags from ChIP-seq experiments (−1 to +1 kb TSS) was calculated. (B) Validation by quantitative real-time PCR of mRNA expression changes mediated by hyperglycemia. Values are displayed as % expression of the housekeeping gene HPRT1, which does not change significantly in response to hyperglycemia. Empty bars represent LG-treated HAECs; black bars represent HAECs exposed to HG. (*) P < 0.05, paired t-test; (**) P < 0.01, paired t-test; (ns) not significant. n ≥ 3. Quantitative real-time PCR reactions for each independent replicate were performed in duplicate. (C) Gene ontologies defined for the subset of up-regulated (20, 60, and 78) genes and down-regulated (20, 40, and 59) genes mediated by hyperglycemia.
Table 3.
Microarray analysis of gene regulating hyperglycemic events
Distinguishable H3K9/K14 acetylation and DNA methylation patterns in response to hyperglycemia
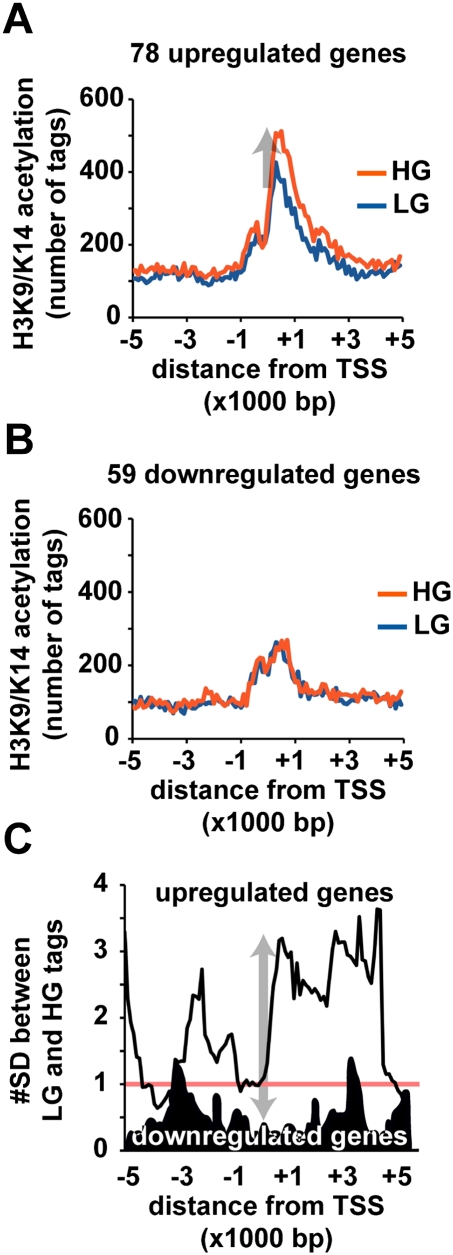
To investigate the association between hyperglycemia-induced gene-activating hyperacetylation events, we analyzed the cumulative number of tags, within the 100-bp region windows for the 78 up-regulated and 59 down-regulated genes. Hyperglycemia increased the number of tags for up-regulated genes (Fig. 4A). In contrast, no acetylation differences were identified for down-regulated loci (Fig. 4B). The significance of these differences were analyzed and indicate the HG/LG ratio for the 78 up-regulated genes is over two standard deviation differences in the 10-kbp window, whereas for the 59 down-regulated genes, the average ratio was below one standard deviation (Fig. 4C). These data demonstrate correlation between histone hyperacetylation and increased gene expression (Table 3).
Figure 4.
Hyperglycemia confers genome-wide H3K9/K14 acetylation patterns close to the transcription start site of promoters in human vascular cells. (A) Genomic positions of sequenced tags are grouped in 100-bp windows relative to the 78 genes with expression values >1.5-fold. (B) Analysis of 59 down-regulated genes with expression values >1.5-fold. Plots are expressed relative to transcription start sites determined on the human NCBI genome build 36.1. The cumulative number of tags within each 100-bp window is plotted relative to the transcription start site. (Blue line) LG exposed HAECs; (red line) HG exposed HAECs. (C) Sliding window calculation demonstrates significant difference between HG and LG traces relative to the pool of up-regulated genes (light trace) and down-regulated genes (dark trace).
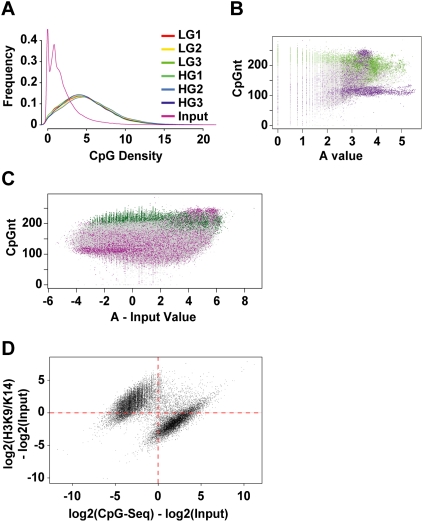
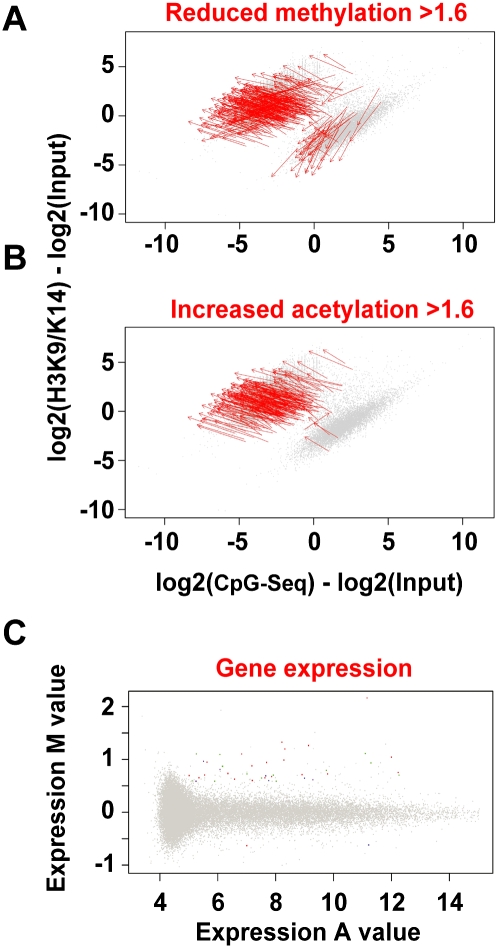
DNA methylation is a major determinant of gene suppression, which is inversely associated with histone hyperacetylation (El-Osta and Wolffe 2000). To examine the role of hyperglycemia-induced DNA methylation changes, we used methyl-capture and CpG-seq procedures. Enrichment of sequences was determined by calculating the number of CpGs in 300-bp bins (Statham et al. 2010). We identified fivefold enrichment in CpG counts in reads when compared with input DNA, which was consistent across all samples (Fig. 5A). In addition, we analyzed the enrichment of methylated CpG Islands and mapped repeat sequences using a M versus A plot (Dudoit and Fridlyand 2002). The distribution of mapped methylated DNA in these experiments shows enrichments for CpG Islands (shown in green) and repeat sequences (shown in purple) (Fig. 5B). Because repeat sequences were represented in the input DNA as well, these were omitted from the overall analyses (Fig. 5C). Histone acetylation is inversely correlated with genome methylation (Jones et al. 1998; Nan et al. 1998). To test this prediction we plotted hyperacetylation derived by ChIP-seq against uniquely mapped CpG-seq reads. We identified a distinct distribution of histone acetylation (y-axis) and CpG methylation (x-axis) with exclusivity for hyperacetylated and unmethylated sequences (upper-left quadrant) contrasted with CpG methylation and hypoacetylated regions (lower-right quadrant) (Fig. 5D). The Fisher exact probability for this distribution is estimated at P = 10−75.
Figure 5.
Distribution of CpG-seq tags in human vascular cells. (A) Enrichment of CpG sequences determined by comparing methyl-capture against the input. Shown here are three experiments of libraries prepared for CpG-seq derived from vascular endothelial cells exposed to low-glucose (LG) or high-glucose (HG) conditions. (B) Distribution of CpG-seq-enriched DNA with respect to C+G counts in 300-bp bins. Green indicates reads localized to CpG islands and purple indicates reads mapping to repeat sequences. (C) Representation of methylated DNA distributed between CpG islands (green) and repeat sequences (purple). Read counts are normalized for 300 bp and subtracted from the same count of input samples. (D) Relationship between the normalized and input-subtracted values of histone acetylation (y-axis) and CpG methylation (x-axis). The plot shows a mutually exclusive distribution of hyperacetylated and unmethylated regions (upper left) distinguished from methylated CpGs and hypoacetylated sites (lower right). The Fisher-exact probability for this quadrant distribution shown by the red dotted axes is estimated at P = 10−75.
Hyperglycemia confers distinguishable acetylation and methylation changes
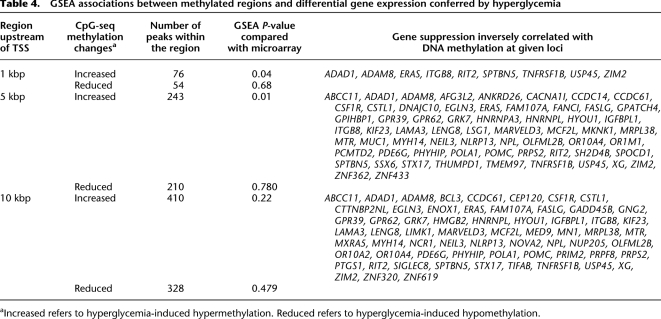
Exposure of vascular endothelial cells to hyperglycemia clearly distinguished methylated sequences by gene set enrichment analysis (GSEA). Hyperglycemia was associated with hypermethylation changes localized to regions close to transcription start sites (∼5 kbp). Interestingly, hyperglycemia was not generally associated with increased gene expression and genomic hypomethylation (Table 4). Our analyses integrated gene expression and changes in histone acetylation and genomic methylation in response to hyperglycemia. First, we distinguished regions in the genome with reduced methylation values >1.6-fold. Figure 6A illustrates that the changes conferred by hyperglycemia are associated with reduced genomic methylation (red arrows) and are inversely correlated with H3K9/K14 hyperacetylation (Meissner et al. 2008). Next, we determined whether this change was also consistent with increased histone hyperacetylation. Figure 6B shows that the great majority of genes with increased acetylation were inversely correlated with CpG methylation. Integrating methylation and acetylation data indicates that these changes could regulate gene expression. When analyzed in this way, a significant fraction of gene expression changes reported in Figure 3 were also associated with acetylation and methylation (Fig. 6C). Collectively, the data presented demonstrate that hyperglycemia mediates gene-activating epigenetic changes that are associated with increased hyperacetylation, which are distinguishable from DNA methylation and gene suppression.
Table 4.
GSEA associations between methylated regions and differential gene expression conferred by hyperglycemia
Figure 6.
Hyperglycemia confers distinguishable gene-activating histone hyperacetylation and DNA methylation changes in the human vascular cell. (A,B) Red arrows represent regions in the genome with changes in values >1.6-fold mediated by hyperglycemia. (A) Decreased CpG-seq counts mediated by hyperglycemia. (B) Increased acetylation ChIP-seq counts conferred by hyperglycemia. Genes were associated with increased CpG methylation (green), decreased methylation (purple), increased acetylation (red), and decreased acetylation (blue). (C) Integration of H3K9/K14 hyperacetylation and CpG methylation associated with gene expression changes.
Constructing the epigenomic landscape of the primary vascular cell
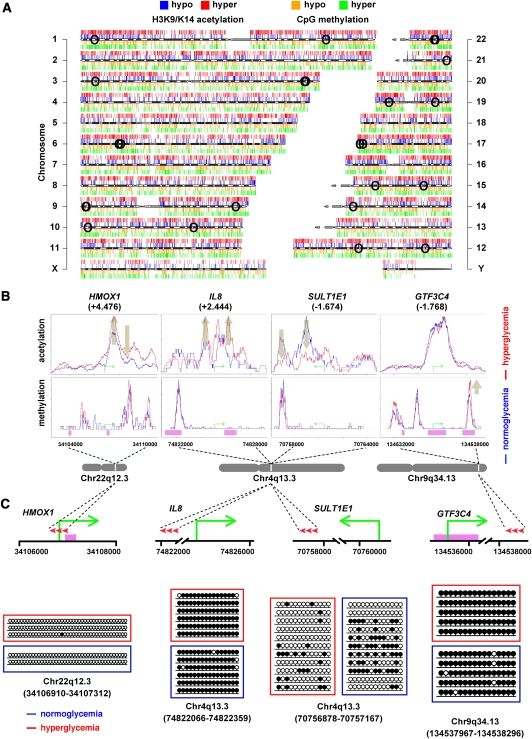
Genome-wide association of histone acetylation and genomic methylation in vascular cells has not been previously examined. A major conclusion of these experiments is that subtle environmental cues such as hyperglycemia are sufficient to drive gene-activating changes in the aortic endothelial cell. Hyperacetylation is tightly correlated with gene expression; our results also demonstrate that hyperglycemia could confer remarkable genome-wide methylation changes that were inversely correlated with histone hyperacetylation. The relationship between genome-wide correlations of chromatin modifications were examined and mapped on the human genome (Fig. 7A). We assessed reduced (hypo-) and increased (hyper-) H3K9/K14 acetylation, which are mapped above each chromosome and shown in blue and red, respectively. Corresponding changes were also charted for genomic methylation as illustrated below each chromosome and shown as orange (hypomethylation) and green (hypermethylation) regions. We observe broad changes to H3K9/K14 acetylation and genomic methylation; specifically, we identify regionalized hyperacetylation coexisting with hypomethylation in response to hyperglycemia. The evidence for this has come from several key observations using massive parallel sequencing. First, we observed good inverse correlation between genomic methylation and histone modification associated with gene expression, and this robustness has been described previously for embryonic stem cells (ES) and ES-derived cells (Meissner et al. 2008). In general, hyperglycemia confers DNA methylation changes associated with increased H3K9/K14 acetylation. This is exemplified on chromosome 22q12.3, with increased acetylation correlated with distinct methylation patterns mapped to the HMOX1 gene (Fig. 7B). Second, sequence context was not always a predictor of CpG Island methylation, and this is also illustrated clearly with HMOX1 (CpG Islands shown in pink). Reduced H3K9/K14 acetylation was often inversely associated with increased genomic methylation peaks outside of the CpG Islands. For example, the expression of the genome at q13.3 of chromosome 4 is distinguished by CpG Island methylation outside of the IL8 promoter with robust hyperacetylation in response to hyperglycemia (+2.444-fold increased in expression). In contrast, reductions in H3K9/K14 acetylation are tightly associated with SULT1E1 gene suppression. As noted above, massive parallel sequencing identified DNA methylation changes often outside of the promoter regions and CpG Islands, and this was sampled at enriched sites of H3K9/K14 acetylation such as GTF3C4 (−1.768-fold reduction in expression). Transient hyperglycemia enriched genomic methylation levels, which were positively associated with gene suppression outside of the CpG Islands of the GTF3C4 promoter. Another clear feature identified are the large domains of hyperacetylation which are inversely correlated with genomic methylation content. To validate identified CpG-seq tags, we used bisulfite sequencing of HMOX1 (position chr22:34106910–34107312, region size 403 bp), IL8 (position chr4:74822066–74822359, region size 294), SULT1E1 (position chr4:70756878–70757167, region size 290), and GTF3C4 (position chr9:134537967–134538296, region size 330) shown in Figure 7C. The absence of CpG-seq tags at the HMOX1 transcription start site agrees with independent bisulfite sequencing results showing hypomethylation of the region (Chr 22q12.3). Bisulfite sequencing confirmed hypermethylation of IL8 at position chr4:74822066–74822359 (Fig. 7C). Also consistent was the hypomethylated region at the TSS (amplicon size 359 bp) of the IL8 promoter (position chr4:74825033–74825391) in both LG and HG groups (data not shown). We validated the presence of CpG-seq tags upstream of the SULT1E1 (Chr 4q13.3) promoter using bisulfite sequencing, showing moderate methylation levels correlating with intermediate tag counts identified by CpG-seq. We determined that CpG-seq tags for GTF3C4 and IL8 were in agreement with bisulfite sequencing results demonstrating hypermethylation of the analyzed regions. In separate experiments, bisulfite sequencing confirmed the hypomethylated CpG-rich region at the TSS (amplicon size 304 bp) of the GTF3C4 promoter (chr9:134535510–134535813) in primary human vascular cells (data not shown). Experimental validation for CpG-seq data using bisulfite sequencing for these genes is summarized in Figure 7C.
Figure 7.
Genome-wide changes conferred by hyperglycemia in the human vascular cell. (A) Human ideogram illustrating the correlation between histone acetylation and the methylome mediated by hyperglycemia. ChIP-seq tracks for hypo- (blue) and hyper- (red) acetylated histone H3K9/K14 are shown above each individual chromosome. CpG-seq tracks for hypo- (orange) and hyper- (green) methylated CpG sequences are shown below each chromosome. (○) Genome localities of individual SNPs associated with diseases listed in Table 5. (B) Mapping specific hyperglycemia-induced histone acetylation and genomic methylation signatures associated with changes in gene expression. H3K9/K14 acetylation and CpG methylation signatures are shown for normoglycemia (blue) and hyperglycemia (red) relative to the transcription start site shown by a green arrow for HMOX1, IL8, SULT1E1, and GTF3C4 genes. Pink regions represent CpG Islands. (C) Experimental validation of CpG-seq tags using bisulfite sequencing of HMOX1 (position chr22:34106910–34107312, region size 403 bp), IL8 (position chr4:74822066–74822359, region size 294), SULT1E1 (position chr4:70756878–70757167, region size 290), and GTF3C4 (position chr9:134537967–134538296, region size 330). (•) Methylated CpG sites; (○) unmethylated CpG sites. Red arrowheads represent the relative positions of bisulfite sequenced amplicons.
Intersecting GWAS with chromatin modifications and pathophysiologies
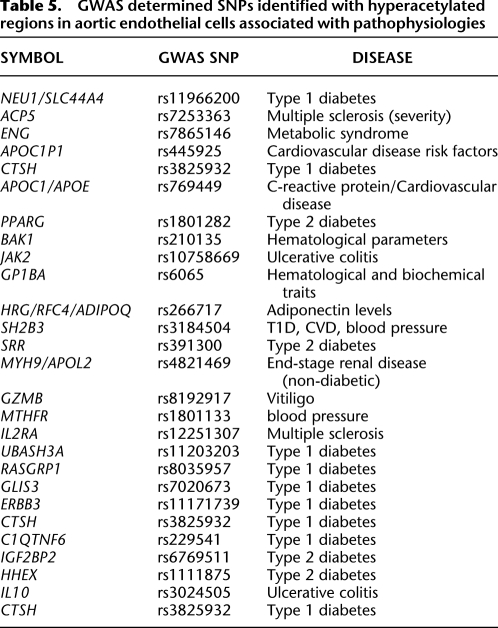
We identified locations in the genome that are implicated with predisposition to diabetes-related disease. The NHGRI GWAS catalog lists 4562 SNPs associated with different human traits and diseases, of which 4270 overlapped with glucose-induced H3K9/K14 acetylation sites. The SNPs that overlapped with histone acetylation were ranked by distance to the nearest loci, and gene set enrichment analysis identified diabetes and other related pathophysiological diseases. The frequencies of H3K9/K14 acetylation in gene promoters contained within linkage disequilibrium intervals of the human germ-line variation sequences associated with disease predisposition were determined and are illustrated in Figure 7A (the location of SNPs are shown as open circles on the chromosomes). Twenty-seven disease-associated SNPs are located in promoters with H3K9/K14 hyperacetylation, and 16 were associated with metabolic syndrome, T1D, T2D, or CVD (Table 5). These represent 7% of the 228 SNPs reported for these traits in the GWAS catalog. This enrichment of disease-associated SNPs are associated with chromatin modifications in response to extracellular glucose (Macintyre et al. 2010).
Table 5.
GWAS determined SNPs identified with hyperacetylated regions in aortic endothelial cells associated with pathophysiologies
Discussion
When the groups of Roy and Lorenzi had originally described that chromatinization of the genome might confer gene-activation events mediated by hyperglycemia, the significance of these findings and the investigator's original hypotheses were underappreciated (Roy et al. 1990). More than two decades later, research examining the fundamental transcriptional changes associated with glucose and expression of the genome have identified some key aspects associated with chromatin modifications (Miao et al. 2007, 2008; El-Osta et al. 2008; Villeneuve et al. 2008; Brasacchio et al. 2009). However, the genome-wide association of these changes with histone acetylation and CpG methylation still remains poorly understood. In this article we describe three sets of experimental data which demonste that transient hyperglycemia per se confers distinguishable chromatin modification. First, we observe specific transcriptional changes associated with diabetes and vascular complications. For example, genes for the signaling molecules HMOX1, IL8, and MMP10 are rapidly transcribed following hyperglycemia, and this response to subtle and transient stimulatory cues is closely associated with histone acetylation. Second, gene expression is often associated with hyperacetylation of histone H3K9/K14 within the promoter of genes. For example, the H3K9/K14 acetylation content of sequences for HMOX1, IL8, and MMP10 were highly responsive to glucose, which is consistent with gene expression changes. The retention of hypoacetylated sequences were often mapped to areas of methyl-CpG content within immediate promoters, which also appeared in surrounding genomic regions. Third, we exemplify the technology demonstrating the validity of distinguishable chromatin signatures; illustrating that the appearance of H3K9/K14 acetylation is predictive of unmethylated genomic sequences. The evidence for this came partly from ex vivo experiments of vascular endothelial cells and transient glucose exposure, identifying genes that were rapidly transcribed. Indeed, our ability to distinguish specific histone and genomic modifications allowed us to identify several properties previously undescribed: (1) An inverse correlation emerges with distinguishing histone acetylation peaks mapped to reduced methyl-CpG content on the HMOX1 locus, and (2) these tightly regulated marks are tailored for HMOX1 gene expression and show the greatest changes in H3K9/K14 acetylation in regions with a dynamic shift in CpG methylation. The findings presented here indicate a remarkable complexity of chromatin change in response to hyperglycemia, with distinct associations explaining altered gene transcription. The data presented also highlight a diversity of chromatin modifications regulating gene expression, indicating that gene-activating H3K9/K14 acetylation events can act independently of methylation at promoter sites such as IL8 on chromosome 4q13.1. In contrast, histone acetylation patterns are reduced on the suppressed SULT1E1 locus also located on 4q13.1. These examples indicate that distinguishable modifications conferred by transient hyperglycemia can explain differential gene regulation.
Building the first installment of a hyperglycemic atlas of the vascular epigenome
The nature of the sugar stimulus is interesting because it is either too weak or transient to result in full activation of target genes (Table 3); nonetheless, our results indicate that hyperglycemia is indeed associated with gene-activating hyperacetylation events. How vascular endothelial cells interpret specific environmental cues with respect to prior exposure and current contextual conditions is not well understood. We propose that hyperglycemia could drive vastly different transcriptional responses, which are also influenced by extracellular concentration (dose dependency). This signaling intensity is demonstrable and strikingly reminiscent of recent experimental results, with primary human aortic endothelial cells exposed to increasing glucose concentrations, and is associated with gene-activating histone methylation events (El-Osta et al. 2008). Clearly, this explains in part the vascular complexity (St Croix et al. 2000) that is associated with transcriptional plasticity, because the intensity of gene activation is also a determinative of different signaling cues. This is indeed the case for stronger extracellular signals that are either proinflammatory or antiproliferative, such as LPS-lipopolysaccharides (Zhao et al. 2001) and TGFB (Benton et al. 2009). For example, human aortic endothelial cells exposed to LPS demonstrate a stronger transcriptional response with greater intensity than that compared with transient hyperglycemia (A Balcerczyk and A El-Osta, unpubl.). These results provide evidence that endothelial cells do perceive short glucose exposure, evidenced by chromatin modifications and regulation of distinct gene expression signatures. Taken together, we suggest that endothelial cell dysfunction and the complications associated with hyperglycemia can be partly explained by altered changes in gene expression conferred by modifications to chromatin. The contrasting histone acetylation and DNA methylation changes regulating the expression of genes are associated with atherogenic effects and vascular disease. In the context of these modifications it will be interesting to determine the significance of different histone modifications to further expand the molecular basis of hyperglycemia (Meissner et al. 2008; Araki et al. 2009).
Vascular genomic sequences are epigenetically distinguishable
Chromatin structure and function control critical nuclear events, and the results presented here demonstrate that the genome is indeed distinguishable, identifying disturbances in pathophysiology. In the case of vascular complications, recent evidence indicates clear association between gene-activating histone modifications with the expression of proinflammatory molecules. This close correspondence between diabetes and inflammation can be partly explained by histone acetylation of lysine residues 9 and 14 and is demonstrable on the tumor necrosis factor (TNF) and prostaglandin-endoperoxide synthase 2 (PTGS2) genes in blood monocytes (Miao et al. 2004). Transcriptional changes derived after hyperglycemia are not just a consequence of culture conditions of blood monocytes, because vascular endothelial cells also demonstrate persistent gene-activating epigenetic changes (El-Osta et al. 2008). Furthermore, understanding the transcriptional changes of blood cells may hold little predictive value in defining the chromatin modifications of the vascular cell.
The vascular cell is an ideal model system for studying the complex series of proatherogenic events because there is a striking mechanistic commonality among chromatin modifications in the aortic vessel, which parallels the cultured vascular cell. Three key findings follow this precedent; first, experiments on aortic and microvascular endothelial cells have shown that ambient hyperglycemia can induce changes in the expression of proinflammatory molecules. Second, it was possible to induce transient hyperglycemia in nondiabetic mice using a hyperinsulinemic clamp to show gene-activating epigenetic changes in isolated aortic endothelial cells. Finally, in a chronic model of diabetes-associated atherosclerosis, proinflammatory molecules are significantly increased and are featured by plaques (El-Osta et al. 2008; Brasacchio et al. 2009). While these individual studies are not demonstrative of the generality of chromatin changes, taken together they are consistent with the role of gene-activating proatherogenic events. In the context of genome-wide mapping of chromatin determinants, these earlier studies were limited in scale and examinination of independent transcriptional events with specific temporal cues (Brasacchio et al. 2009). In contrast, in this article we used genome-wide sequencing to show a more comprehensive characterization of differentially enriched histone hyperacetylation signatures mediated by hyperglycemia that are inversely associated with methyl-CpG content. The application of massive parallel sequencing and comparative approaches allows for genome-wide identification of chromatin modifications, and, although ChIP and CpG sequencing approaches are affinity-based assays dependent on antibody-antigen recognition, there are critical differences that could explain the discordance in sensitivity identifying histone hyperacetylation and CpG methylation in the pathological process. One distinguishing feature relevant in this study is the biochemical separation, capture, and stepwise salt-elution of double-stranded DNA with varying CpG methylation densities. Affinity capture of methylated CpG sequences under these conditions provides a specific and important snapshot of methylation patterns in the primary endothelial cell. Well-studied in different disease contexts, but of unknown function in diabetes, DNA methylation changes are associated with a complex pattern of differential gene regulation. We identified genome-wide correlations between gene expression with histone acetylation and CpG methylation. The complexity of these changes in response to glycemic variability shows a fine balance of gene-activating changes (positive correlation with acetylation and negative correlation with CpG methylation) in vascular endothelial cells. The diverse degree of methylation changes are either local or specific for CpG sites, while broader methylation changes are involved with distinct signal intensities that contribute to overall transcriptional responses. For example, analysis of CpG sequences that contain at least 20 reads identified 42 CpG Islands with almost twofold reduction. These results indicate that demethylation is also likely to be associated with distinct transcriptional responses mediated by hyperglycemia that direct endothelial cell function. Further studies will be necessary to directly test the role of genomic demethylases in the regulation of transcriptional events critical for mediating extracellular responses such as high glucose.
Establishing the H3K9/K14 acetylation signature of hyperglycemic damage
Genomic regions with high amounts of hyperacetylation in hyperglycemic cells represent 0.3% of the genome. When we scanned across TSS regions (2 kb upstream and 5 kb downstream), hyperacetylation represented <10% of RefSeq genes contributing ∼3.5%–5.4% of acetylation changes. These results are in agreement with the functions of genes identified, which include pathophysiology of diabetic disease and involvement of NF-κB signaling pathways.
Hyperactylation downstream from the TSS is associated with actively transcribed genes (Wang et al. 2008, 2009). To explore the possibility that acetylation changes occur within promoters other than RefSeq, we screened the genome and found that of the 5076 regions that exhibited increased amounts of histone acetylation in hyperglycemic cells, 4595 regions overlapped with the Fantome 4 CAGE promoter data (which has 3.7 million empirically determined TSS sites). While the expected intersection between the hyperacetylated intervals in the genome and the CAGE-TSS is expected to be 0.002%, we observed an intersection of 89%. This represents a remarkable enrichment of 41,000-fold of transcription start sites (CAGE), indicating the contribution of hyperacetylation at transcription start sites.
Ingenuity Pathway Analysis identified gene subsets to pathological conditions such as type 1 and type 2 diabetes, cardiovascular disease, and metabolic disorders, indicating hyperglycemia, per se, with the clinical condition. Gene ontology (GO) analysis of up to 100 of the most hyperacetylated regions identified genes involved in the regulation of apoptosis and protein kinases. In particular, the GO annotation to apoptotic response recapitulates the proapoptotic effect of hyperglycemia on multiple vascular cell types (Tomas et al. 2002). Expansion of the same analysis of hyperacetylated regions indicate novel processes leading to hyperglycemic damage, including small GTPases dependent signaling pathways, cytoskeletal alterations, and dysfunctional cell-junction assembly. In a similar manner, gene ontology analysis performed on the set of transcriptionally active genes (values >1.5-fold) show increased expression of amino acid transporters and enzymes implicated in oxidoreductive processes that are indicative of mitochondrial oxidative stress and hyperglycemic damage (Brownlee 2001). Gene ontology analyses of hypoacetylated genes suggest that deacetylation and transcriptional suppression may be less important in the response to hyperglycemic damage. This hypothesis is strengthened by the observation that hyperglycemia-induced gene activation events are strongly correlated with increased H3K9/K14 acetylation at transcription start sites, and this is clearly distinguished from events associated with the suppression of gene expression that is not correlated with H3K9/K14 deacetylation.
The complexity of transcriptional regulation in the human cell is exquisite not only because of the multiplicity of post-translational modifications to histone tails that underlies the covalent language, which we now collectively refer to as the histone code (Strahl and Allis 2000). In fact, it is because of the intricate signatures that segregate regulatory determinants, which recognize these chemical variations into chromatinized substrates, that investigators have become interested in examining and distinguishing these in models of diabetes (Cooper and El-Osta 2010). Previous experiments performed in our laboratory indicate that hyperglycemia can mediate specific and persistent chromatin changes in the vascular endothelial cell (El-Osta et al. 2008). Strikingly, prior hyperglycemia is associated with gene-activating marks linked with histone methylation (Brasacchio et al. 2009). In this context, perhaps the most striking mechanistic parallel described by recent studies in the literature is the experimental and clinical observation that hyperglycemia can induce changes such as histone methylation (Yerneni et al. 1999; Villeneuve et al. 2008). Taken together, the results of our studies and this first installment suggest that hyperglycemia confers gene-activating events in the vascular endothelium, indicating chromatinization is distinguishable, and confirming that the associations for acetylation and methylation can be gene specific and may serve as a basis for endothelial cell dysfunction.
Methods
An expanded Methods section is available as a Supplemental section.
Cell culture
Primary HAECs (human aortic endothelial cells) from Lonza were cultured in EBM-2 medium (Lonza) containing EGM-2 growth factors and supplements (Lonza) and 10% fetal bovine serum (GIBCO). HAECs were between passages 3 and 6 for these experiments. Glucose response was analyzed in cells exposed for 48 h with medium containing 30 mM high glucose (HG) or 5.5 mM low glucose (LG). Mannitol did not change the expression of CCL2, ICAM1, and IL8 genes.
Isolation of hyperacetylated histones using chomatin immunoprecipitation (ChIP)
ChIP on ∼1 million cells was performed as described in the Supplemental Methods.
Enrichment of differentially methylated regions
Methylated DNA from HAECs was recovered using MethylMiner (Invitrogen) as described recently (Gebhard et al. 2006; Brinkman et al. 2010; Harris et al. 2010; Nair et al. 2011).
ChIP-seq and CpG-seq
ChIP-seq libraries were prepared according to the protocols described in the Illumina ChIP-seq library preparation kit with some modifications. Briefly, 15 ng of immunopurified DNA or 30 ng of genomic DNA from an input sample was end-repaired, followed by the 3′ addition of a single adenosine nucleotide and ligation to universal library adapters. Ligated material was separated on a 2.0% agarose gel, followed by the excision of a 250–350-bp fragment and column purification (QIAGEN). DNA libraries were prepared by PCR amplification (18 cycles). Column purified libraries were quantified fluorometrically and visualized by MultiNA electrophoresis for quality control (Shimadzu). Libraries were diluted to 10 nM and stored at −20°C prior to cluster generation and sequencing on a GAIIx, performed according to the manufacturer's protocols (Illumina). Sequencing depth ranged from ≈5 × 106 to ≈2 × 107 informative sequence tags per lane (i.e., 35-bp reads that could be uniquely aligned on the human genome, Supplemental Table 1). ChIP-seq reproducibility among different lanes analyzing the same experimental condition was demonstrated by principal component analysis, in which the LG and HG conditions segregated separately (data not shown). Reproducibility was also demonstrated by scatter plots of three independent repeats of LG and HG exposure, which displayed a correlation coefficient, (r), of 0.907 and 0.910, respectively (data not shown). CpG-seq data analysis is described in the Supplemental Methods section. Sequencing images were processed through Pipeline 1.4 for signal extraction, base calling, and genome alignment of the human genome (annotation Hg18, NCBI36), using the Burrows-Wheeler Aligner (Li and Durbin 2009). Sorted text files representing uniquely mapped reads (tags) passing filter were used for downstream peak-finding analysis and statistical comparison between groups.
To identify regions significantly enriched for acetylated H3K9/K14 (Table 1), we used a sliding window approach (Guttman et al. 2009). The genome was divided into nonoverlapping windows (100 bp) and the number of tags was counted. The distribution of tags within windows is found to be a Poisson distribution with λ (expected number of occurrences within a window) equaling the mean genome background (the number of reads in the library divided by the number of nonoverlapping windows). We applied a Poisson distribution using the genome background as the null model to determine specific thresholds for statistical significance (set at P < 10−4). Contiguous windows below the threshold significance were collated, accounting for the different size of region lengths observed (Table 2). Calculations used were recently described in Kowalczyk et al. (2011).
RNA isolation, cDNA synthesis, and RT–qPCR
Total RNA was isolated using TRIzol (Invitrogen). Genomic DNA contamination was removed by DNAse treatment (QIAGEN). First-strand cDNA synthesis was performed using a high-capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. PCR amplification was performed using an Applied Biosystems 7500 Fast Real-Time PCR System. A total of 5 pmoles of forward and reverse primer, cDNA template, and FAST SYBR Green Master Mix (Roche) were added. Reactions were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. Analyzed genes were normalized against HPRT1.
Microarray hybridization and bioinformatic analysis
These procedures are described in the Supplemental Methods.
Bisulfite sequencing
Bisulfite conversion was performed as recently described in Harikrishnan et al. (2010). In brief, genomic DNA was converted using the EZ DNA Methylation Kit (Zymo Reserch). One microgram of DNA was used in the conversion reaction, and the DNA was eluted from the carrier by heating to 60°C in Tris-EDTA buffer and stored at −20°C prior to PCR amplification. Competent cells were supplied by Invitrogen (ONE Shot top 10 F′ kit). DNA was cloned using the topoisomerase TA vector (Invitrogen), and blue colonies were selected and grown in LB broth. Plasmid was purified using a miniprep kit (Promega), and DNA was sequenced using ABI PRISM Big dye 3.1 Terminator (PE-Applied Biosystems). Primers used are the following: HMOX1_fwd_BS AGATTTTGTTTTTTAAGGGTTATAT and HMOX1_rev_BS CAAAACCCAAATTTCTAACTC (amplicon size 403 bp), IL8_Peak_1_F_BS TTTTTTGAGTAGTTGGGATTATAGG and IL8_Peak_1_R_BS CTCCTAAACCATAAAAATCATTTAATAAAC (amplicon size 294 bp), IL8_2_fwd_BS GGGATGGGTTATTAGTTGTAAA and IL8_2_rev_BS TTATTACCAAAACATCAAAAATAACT (amplicon size 359 bp), SULT1E1_2_fwd_BS TAGATGATTGGTATTGTGTTGTTAT and SULT1E1_2_rev_BS TAAATTCCTACCATTCTCCTTC (amplicon size 290 bp), GTF3C4_1_fwd_BS GAATTAAAGTTTGTTTGGGTAGT and GTF3C4_1_rev_BS TATTCATCTTCTCTCAAATACAAAA (amplicon size 304 bp), GTF3C4_2_5_F_BS TTGGGTTTTGGTATAAGATAGGT and GTF3C4_2_5_R_BS ACCTCAACCTCCCAATAACTA (amplicon size 330 bp). All clones were sequenced using the ABI 3100 Genetic Analyzer. Sequences were quality controlled, trimmed, and aligned using DNAStar Lasergene SeqMan software. Methylation status of clones were determined and diagrams created using BiQ Analyzer software (Bock et al. 2005).
Data access
ChIP-seq, CpG-seq, and microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) data sets under accession numbers GSE29251 and GSE30780.
Acknowledgments
We acknowledge grant and fellowship support from the Juvenile Diabetes Research Foundation International (JDRF), the Diabetes Australia Research Trust (DART), the National Health and Medical Research Council (NHMRC), the National Heart Foundation of Australia (NHF), the INSERM-NHMRC Cooperation Programme, the Foundation for Polish Science (FNP), and the National Research Program for Basic Research of China. Supported in part by the Victorian Government's Operational Infrastructure Support Program.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116095.110.
References
- Araki Y, Wang Z, Zang C, Wood WH 3rd, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30: 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA 2010. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics 3: 33 doi: 10.1186/1755-8794-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton RL, Maddie MA, Dincman TA, Hagg T, Whittemore SR 2009. Transcriptional activation of endothelial cells by TGFβ coincides with acute microvascular plasticity following focal spinal cord ischaemia/reperfusion injury. ASN Neuro 1: e00015 doi: 10.1042/AN20090008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A 2009. An operational definition of epigenetics. Genes Dev 23: 781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T 2005. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21: 4067–4068 [DOI] [PubMed] [Google Scholar]
- Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El-Osta A 2009. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG 2010. Whole-genome DNA methylation profiling using MethylCap-seq. Methods 52: 232–236 [DOI] [PubMed] [Google Scholar]
- Brownlee M 2001. Biochemistry and molecular cell biology of diabetic complications. Nature 414: 813–820 [DOI] [PubMed] [Google Scholar]
- Chalmers J, Cooper ME 2008. UKPDS and the legacy effect. N Engl J Med 359: 1618–1620 [DOI] [PubMed] [Google Scholar]
- Cooper ME, El-Osta A 2010. Epigenetics: mechanisms and implications for diabetic complications. Circ Res 107: 1403–1413 [DOI] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J 2002. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol 3: RESEARCH0036 doi: 10.1186/gb-2002-3-7-research0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein F, Thompson RF, Bhagat TD, Fazzari MJ, Verma A, Barzilai N, Greally JM 2010. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS ONE 5: e8887 doi: 10.1371/journal.pone.0008887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Osta A, Wolffe AP 2000. DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Expr 9: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M 2008. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205: 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM 2010. Introduction to epigenomics and epigenome wide analysis. Methods Mol Biol 620: 243–265 [DOI] [PubMed] [Google Scholar]
- Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R, Rehli M 2006. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res 66: 6118–6128 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan KN, Bayles R, Ciccotosto GD, Maxwell S, Cappai R, Pelka GJ, Tam PP, Christodoulou J, El-Osta A 2010. Alleviating transcriptional inhibition of the norepinephrine slc6a2 transporter gene in depolarized neurons. J Neurosci 30: 1494–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28: 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MA, Schiekofer S, Kanitz M, Klevesath MS, Joswig M, Lee V, Morcos M, Tritschler H, Ziegler R, Wahl P, et al. 1998. Insufficient glycemic control increases nuclear factor-κB binding activity in peripheral blood mononuclear cells isolated from patients with type 1 diabetes. Diabetes Care 21: 1310–1316 [DOI] [PubMed] [Google Scholar]
- Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA 2008. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 359: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19: 187–191 [DOI] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C 2009. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 18: 3769–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A, Bedo J, Conway T, Beresford-Smith B 2011. The poisson margin test for normalization-free significance analysis of NGS data. J Comput Biol 18: 391–400 [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. 2006. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929–1935 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Groop L 2009. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 58: 2718–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre G, Bailey J, Haviv I, Kowalczyk A 2010. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics 26: i524–i530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F, Gonzalo IG, Lanting L, Natarajan R 2004. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 279: 18091–18097 [DOI] [PubMed] [Google Scholar]
- Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R 2007. Genome wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem 282: 13854–13863 [DOI] [PubMed] [Google Scholar]
- Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R 2008. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes 57: 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ 2011. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389 [DOI] [PubMed] [Google Scholar]
- Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, Raskin P, Zinman B 2005. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med 353: 2643–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DM, Zinman B, Cleary PA, Backlund JY, Genuth S, Miller R, Orchard TJ 2009. Modern-day clinical course of type 1 diabetes mellitus after 30 years' duration: the diabetes control and complications trial/epidemiology of diabetes interventions and complications and Pittsburgh epidemiology of diabetes complications experience (1983-2005). Arch Intern Med 169: 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ 2009. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper GM, Riaz ul H 1997. Activation of nuclear factor-κB in cultured endothelial cells by increased glucose concentration: prevention by calphostin C. J Cardiovasc Pharmacol 30: 528–532 [DOI] [PubMed] [Google Scholar]
- Piga R, Naito Y, Kokura S, Handa O, Yoshikawa T 2007. Short-term high glucose exposure induces monocyte-endothelial cells adhesion and transmigration by increasing VCAM-1 and MCP-1 expression in human aortic endothelial cells. Atherosclerosis 193: 328–334 [DOI] [PubMed] [Google Scholar]
- Roy S, Sala R, Cagliero E, Lorenzi M 1990. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci 87: 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Zhao K 2008. Genome wide approaches to studying chromatin modifications. Nat Rev Genet 9: 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, et al. 2000. Genes expressed in human tumor endothelium. Science 289: 1197–1202 [DOI] [PubMed] [Google Scholar]
- Statham AL, Strbenac D, Coolen MW, Stirzaker C, Clark SJ, Robinson MD 2010. Repitools: an R package for the analysis of enrichment-based epigenomic data. Bioinformatics 26: 1662–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM 2010. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol 11: R36 doi: 10.1186/gb-2010-11-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF, Fazzari MJ, Niu H, Barzilai N, Simmons RA, Greally JM 2010. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J Biol Chem 285: 15111–15118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas E, Lin YS, Dagher Z, Saha A, Luo Z, Ido Y, Ruderman NB 2002. Hyperglycemia and insulin resistance: possible mechanisms. Ann N Y Acad Sci 967: 43–51 [DOI] [PubMed] [Google Scholar]
- UK Prospective Diabetes Study (UKPDS) Group. 1998. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet 352: 837–853 [PubMed] [Google Scholar]
- Vaquero A, Reinberg D 2009. Calorie restriction and the exercise of chromatin. Genes Dev 23: 1849–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R 2008. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci 105: 9047–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Writing Team for the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group 2002. Effect of intensive therapy on the microvascular complications of type 1 diabetes mellitus. JAMA 287: 2563–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerneni KK, Bai W, Khan BV, Medford RM, Natarajan R 1999. Hyperglycemia-induced activation of nuclear transcription factor κB in vascular smooth muscle cells. Diabetes 48: 855–864 [DOI] [PubMed] [Google Scholar]
- Zhao B, Bowden RA, Stavchansky SA, Bowman PD 2001. Human endothelial cell response to gram-negative lipopolysaccharide assessed with cDNA microarrays. Am J Physiol Cell Physiol 281: C1587–C1595 [DOI] [PubMed] [Google Scholar]