Abstract
Immense populations of viruses are present in the human gut and other body sites. Understanding the role of these populations (the human “virome”) in health and disease requires a much deeper understanding of their composition and dynamics in the face of environmental perturbation. Here, we investigate viromes from human subjects on a controlled feeding regimen. Longitudinal fecal samples were analyzed by metagenomic sequencing of DNA from virus-like particles (VLP) and total microbial communities. Assembly of 336 Mb of VLP sequence yielded 7175 contigs, many identifiable as complete or partial bacteriophage genomes. Contigs were rich in viral functions required in lytic and lysogenic growth, as well as unexpected functions such as viral CRISPR arrays and genes for antibiotic resistance. The largest source of variance among virome samples was interpersonal variation. Parallel deep-sequencing analysis of bacterial populations showed covaration of the virome with the larger microbiome. The dietary intervention was associated with a change in the virome community to a new state, in which individuals on the same diet converged. Thus these data provide an overview of the composition of the human gut virome and associate virome structure with diet.
Bacteriophages are the most abundant biological entities on Earth, with an estimated population of ∼1031 total particles (Weinbauer 2004; Suttle 2005), but their roles in human health are only beginning to be studied (Edwards and Rohwer 2005; Breitbart et al. 2008; Reyes et al. 2010). Phage model systems were pivotal in the early development of molecular biology (Cairns et al. 1966; Judson 1996). Today, much of phage research is focused on phage in their natural environments, including the viral component of the human microbiome (Dinsdale et al. 2008; Willner et al. 2009, 2010; Reyes et al. 2010). The new emphasis on studies of whole populations has been made possible in part by the development of “next generation” sequencing methods, which allow quantification of the types and proportions of phages in complex mixtures by deep sequencing of environmental samples (Thurber et al. 2009).
Lysogenic or temperate phages are able to integrate their chromosomes into the bacterial genome (Hendrix et al. 1983; Ptashne 1992), and so can alter the phenotype of the host bacterium by lysogenic conversion (Brussow et al. 2004). Transduction of genes for toxins by phage is well known, as in the case of cholera (Waldor and Mekalanos 1996) and Shiga toxin (Brussow et al. 2004). Additional functionality, identified more recently, may promote bacterial adaptation to the host environment—genes for functions involved in energy harvest (Reyes et al. 2010) and platelet adhesion (Willner et al. 2010) have been identified in viral metagenomic data, and cryptic prophages of E. coli have been shown to encode genes for resistance to antibiotics and other environmental stresses (Wang et al. 2010). The contributions of these and other phage genes to microbiome function are just beginning to be studied.
Diet is expected to alter the composition of the human microbiome, and specific microbiome assemblages, in turn, are expected to affect the welfare of the human host, but interactions between phage and diet in the human microbiome are mostly unexplored. One recent study used next-generation sequencing to characterize human gut viruses from four twin pairs and their mothers (Reyes et al. 2010) and found similarity of communities between twins and their mothers, and stability of viral communities over time. Dynamics in this study did not show cyclic changes in phage and bacterial abundance as would be expected for Lotka-Volterra predator–prey relationships (Bohannan and Lenski 1997), or episodes of outgrowth of particular bacterial species followed by blooms of their phage as in “kill-the-winner” dynamics (Rodriguez-Valera et al. 2009). The factors responsible for the observed longitudinal stability have not been fully clarified.
Here, we present a study of the dynamics of the human gut virome during a deliberate perturbation by a dietary intervention. We compared shotgun metagenomic sequences from virome samples, as well as metagenomic sequences from bacterial populations. We found that the predominant source of variation was differences among individuals, but that significant changes in viral populations were detectably associated with switching to a defined diet, and that convergence of viral populations was seen for individuals on similar diets.
Results and Discussion
Sampling and sequencing
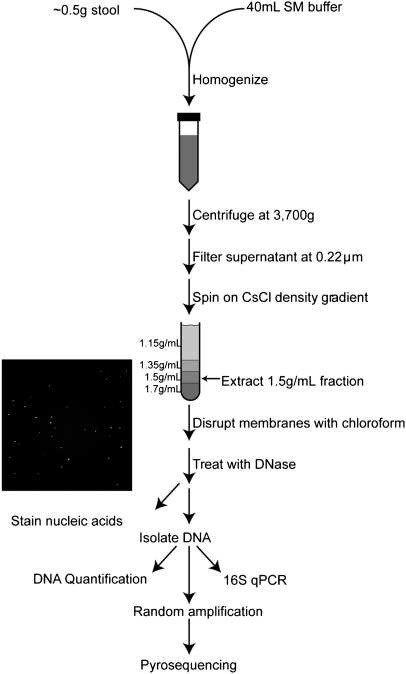
We purified virus-like particles (VLPs) from stool samples collected longitudinally from six healthy volunteers between the ages of 18 and 40 yr who had normal bowel frequency, normal body mass index, no history of chronic intestinal disease, diabetes, or immune deficiency, and who had not been treated with antibiotics for a minimum of 6 mo prior to entering the study. Two individuals were fed a high-fat/low-fiber diet, three were fed a low-fat/high-fiber diet, and one was on an ad-lib diet. Samples were collected at up to four time points (days 1, 2, 7, and 8), with the controlled diet starting after sample collection on day 1. VLPs were purified (Fig. 1) by filtration and CsCl density gradient fractionation. In what follows, we use “VLP” to refer to these preparations. Although we are able to isolate multiple phage types from these preparations, and EM analysis confirms the presence of virus-like particles, the fraction of particles that are replication-competent virions is unknown, so we avoid referring to the full population as “viruses”. VLPs were treated exhaustively with DNase, then deproteinized, and total VLP DNA was purified (Thurber et al. 2009). The VLP-associated DNA was randomly amplified by Phi29 polymerase and shotgun sequenced using the 454 Life Sciences (Roche) GS FLX Titanium platform. Amplification with Phi29 polymerase can distort the ratios of different members of the community (Yilmaz et al. 2010), but all samples studied here were processed similarly, allowing consistent comparisons between samples. After filtering, the VLP data set yielded 936,213 high-quality sequences with a mean length of 359 nt (336 Mb total; Supplemental Table S1). Initial analysis of individual reads showed that 98% of these sequences had no significant match to an identified sequence in the nonredundant database (E-value < 10−5) when analyzed individually, consistent with previous studies of similar preparations (Breitbart et al. 2003, 2008; Reyes et al. 2010).
Figure 1.
Purification of VLP DNA. Stool was homogenized in SM Buffer; particulate matter was spun down; supernatant was filtered at 0.22 μm to remove cells; VLPs were purified on a CsCl density gradient and treated with nuclease to eliminate unprotected DNA. The absence of bacterial cells was confirmed by staining VLP preparations for nucleic acids. VLP DNA was quantified, amplified, and pyrosequenced.
To track bacterial populations, total DNA was isolated from the same stool samples and analyzed using deep sequencing of 16S rDNA amplicons and shotgun sequencing of total DNA (Hoffmann et al. 2009). The 16S rDNA sequence-tag data set contained 63,405 reads, with a mean length of 268 nt. Sequences were filtered using QIIME (Caporaso et al. 2010) and assigned to bacterial lineages using RDP (Wang et al. 2007). Bacterial communities were compared using UniFrac (Lozupone and Knight 2005). Shotgun sequencing of total stool DNA (mostly from bacteria) yielded 1,007,534 reads with a mean length of 344 nt.
To quantify the purity of our VLP preparations, we checked bacterial 16S sequences in the VLP DNA. VLP DNA preps were confirmed to be at least 10,000X reduced in bacterial 16S DNA by Q–PCR (data not shown), and VLP DNA samples contained only 21 reads with similarity to bacterial 16S, a 35-fold reduction compared with bacterial shotgun sequencing (P < 10−15, χ2 test). Bacterial sequences could be present in the VLP preparations as contamination, or as a result of generalized transduction, specialized transduction, or incorporation in Gene Transfer Agents (GTAs) (Bushman 2001; McDaniel et al. 2010). Thus, the origin of the low-level bacterial sequences in our data set is uncertain.
Assembly and initial analysis
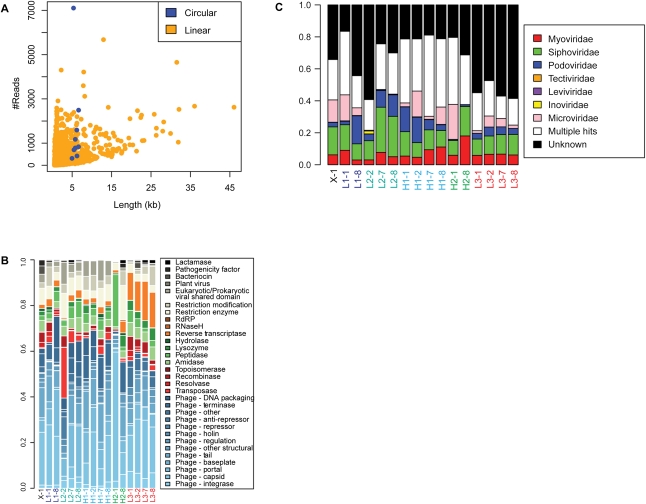
The average read length in our VLP data set was longer than in most previous metagenomic studies of viral DNA, allowing extensive assembly of individual reads into contigs. We assembled VLP sequences using the Newbler assembler (Miller et al. 2010) (40 bp overlap, 90% identity), which yielded 7175 contigs at least 500 bp in length. Fully 86.6% of the sequence reads were recruited into these contigs (Fig. 2A). The longest contig was 46 kb, 73 contigs were longer than 10 kb, 279 contigs were longer than 5 kb, and 3028 contigs were longer than 1 kb.
Figure 2.
Assembly and functional annotation of shotgun metagenomic sequences from the human gut virome. (A) Analysis of recruitment of VLP sequence reads into contigs. The y-axis shows the number of sequence reads, the x-axis shows contig length. Pyrosequencing data from the human virome were assembled into 7147 contigs up to 47.8 kb in length. Linear contigs are shown in yellow, circular contigs are shown in blue. (B) Analysis of protein functions in VLP contigs. The functions encoded in VLP contigs were predicted using the Pfam database, then grouped using custom database-relating Pfam domain identifiers to phage functions (Supplemental Table S4). The relative proportions of pyrosequencing reads falling within ORFs of different annotations were plotted according to their sample of origin (y-axis). Each bar is indicated by the sample code, where L or H indicates low- or high-fat diet, the adjacent number indicates the subject number, and the number after the hyphen indicates the day of the study. “X-1” indicates ad-lib diet. (C) Taxonomic classification of VLP communities is consistent across samples. Samples (in columns, labeled as in Figs. 1, 5) are characterized according to the number of sequences from each sample that are assembled into a contig that is classified by taxonomic family. Phage families are indicated by the color code to the right. “Unknown” (black) indicates contigs that cannot be classified in any way. “Multiple hits” (white) indicate contigs that have proteins that are similar to multiple families.
The approximate size of the VLP community can be estimated by PHACCS (PHAge Communities from Contig Spectrum) (Angly et al. 2005), which calculates the degree to which a group of sequences coassemble into contigs, and compares them with simulated communities of different sizes. Similar to what has been seen recently in the human gut virome (Reyes et al. 2010), the median richness (number of species) of the 16 samples was 44 (range = 19–785), and the average Shannon Diversity was 3.46 (SD = 0.59). The most abundant genotype was predicted to account for 16.20% of the total (SD = 2.09%), predicting the complete assembly of the most abundant VLP genomes in our study.
Analysis of gene content
To characterize these VLP contigs, we compared both nucleotide sequences and open reading frames [ORFs] with (1) the NCBI nonredundant database, (2) the Pfam database of conserved amino acid motifs (Sonnhammer et al. 1998), (3) the Clusters of Orthologous Groups (COG) database of annotated bacterial protein families (Tatusov et al. 2003), (4) A CLAssification of Mobile genetic Elements (ACLAME) (Leplae et al. 2009), (5) the Antibiotic Resistance Genes Database (ARDB) (Liu and Pop 2009), and (6) the Virulence Factors Database (VFDB) (Yang et al. 2008). All VLP contigs and associated annotation can be viewed using a web-based interface at http://microb215.med.upenn.edu/cgi-bin/gb2/gbrowse/phage_metagenomics/. A total of 22% of ORFs contained recognizable Pfam motifs. Essential bacteriophage functions were well represented, including functions required for both lytic and lysogenic growth (Fig. 2B).
VLP contigs were classified according to their similarity to ICTV-defined bacteriophage families (Fig. 2C). There were 1268 contigs with amino acid similarity to members of the Siphoviridae family (18% of the total), 686 (10%) to Myoviridae, 344 (4.8%) to Podoviridae, 68 (0.9%) to Microviridae, and 0.4% to other families. Of the remaining contigs, 813 (11%) had amino acid similarity to multiple bacteriophage families, and 3969 (55%) did not have significant similarity to any bacteriophage families. Membership in these families was similar among the subjects studied (Fig. 2C). No strong candidates for eukaryotic viruses were detected either through nucleotide comparison to known eukaryotic viral genome sequences, or through similarity to conserved eukaryotic protein domains. In a few cases, a gene was annotated as similar to a eukaryotic virus, but for each of these genes contigs could be identified that also encoded multiple phage genes, suggesting that these proteins contain motifs common to both bacterial and eukaryotic viruses. Thus, this classification indicates that the viruses studied here were comprised primarily of tailed bacteriophage, with some representation of additional DNA phage families.
Analysis showed that nine VLP contigs formed closed circles, 4.5–6 kb in length, suggestive of completion of these genome sequences. None of the nine had significant nucleotide similarity to any sequence in the NCBI nonredundant database (E-value < 10−3). However, eight of the nine contigs contain an ORF that aligns to the capsid F protein sequence from Microphages (Rohwer and Edwards 2002), the family of 4.5–6-kb circular ssDNA phage containing the prototype bacteriophage φX174. Three of the eight contigs were closely related to the chp1-like Microphage. Those eight genomes also contain proteins similar to phage proteins involved in replication (n = 4), proteolysis (n = 2), and scaffolding assembly (n = 2). Only two of these genomes have significant sequence similarity to each other—contigs c03390 and c04421 are 93% identical, but were too diverse to coassemble. The ninth genome (contig c04570) contains proteins that resemble those found on plasmids from a wide range of bacteria, primarily Firmicutes, and may represent a temperate phage that maintains itself as a plasmid (Sternberg and Austin 1981).
Comparison of total community and VLP metagenomes
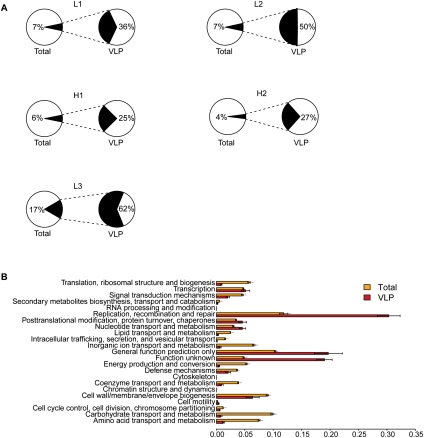
We next investigated the proportion of phage DNA in our total (bacterial) metagenomic data set. We calculated the proportion of VLP sequences that have a significant match in the total shotgun data set (E-value < 10−5), and vice versa (Fig. 3A). As expected (Reyes et al. 2010), the VLP sequences represent a minority of the total DNA from stool, in the range of from 4% to 17%, although this value is dependent on sampling depth, because the VLP community is a subset of the total community. We also assembled and annotated the total community shotgun sequence data set (347 Mb) using COG (Tatusov et al. 2003), then compared it with the VLP data set. The proportions of COG classes present in the total community (mostly bacteria) and VLPs were quite different, as expected (Fig. 3B). Bacteria were significantly enriched in genes for synthesis of amino acid and carbohydrate precursors, ion transport and metabolism, translational machinery, and cell wall/membrane biogenesis, while VLP contigs were enriched in genes for replication, recombination and repair, and unknown functions (P < 0.05, t-test). Thus, the profile indicates that viruses recruit host cell machinery for translation, energy production, and synthesis of macromolecular precursors while using their own coding capacity to encode functions for replication.
Figure 3.
Comparison of gene content in total microbial communities and VLP communities. (A) The proportions of total genes identified in shotgun metagenomic analysis of total stool DNA (left) is compared with genes in VLP communities (right). (B) VLP DNA encodes biochemical functionality that is markedly different from the total microbiome. Function annotation was performed according to comparison to the COG database (Tatusov et al. 2003). The proportion of reads from each assembled data set that fall within an ORF of the indicated annotation are plotted on the x-axis (mean ± SE).
Prophage abundance
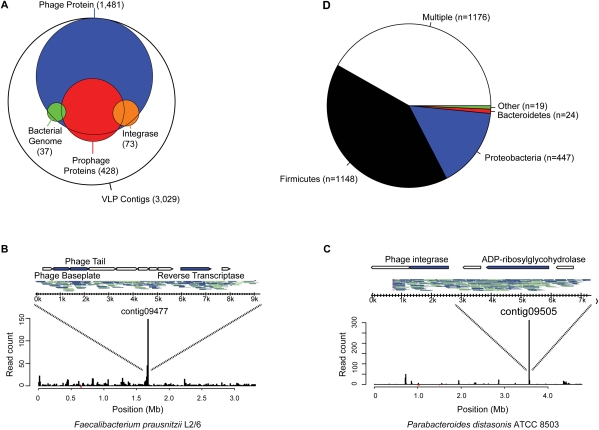
A key question in investigating the gut virome centers on what fraction of phages are temperate, since this group can install new genes in bacteria and alter phenotype via lysogenic conversion (Brussow et al. 2004; Reyes et al. 2010). We used three indications of a temperate lifestyle to annotate the VLP contigs: (1) nucleotide identity to sequenced bacterial genomes, which is indicative of prophage formation (90% of bases aligned at 90% identity), (2) presence of integrase genes (according to Pfam) (Sonnhammer et al. 1998) and COG (Tatusov et al. 2003) annotations, and (3) significant similarity of multiple proteins to prophages annotated in the ACLAME (Leplae et al. 2009) database of mobile genetic elements (E-value < 10−5). This strategy provides a minimal estimate of the number of temperate phages in our data set, since authentic temperate phages may not be positive by any of these criteria. Of the 3029 VLP contigs of at least 1 kb in length, 428 (14%) had multiple proteins that significantly resembled an annotated prophage, 73 (2.4%) contained an integrase gene, and 37 (1.2%) aligned to a sequenced bacterial genome (Fig. 4A). Of the 505 contigs that fit at least one of these three criteria, 442 (88%) also contained an annotated bacteriophage gene. When individual VLP reads were mapped to known bacterial genomes, only 1.5% (13,808) aligned at 90% identity or higher. Of those reads, 74% map to just five genomes, Bacteroides vulgatus ATCC 8482 (n = 4555), Eubacterium eligens ATCC 27750 (n = 2147), Faecalibacterium prausnitzii L2/6 (n = 2081), Parabacteroides distasonis ATCC 8503 (n = 785), and Bacteroides thetaiotaomicron VPI-5482 (n = 583). These reads aligned to short, mostly contiguous regions of each genome and assembled into contigs that encode bacteriophage proteins (Fig. 4B,C), as expected for prophages. All of the reads matching Eubacterium eligens (n = 2147) mapped to its plasmid, which also contains annotated phage proteins, suggesting that this plasmid may contain an integrated prophage or itself be a nonintegrating prophage, potentially analogous to phage P1 (Sternberg and Austin 1981). Of our 3029 VLP contigs over 1 kb, a minimum of 505 (or 17%), can be tentatively classified as temperate phages by at least one of the above criteria.
Figure 4.
Analysis of temperate bacteriophages in the human gut virome. (A) Venn diagram indicating frequency of functions associated with temperate phages. VLP contigs were annotated according to the presence of integrase-like sequences (orange), BLAST alignment to sequenced bacterial genomes, suggestive of prophage formation (90% length at 90% identity; green), and presence of multiple genes with significant similarity to a prophage element within the ACLAME database (red). Map of VLP pyrosequencing reads aligning to the genomes of (B) Faecalibacterium prausnitzii L2/6 and (C) Parabacteroides distasonis ATCC 8503 (90% length at 90% identity). (Inset) Contigs that correspond to each peak, the reads that make up each contig, and their annotated genes. Top strand reads are indicated by blue, bottom strand reads by gray. (D) Prevalence of prophage sequences according to bacterial phylum of origin. Contigs with amino acid similarity (E-value < 10−5) to prophage sequences within the ACLAME databases are shown according to which bacterial phylum they match. Any contig with similarity to more than one phylum is classified as “Multiple.” Roughly 39% of VLP contigs (n = 2814) did not have significant similarity to any prophage sequence in this database.
Variation among animals in prophage induction has been previously shown in mouse models (Reyes et al. 2010), leading us to investigate induction here. We probed the level of induction of the above five prophages indirectly by comparing shotgun sequences from total community DNA (mostly bacterial), with VLP sequences (Supplemental Fig. S2). In only one case (F. prausnitzii) did the abundance of the bacterial host correlate with the abundance of its corresponding prophage. The lack of correlation of the other four prophages suggests the possibility that the degree of induction varied among the individuals studied.
We were able to infer the groups of bacteria infected by these putative temperate phage by comparing the VLP contigs to prophage sequences in the ACLAME database. Of the 2814 contigs that had significant amino acid similarity (E-value < 10−5) to an ACLAME prophage sequence, the contigs that were assigned exclusively to the Firmicutes accounted for fully 41% of the total (n = 1148) (Fig. 4D). Surprisingly, similarity to prophage from the other common gut resident, Bacteroidetes, accounted for <0.9% of the assigned contigs (n = 24). While Proteobacteria account for only a modest fraction of the total gut bacteria, contigs with similarity to Proteobacteria accounted for 16% (n = 447) of the total. This pattern also extended to contigs that were similar to multiple bacterial phyla (“Multiple” in Fig. 4D). Of the 1176 VLP contigs that were similar to more than one bacterial Phylum, 34% had a match to Firmicutes, 0.8% had a match to Bacteroides, and 13% had a match to Proteobacteria. This suggests that the prevalence of temperate bacteriophage in the gut may differ among bacterial phyla, though the conclusions depend on representation of phage sequences in the databases used for analysis, and so conclusions may evolve as further sequence data accumulates.
Functionality in the gut virome: CRISPRs
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) are mediators of a recently described form of bacterial adaptive immunity. Short DNA sequences (26–72 nt in length) are captured from invading DNA elements and installed as CRISPR spacer sequences in bacterial chromosomes. Multiple spacer sequences are separated by partially palindromic repeats within CRISPR arrays. After transcription of CRISPR arrays, RNA copies of the CRISPR spacer sequences act as recognition modules in nucleoprotein complexes, which bind to targeted nucleic acid from genomic invaders such as phage or plasmids, and program their degradation (Brouns et al. 2008; Sorek et al. 2008). Thus, CRISPR spacers provide a record of the genomic parasites that bacteria have encountered.
Analysis of CRISPRs in our assembled data provided a detailed record of phage–host and phage–phage competition. A total of 38 CRISPR arrays were found in shotgun metagenomic assemblies for our total community data set. The 38 contigs containing these CRISPRs aligned primarily to genomes of gut Bacteriodetes (n = 16, E-value < 10−5). Of the 45 spacer sequences found within those 38 CRISPR arrays, fully 80% (n = 36) showed no matches to any known sequence, but one spacer perfectly matched a VLP contig sequence (contig c05189; 100% identity over 43 bases). Both the bacterially encoded CRISPR spacer and VLP target were found within a single individual, suggesting that the CRISPR spacer sequence may have restricted phage replication within that subject.
Unexpectedly, we also found 22 CRISPR arrays in our VLP contigs. CRISPRs have not previously been reported from free phage particles, but one report did identify CRISPR sequences in potential prophages of Clostridium difficile (Sebaihia 2006). A related CRISPR array was found in our virome samples (with >95% identity in repeat regions to the annotated C. difficile CRISPR5). Analysis of novel CRISPR spacer sequences showed one high-stringency match between a VLP spacer (on contig c05834) and a separate VLP contig (c02690) (95% identity over 39 bases). The CRISPR spacer–target pair were isolated from the same individual and both were detected at the same two time points. Phage are well known to encode an extensive set of functions for competing with other phage (Calendar 1988; Refardt 2011)—these data indicate that CRISPRs may mediate phage–phage competition as well. It will be valuable to assess CRISPR evolution and targeting in further phage metagenomic data sets.
Functionality in the gut virome: Antibiotic resistance
To interrogate antibiotic resistance in the VLP metagenomic data set, the ORFs found on contigs and unassembled reads were compared with known antibiotic resistance genes (BLASTp against the ARDB database) (Liu and Pop 2009) E-value < 10−5, yielding 614 matches to antibiotic resistance genes. These included multidrug efflux transporters (n = 355), vancomycin resistance genes (n = 129), tetracycline resistance genes (n = 18), and beta-lactamases (n = 16). The 33 highest quality matches to assembled contigs were examined further (Supplemental Table S2). Some of these encoded clear antibiotic resistance genes (e.g., beta-lactamase, drug efflux pumps, streptogramin acetyltransferase), while others encoded relatives of antibiotic resistance proteins that are of uncertain importance to resistance (e.g., ABC transporters and the VanRS two-component signaling system). Five of the contigs also contained identifiable phage genes. Transmissible antibiotic resistance is believed to involve primarily mobilization by plasmids and transposons (Barlow 2009; Juhas et al. 2009)—our results indicate that the role of mobilization by phage may deserve further investigation.
Variation of the gut virome among individuals and during dietary intervention
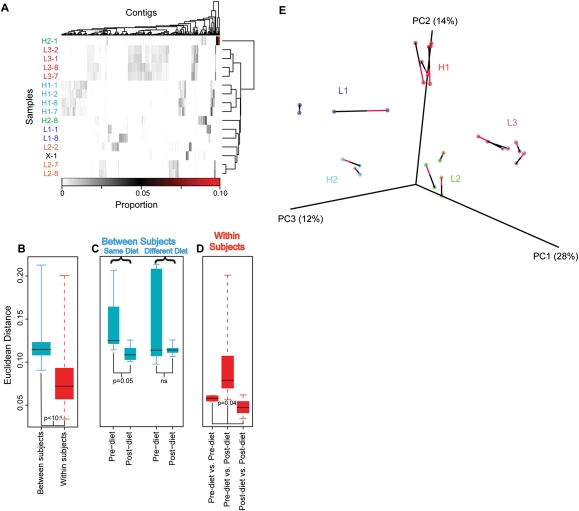
We next asked how viromes varied among individuals and how the dietary intervention affected virome structure. We characterized each virome sample by enumerating the proportion of sequence reads recruited into each VLP contig over the full contig data set. In Figure 5A the proportional abundance of each VLP contig is shown by the color code. The Euclidean distance was then computed between all pairs of virome samples and used for statistical analysis. Comparison of the collection of within-subject distances among time points to between-subject distances showed that between-subject distances were significantly greater (Fig. 5B; P < 0.001, significance assessed by permutation of sample labels). Thus, each individual contained a unique virome that was globally stable over the 8-d time course. A previous study showed individual distinctiveness and stability of the human virome over a year (Reyes et al. 2010).
Figure 5.
Alterations in VLP contig abundance associated with diet. (A) Proportions of VLP sequence reads in contigs from different subjects and time points. Vertical bars indicate the proportion of reads within each contig. The contigs are shown in columns, and subject/time-point combinations in rows. Hierarchical clustering was performed on both rows and columns according to Euclidean distance and complete distance agglomeration. Samples are labeled by subject according to diet: high-fat (H1, H2), low-fat (L1, L2, L3), and ad-lib (X), as well as day of dietary intervention (days 1, 2, 7, or 8). The proportion of all VLP reads in each contig are shown by the scale at the bottom. (B,C,D) The Euclidean distance between samples is shown according to median (line), quartile (box), and range (whisker). (B) Between-subject variation is significantly greater than within-subject variation (P < 10−4, subject label permutation). (C) The distance between the gut viromes of individuals on the same diet (left) was significantly smaller at the end of their dietary treatment than it was at the start (P = 0.05, permutation of diet labels), while there was no increase in similarity for individuals on different diets (right). (D) The distances within subjects that was measured between samples taken on the first 2 d of the timecourse (left) and the last 2 d of the timecourse (right) were significantly lower than the distances between those two sets of days (middle; P = 0.04, permutation of day labels). (E) Covariation of bacterial and VLP community diversity. Distances between pairs of bacterial and VLP communities were calculated as described in the Methods. The similarity of bacterial and VLP communities is shown through PCoA analysis, where each data set was rotated and scaled for maximum superimposition. Each circle represents a sample, either bacterial or VLP. The bacterial and VLP communities from the same sample are connected by a line, where the red half of the line touches the VLP community, and the black half touches the bacterial community. The percent of total variation accounted for by each axis is shown in the axis label. The alignment of bacterial and VLP communities was highly significant (P = 0.004).
In order to test the effect of diet on the composition of the gut virome, we compared the distance between VLP communities in individuals both before and after they started their controlled diet (Fig. 5C). The distance between the gut viromes of individuals on the same diet (Fig. 5C, left) was significantly smaller at the end of their dietary treatment than it was at the start (P = 0.05, significance assessed by permutation of diet labels). There was no increase over time in virome similarity for individuals on different diets (Fig. 5C, right). A group of 39 VLP contigs were identified that changed in association with the dietary intervention (Supplemental Table S3). In comparison to the full data set of 7175 contigs, this subset showed a trend toward enrichment in Siphoviridae (P < 0.06) and depletion of Myoviridae (P < 0.08), which is suggestive of a phylogenetically distinct diet responsive bacteriophage population.
This dietary effect was also seen in a subject-by-subject analysis. For each subject, the distances between samples taken before the defined diet were compared, and distances between samples taken after defined diet were compared (Fig. 5D). As a contrast, distances between samples, where one was before-diet and the other was after-diet, were also compared. The distances were greatest for comparisons between before-diet and after-diet samples (Fig. 5D; P < 0.05, significance assessed by permutation of day labels). Thus, over the period of the dietary intervention, the viral community changed detectably to a new state.
The gut virome is dominated by prokaryotic viruses that prey on gut bacteria, raising the question of how changes in bacterial and viral communities are linked. We tested covariation by comparing 16S rDNA amplicon data from the gut DNA samples, which measured bacterial diversity with the VLP shotgun metagenomic data, which measured viral diversity (Fig. 5E). Variation was significantly correlated between bacterial and VLP communities (Mantel test, R = 0.40, P < 0.001). This correlation was significant when tested by subject label permutation (P < 0.001), indicating that interpersonal variation is correlated between these communities.
Conclusions
In summary, the VLP sequence data presented here could be assembled into contigs that recruited fully 86.6% of sequence reads, providing a resource of over 7000 complete and partial phage genomes and making possible extensive interpretation of ORF function. A substantial portion of the phages in our samples are likely temperate, so that genes contained within phage may alter phenotype of the bacterial host by lysogenic conversion. Novel functionalities emphasized here include phage-encoded CRISPR arrays and antibiotic resistance genes. Also seen were the expected diversity of lysins (Seo et al. 2010), holins (Wang et al. 2000), bacteriocins (Dawid et al. 2007), restriction/modification systems (Kobayashi 2001), and virulence factors (O'Brien et al. 1984; Lainhart et al. 2009; Campos et al. 2010).
The major determinants of microbiome community composition and dynamics remain to be fully clarified. Bacteriophage and bacterial abundances did not oscillate detectably over the time period studied, as would be predicted by Lotka-Volterra predator–prey dynamics, nor did we detect boom and bust outgrowth of phage-host pairs, indicative of kill-the-winner dynamics, consistent with (Reyes et al. (2010) and Rodriguez-Brito et al. (2010). A number of factors may confound the detection of competition in natural populations. The rate at which “winning” bacterial clones (Rodriguez-Valera et al. 2009) arise and are preyed upon may be too fast or too slow to be apparent in this set of samples. The sample set may not be dense enough to detect rapid changes. In addition, phage types that infect different bacterial hosts may be indistinguishable at the current depth of sequencing and with available annotation (Rodriguez-Brito et al. 2010). Viromes were relatively stable within each individual, and interpersonal variation was the largest source of variance observed, even when individuals were on the same diet. This allows us to rule out the idea that phage populations are predominantly acquired on a daily time scale as transients in food, because individuals eating the same food did not come to harbor identical viromes. However, the gut virome changed significantly during the change in diet by alteration of the proportions of pre-existing populations, so that subjects on the same diet showed more similar, though not identical, virome composition. Whether the changes in phage abundance are simply a result of changes in abundance of their hosts, or whether additional mechanisms are involved will require further work to clarify, though initial data suggests a possible contribution of lysogenic induction. It will be valuable going forward to develop improved methods for quantifying phage induction in vivo. Considerable further study will be required to understand the acquisition of gut viral communities and the factors mediating the balance between long-term stability and dynamic response to the environment.
Methods
Sample collection
Six healthy adult volunteers (at least 18 yr old) were recruited to provide stool samples within the Center for Clinical and Translational Research at the Hospital of the University of Pennsylvania. Exclusion criteria included having had diarrhea within 1 wk prior to the sample collection, abnormal bowel movement frequency (at least once every 2 d and no more than three times a day), consumption of any antibiotics within 6 mo prior to sample collection, or any prior diagnosis with inflammatory bowel disease, irritable bowel syndrome, celiac sprue, or other chronic inflammatory diseases of the intestines, or BMI outside the range of 18.5 and 35. All collection was carried out after subjects provided informed consent under an approved IRB protocol.
DNA isolation, PCR amplification, and purification
VLP DNA was isolated in the following manner (Fig. 1). Approximately 0.5 g of stool was homogenized in 40 mL of SM buffer (Sambrook and Russell 2001). Particulate matter was spun down at 4700g for 30 min and supernatant was filtered at 0.22 μm (PES filter, Nalgene). The filtrate was centrifuged on a CsCl step gradient (described in detail in Sambrook and Russell 2001; Thurber et al. 2009), and the 1.35–1.5 g/mL fraction was extracted from the column. We note that some viruses are known to have densities outside of this range (e.g., Tectiviridae, Poxviridae, Herpesviridae), and so would be lost during purification (Thurber et al. 2009); no attempt was made to isolate RNA viruses. Samples were treated with chloroform for 10 min, treated with DNase (Invitrogen) for 10 min at 37°C, and then extracted with the DNeasy Blood and Tissue Kit (Qiagen). VLP DNA was amplified with Genomiphi V2 polymerase (GE Healthcare).
Total stool DNA was isolated using the QIAamp DNA Stool Mini Kit (Qiagen), as described previously (Wu et al. 2010).
454/Roche sequencing methods
Both total DNA and amplified VLP DNA were randomly sheared and ligated to adapters using the 454 GS Titanium Rapid Library Preparation Kit with MID adaptors (454). Bacterial 16S amplicons were amplified with primers BSR 357-A (bar coded) and BSF8-B (not bar coded), which anneal to the V1–V2 region of the bacterial 16S rRNA gene, and include 454 sequencing adaptors (Wu et al. 2010). Samples were pooled and sequenced using GS FLX Titanium chemistry on a 454 Genome Sequencer (Margulies et al. 2005).
16S sequence analysis
Bacterial 16S sequences were analyzed using Pyronoise, assigned by comparison to the RDP database (Wang et al. 2007), and communities compared with each other using UniFrac (Lozupone and Knight 2005) scores, all within the Qiime software package (Caporaso et al. 2010). Sequences were filtered according to size (between 200 and 800 nt), ambiguous bases were removed (max = 2), homopolymer runs were truncated (max = 20), and primer mismatches removed (max = 0).
VLP contig sequence analysis
Quality control
454 pyrosequencing yielded a total of 1,052,246 VLP sequences. These reads were filtered for quality using an in-house pipeline. This pipeline removed sequences sequentially due to (1) incorrect bar codes (n = 23,290); (2) length <50 nt (n = 16,939); and (3) multiple ambiguous bases (n = 19,708). For removing the human reads from the data, the sequences were aligned to the human genome using BLAT (Kent 2002). A total of 476 reads (∼0.05% of the data) had >70% similarity to the human genome and were removed. QIIME (Caporaso et al. 2010) was used to remove duplicate reads from the data set in the following manner. First, five bases were trimmed off from the 3′ end of each of the sequences. The trimmed reads were then used to form OTUs using the TRIE algorithm at 97% identity. A total of 55,620 duplicate reads were discarded and the final data set of 936,213 sequences (with the final five bases preserved) was used for further analysis. The size distribution and quality scores of these high-quality reads are shown in Supplemental Figure 1.
Assembly
The 454 Newbler Assembler (Miller et al. 2010) was used to de novo assemble VLP sequences. Default parameters were used to obtain contigs, with minimum overlap length of 40 bp and minimum overlap identity of 90%. One sequence was only allowed to be assigned to one contig. The Newbler assembler uses the overlap-layout consensus method to assemble sequences, where overlaps are computed by pairwise sequence alignments and then multiple sequence alignment is used to derive the consensus sequence. Similarity between VLP contigs was assessed using YASS (Noe and Kucherov 2005), a DNA local alignment tool.
Analysis of VLP genomes and visualization
The contigs generated from assembly were compared (using BLASTn) to the NCBI nonredundant nucleotide database, ACLAME (A CLAssification of Mobile genetic Elements) (Leplae et al. 2009) and VFDB (Virulence Factors Database) (Yang et al. 2008). In-house scripts were used to derive ORFs from contigs by translating the reads in all six reading frames using NCBI's Bacterial, Archaeal, and Plant Plastid Code. Only ORFs at least 100 aa were considered in further analysis. The ORFs were compared (BLASTp) with the NCBI nonredundant amino acid sequence database, Pfam (Sonnhammer et al. 1998), COG (Tatusov et al. 2003), Antibiotic Resistance Genes Database (ARDB) (Liu and Pop 2009), ACLAME protein, and VFDB protein databases.
The Generic Genome Browser (GBrowse) (Podicheti et al. 2009) was configured on the local server (http://microb215.med.upenn.edu/cgi-bin/gb2/gbrowse/phage_metagenomics/) to visualize assembled VLP contigs and singletons. MySql adaptor was used to set up the databases. The “Phage metagenomics” database contains the contigs, ORFs, and hits obtained by querying the reads using BLAST. Another database, “Phage Circular Genomes” was created to view the nine complete circular phage genomes, the ORFs from the genomes, and BLASTn/BLASTp/BLAST-rps hits from the databases listed above. The contigs, ORFs, and the BLAST results information were converted to the (GBrowse-compatible) gff3 file format using in-house scripts, which were then configured as MySql tables to view in GBrowse.
Taxonomic classification
Contigs were assigned to different bacteriophage families according to their amino acid similarity to the genomes of ICTV-defined groups. To be placed in a certain group, a contig must have amino acid similarity (E-value < 10−3) to a protein from the genome of a member of that group. Contigs were excluded that had a single ORF that was similar to members of multiple families (cutoff at 90% of the top score), or had multiple ORFs that were similar to different families. Samples were characterized according to their membership in these taxonomic families by counting the number of reads from each sample that was assembled into a contig with a given classification.
CRISPR identification and spacer similarity
CRISPR arrays were identified in both the VLP contig and total DNA contig data sets using the CRISPRfinder utility (Grissa et al. 2007b). Both direct repeat and spacer sequences were compared with previously sequenced CRISPR spacers and direct repeats (from CRISPRdb) (Grissa et al. 2007a) using BLAT (90% aligned at 90% identity), and to the nonredundant database (from NCBI) using BLASTn (90% aligned at 90% identity). Spacers from CRISPRdb (Grissa et al. 2007a) were also compared with the VLP contig data set using BLAT (90% aligned at 90% identity).
Functional comparison of VLP DNA and total DNA
Biochemical functionality was predicted according to similarity of ORFs to proteins within the COG database (Tatusov et al. 2003). Membership in a given COG category was determined by the proportion of reads (out of a given data set) that fell within ORFs that had significant similarity (E-value < 10−5) to a protein in the COG database in that category. Differences between VLP and total DNA were calculated via a two-sample t-test.
PHACCS analysis
Community structure was estimated by using PHACCS (v1.1.2) (Angly et al. 2005), implemented with the Octave (v3.2) environment using contig spectra as input, which were generated by Circonspect (v0.2.4). The contig spectra were generated by randomly sampling 10,000 sequences truncated to 100 bp in length, assembling at 98% identity and 35 bp overlap, and counting the number of contigs with one member, two members, etc. PHACCS was run on those spectra using a genome size of 50 kb, under a power law scenario.
Statistical methods
Euclidean distances were calculated by the R function dist. This function takes vectors as inputs, where each element was the number of sequences from a given sample that assembled into a given contig.
To assess the significance of diet effect and association between two distance matrices, day labels were permuted for samples from the same subject, the test statistic (t-statistic and Pearson correlation, respectively) was recalculated using the permuted distance matrix and compared with the observed test statistic to yield P-values. To visualize this covariation, we used Procrustes (part of the Qiime software package) (Caporaso et al. 2010), which rotates two distance matrices to maximize superimposition, and then plotted using KiNG 2.16. Significance was called at a P-value of 0.05 or below, though we note it would be helpful to have more samples and use a lower cut value. To assess the difference in taxonomic categorization within the contigs that were accountable for the effect of diet (above), we took a permutation approach. The complete set of contigs was randomly sampled 10,000 times, and that was compared with the observed distribution of the diet-associated contigs. The probability of obtaining the observed proportions was estimated by the number of permutations that had a proportion that was no less extreme.
Data access
All sequence reads have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession numbers SRX020379, SRX020378, SRX020505, SRX020504, and SRX020587.
Acknowledgments
We thank members of the Wu, Lewis, and Bushman laboratories for help and suggestions. This work was supported by the Human Microbiome Roadmap Demonstration Project UH2DK083981 (G.D.W., F.D.B., J.D.L., Co-PIs). We also acknowledge the Penn Genome Frontiers Institute and a grant from the Pennsylvania Department of Health (the Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions); NIH AI39368 (G.D.W.); the Molecular Biology Core of The Center for Molecular Studies in Digestive and Liver Diseases (P30 DK050306); and The Joint Penn-CHOP Center for Digestive, Liver, and Pancreatic Medicine. We also acknowledge NIH instrument grant S10RR024525 and NIH CTSA grant UL1RR024134 from the National Center for Research Resources, and the Crohn's and Colitis Foundation of America. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122705.111.
References
- Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, Felts B, Nulton J, Mahaffy J, Rohwer F 2005. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6: 41 doi: 10.1186/1471-2105-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow M 2009. What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol 532: 397–411 [DOI] [PubMed] [Google Scholar]
- Bohannan BJM, Lenski RE 1997. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 78: 2303–2315 [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F 2003. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185: 6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, et al. 2008. Viral diversity and dynamics in an infant gut. Res Microbiol 159: 367–373 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, Hardt WD 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68: 560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD 2001. Lateral DNA transfer: Mechanisms and consequences. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Cairns J, Stent GS, Watson JD 1966. Phage and the origins of molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Calendar R 1988. The bacteriophages. Plenum Press, New York [Google Scholar]
- Campos J, Martinez E, Izquierdo Y, Fando R 2010. VEJ{phi}, a novel filamentous phage of Vibrio cholerae able to transduce the cholera toxin genes. Microbiology 156: 108–115 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid S, Roche AM, Weiser JN 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun 75: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452: 629–632 [DOI] [PubMed] [Google Scholar]
- Edwards RA, Rohwer F 2005. Viral metagenomics. Nat Rev Microbiol 3: 504–510 [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C 2007a. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172 doi: 10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C 2007b. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35: W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Roberts JW, Stahl FW, Weisberg RA 1983. Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F 2009. Community-wide response of gut microbiota to enteropathogenic Citrobacter infection revealed by deep sequencing. Infect Immun 77: 4668–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson HF 1996. The Eighth Day of Creation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33: 376–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT–the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res 29: 3742–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainhart W, Stolfa G, Koudelka GB 2009. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J Bacteriol 191: 5116–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Lima-Mendez G, Toussaint A 2009. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res 38: D57–D61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Pop M 2009. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res 37: D443–D447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH 2010. High frequency of horizontal gene transfer in the oceans. Science 330: 50 doi: 10.1126/science.1192243 [DOI] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G 2010. Assembly algorithms for next-generation sequencing data. Genomics 95: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe L, Kucherov G 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33: W540–W543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226: 694–696 [DOI] [PubMed] [Google Scholar]
- Podicheti R, Gollapudi R, Dong Q 2009. WebGBrowse–a web server for GBrowse. Bioinformatics 25: 1550–1551 [DOI] [PubMed] [Google Scholar]
- Ptashne M 1992. A genetic switch. Cell Press and Blackwell Scientific Publications, Cambridge, MA [Google Scholar]
- Refardt D 2011. Within-host competition determines reproductive success of temperate bacteriophages. ISME J doi: 10.1038/ismej.2011.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J 4: 739–751 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836 [DOI] [PubMed] [Google Scholar]
- Rohwer F, Edwards R 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol 184: 4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW 2001. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sebaihia M 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38: 779–786 [DOI] [PubMed] [Google Scholar]
- Seo HS, Xiong YQ, Mitchell J, Seepersaud R, Bayer AS, Sullam PM 2010. Bacteriophage lysin mediates the binding of streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog 6 doi: 10.1371/journal.ppat.1001047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R 1998. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26: 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P 2008. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6: 181–186 [DOI] [PubMed] [Google Scholar]
- Sternberg N, Austin S 1981. The maintenance of the P1 plasmid prophage. Plasmid 5: 20–31 [DOI] [PubMed] [Google Scholar]
- Suttle CA 2005. Viruses in the sea. Nature 437: 356–361 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41 doi: 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F 2009. Laboratory procedures to generate viral metagenomes. Nat Protoc 4: 470–483 [DOI] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914 [DOI] [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R 2000. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54: 799–825 [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK 2010. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1: 147 doi: 10.1038/ncomms1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG 2004. Ecology of prokaryotic viruses. FEMS Microbiol Rev 28: 127–181 [DOI] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 4: e7370 doi: 10.1371/journal.pone.0007370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP Jr, Rohwer F, et al. 2010. Microbes and Health Sackler Colloquium: Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci doi: 10.1073/pnas.1000089107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, et al. 2010. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol 10: 206 doi: 10.1186/1471-2180-10-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen L, Sun L, Yu J, Jin Q 2008. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res 36: D539–D542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S, Allgaier M, Hugenholtz P 2010. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods 7: 943–944 [DOI] [PubMed] [Google Scholar]