Abstract
Copy number variants (CNVs) are increasingly acknowledged as an important source of evolutionary novelties in the human lineage. However, our understanding of their significance is still hindered by the lack of primate CNV data. We performed intraspecific comparative genomic hybridizations to identify loci harboring copy number variants in each of the four great apes: bonobos, chimpanzees, gorillas, and orangutans. For the first time, we could analyze differences in CNV location and frequency in these four species, and compare them with human CNVs and primate segmental duplication (SD) maps. In addition, for bonobo and gorilla, patterns of CNV and nucleotide diversity were studied in the same individuals. We show that CNVs have been subject to different selective pressures in different lineages. Evidence for purifying selection is stronger in gorilla CNVs overlapping genes, while positive selection appears to have driven the fixation of structural variants in the orangutan lineage. In contrast, chimpanzees and bonobos present high levels of common structural polymorphism, which is indicative of relaxed purifying selection together with the higher mutation rates induced by the known burst of segmental duplication in the ancestor of the African apes. Indeed, the impact of the duplication burst is noticeable by the fact that bonobo and chimpanzee share more CNVs with gorilla than expected. Finally, we identified a number of interesting genomic regions that present high-frequency CNVs in all great apes, while containing only very rare or even pathogenic structural variants in humans.
After the discovery of a considerable amount of copy number variants (CNV) in humans (e.g., Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Tuzun et al. 2005), it was a natural step to investigate whether similar structural polymorphism existed in other species. It is clear that copy number variability is a common feature of a wide range of species, from flies (Dopman and Hartl 2007) to maize (Schnable et al. 2009), and including mice (Egan et al. 2007; Graubert et al. 2007; Cahan et al. 2009), rats (Guryev et al. 2008), dogs (Chen et al. 2009), pigs (Ramayo Caldas et al. 2010), goats (Fontanesi et al. 2010), macaques (Lee et al. 2008), and chimpanzees (Perry et al. 2008). CNVs have been associated with traits of evolutionary interest, especially human disease-related traits (see, e.g., Craddock et al. 2010, and references therein), but also traits in other species such as breed-specific features in dogs (Chen et al. 2009), metabolic traits in mice (Orozco et al. 2009), and, possibly, phenotypic differences in inbred lines of maize (Schnable et al. 2009). In addition, in humans and other mammals, CNVs are linked to segmental duplications (SDs) (Eichler 2006), which adds interest to their study in our lineage, especially in light of the outburst of segmental duplication activity that occurred in our common ancestor with the African great apes (Marques-Bonet et al. 2009).
More primate data are needed to build a better picture of structural evolution in the genome of our lineage. A study of structural polymorphism in the genomes of different great ape species can help distinguishing general and species-specific features of copy number variation, as well as ascertaining loci that may be polymorphic exclusively in a given group of species. For example, loci harboring human pathogenic CNVs that present high-frequency structural polymorphism in all great apes may have had special evolutionary relevance in adaptation and health. Previous studies of interspecific comparisons (e.g., Fortna et al. 2004; Newman et al. 2005; Perry et al. 2006; Wilson et al. 2006; Dumas et al. 2007; Armengol et al. 2010) provide no comparative information about species-specific polymorphism, for they only document human-specific losses and gains, while most intraspecific studies, such as that by Perry et al. (2008) in chimpanzees, have so far focused on single species.
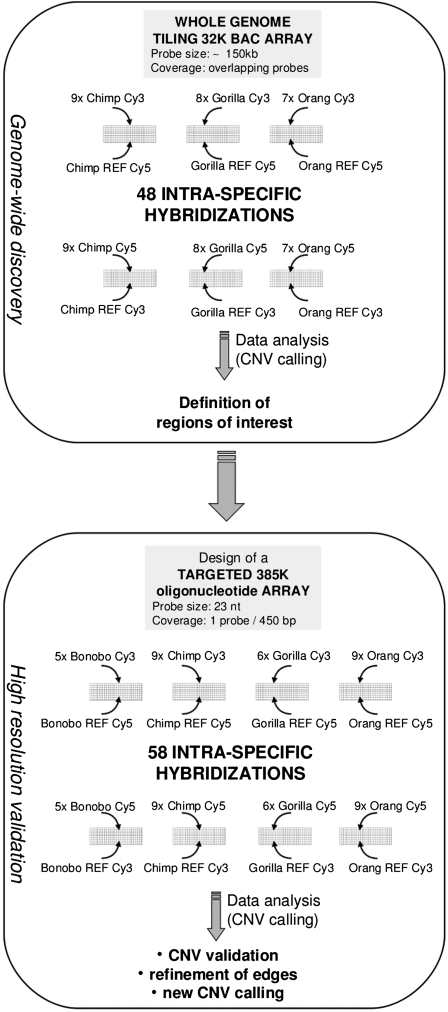
We studied genomic regions of structural polymorphism in all the great apes (bonobo, chimpanzee, gorilla, and orangutan) by means of a two-step approach based on intraspecific aCGH. In a first phase (discovery phase), we used a genome-wide tiling-path 32K BAC array to discover CNV regions in chimpanzees, gorillas, and orangutans. In a posterior Refinement Phase we validated these CNVs, refined their boundaries, and discovered new variants by means of a targeted 385K oligonucleotide array that we had designed using the information gathered during the first phase. We characterized structural polymorphism in each species and compared it among species, contrasting our findings with extant SD maps and studying selective pressures upon structural variation. In addition, we could compare structural variation with polymorphism at the nucleotide sequence level for the same individual bonobos and gorillas. Finally, the mapping of CNVs for the four closest species to humans allowed us to define, for the first time, not only human-specific CNVs, but also human-specific nonpolymorphic regions, that is, genomic regions that present structural polymorphism in all great apes, but are either fixed or present only rare and even pathogenic variants in humans.
Results
Comparing CNVs across species
A total of 51 individuals from the four great ape species were used in a two-phase study (Fig. 1; see Methods for details). The second and final phase consisted of using a targeted oligonucleotide platform to perform aCGH hybridizations with 29 individuals (five bonobos, nine chimpanzees, six gorillas, and nine orangutans). Each individual was hybridized against a member of its own species, and hybridization was performed using reversed-dye labeling of the samples to minimize the effect of dye-specific biases. This procedure allowed us to detect a total of 1170 CNV calls in the five bonobos, 1388 in nine chimpanzees, 1274 in six gorillas, and 1160 in nine orangutans. The average number of calls per individual was 234 in bonobos, 154 in chimpanzees, 212 in gorillas, and 129 in orangutans. All of these calls configure different sets of CNV regions (CNVRs) in different great apes. Bonobos present 505 CNVRs, chimpanzees 404, gorillas 614, and orangutans 399 (Supplemental Table S1).
Figure 1.
Experimental strategy for CNV discovery and validation. The approach is divided into two steps: a first genome-wide discovery phase; and a second targeted validation and refinement phase.
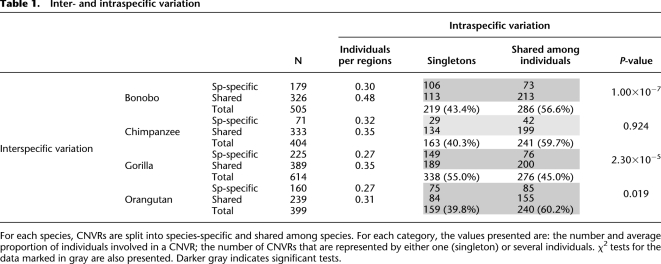
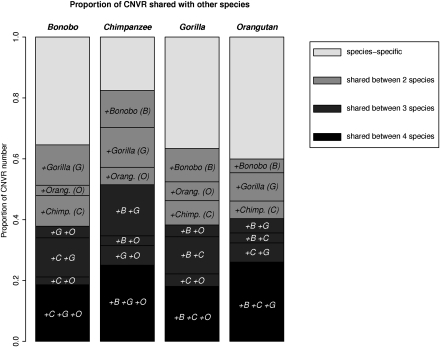
Overall, CNVs are highly shared among species. Table 1 and Figure 2 allow for detailed comparisons (see also Supplemental Fig. S1). The first column in Table 1 contains the absolute numbers of species-specific and shared CNVRs. The first striking observation is that CNVRs shared among species are supported by more individuals than species-specific CNVRs, which tend to be singletons (Table 1), indicating a more recent origin of the latter (see below). Another result is that gorillas, having the largest total number of CNVRs, present both the most species-specific and the most shared CNVRs. To make these figures comparable, we need to consider the proportions of different classes of CNVRs (Fig. 2). Bottom black bands show the proportion of CNVRs that are shared among the four great ape species. Note that the absolute number of CNVRs represented by these black bands is the same for the four species (see Supplemental Table S2), but size varies because it is expressed as a proportion of all CNVRs in each species. The proportion of CNVRs shared between three species (seen as dark-gray bands) are, as expected, roughly proportional to the relative time of divergence in the species tree. Orangutan stands out with the highest proportion of both, species-specific and four-way shared CNVRs. The proportion of species-specific regions is similar in bonobo and gorilla, while chimpanzee harbors relatively few species-specific CNVRs. This could reflect one of two possible scenarios. On one hand, CNVs existing in the common ancestor of bonobo and chimpanzee would have been fixed in chimpanzee. On the other hand, the rate of CNV creation may be lower in chimpanzee.
Table 1.
Inter- and intraspecific variation
Figure 2.
CNVR interspecific comparison. For each species, the proportion of its CNVR shared with none, one, two, or three other species is plotted.
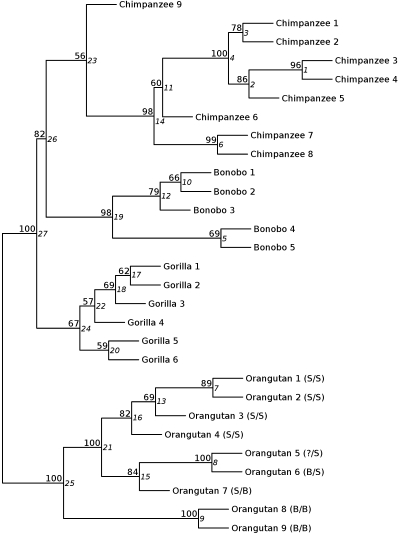
Another interesting finding comes from consideration of the regions shared between pairs of species (light-gray bands in Fig. 2). While the extent of shared CNVRs with orangutan is uniformly lower in the three African great apes, the amount of CNVRs shared between chimpanzee and bonobo is not as expected. Given their very recent divergence time, these two species should share more CNVRs than either of them with gorilla. In contrast, both species share a high proportion of CNVRs with gorilla, roughly as high as between themselves. This is suggestive of homoplasy caused by independent expansions of similar duplicated regions in these species. To rule out that high similarity between all three African apes was due to one or a few individuals with special CNVs, we performed a clustering analysis of all of the individuals according to their participation, or not, in each CNVR. The resulting tree, together with the bootstrap values of every branch is shown in Fig. 3. Edge numbers represent the order in which clusters were built. Given the clustering method (agglomerative hierarchical clustering, see Methods), small edge numbers indicate closer individuals, while higher edge numbers reflect clusters that formed later in the process. Therefore, branch length in this plot is not expected to be directly proportional to genetic distance. However, all of the individuals from each species group together and, as expected, all species-specific clusters are built before the joining of two different species. In addition, the last intraspecific edge is the one between the cluster involving orangutans 8 and 9, and the rest of orangutan samples, as expected from the classification of orangutan samples according to their subspecies.
Figure 3.
CNVR clustering tree. Dendrogram showing individuals clustered on the basis of their CNVR similarities. Numbers on the upper lefthand side of each node indicate bootstrap values. Numbers on the righthand side of each node (in italic) are edge numbers. S and B next to the orangutan individuals stand for Sumatra or Borneo, the ancestral geographical origin of the samples. The first letter represents the origin of the sample and the second letter is for the origin of the reference. Individuals are called by short names that are defined in Supplemental Table S1.
Distribution of structural diversity
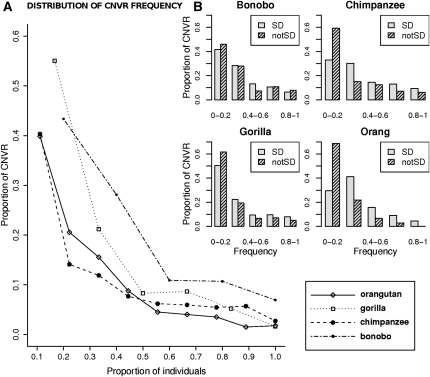
Our data allow us to study interspecific differences in CNV frequency and length. Studies on CNV size are limited by the length of regions tiled in the oligonucleotide array; detailed results can be found in the Supplemental Information and Supplemental Tables S1 and S3. Regarding CNV frequencies, they clearly differentiate the four great apes For example, bonobo is the species with the highest frequency CNVRs (with 42% of individuals supporting the average CNVR), and chimpanzee and gorilla present intermediate frequency CNVRs (34% and 32%, respectively). Again, orangutan is the exception, presenting a lower average CNVR frequency (29%) (see Supplemental Table S1). The proportion of CNVRs as a function of their frequency in each species is shown in Figure 4A. Although these graphs are helpful to get an overview picture of CNV diversity patterns, they cannot represent a formal testing. For example, the values of bonobo are overall higher, because sample size is smaller for this species and, therefore, there are fewer frequency categories and higher proportions of CNVRs in each category. To better compare CNV diversity patterns in the four species, we constructed two variables: πCNV and SCNV (see Methods). SCNV corresponds to the number of CNVRs segregating in our sample (i.e., removing CNVRs that are present in all of the individuals). πCNV is the average number of pairwise differences among individuals in the CNVR complement (average number of differences in the presence/absence of CNVRs). The values taken by the two variables are dependent on sample size (see Supplemental Fig. S2), because fewer individuals provide less power to detect rare CNVs and are correlated to each other, so that πCNV becomes larger with a larger number of segregating CNVRs. For these reasons, we studied the ratio between the two variables (πCNV /SCNV) by means of a resampling strategy. Thus, in what follows, every time we contrast two groups with different sample sizes (for example, CNVs in different species and/or genic CNVs vs. intergenic CNVs), we resample from the largest group to perform meaningful comparisons with identical sample sizes.
Figure 4.
Frequency distribution of CNVRs in the four species. (A) All species' CNVRs are considered together. (B) Species' CNVRs are split according to their overlap to segmental duplications.
We first tested for differences in the πCNV /SCNV ratio between each pair of species, performing resamplings of an equal number of individuals and CNVRs before permuting CNVRs (see Methods). These tests showed that the πCNV /SCNV ratios are roughly similar in all species, with the exception of the bonobo–gorilla comparison, which presents a marginally significant difference in CNV diversity distribution (Supplemental Table S4). We repeated this test separately for genic and intergenic CNVRs, and observed that the differences between bonobo and gorilla are mostly due to CNVs that overlap genes. In genic CNVs, the πCNV /SCNV ratio is significantly different in the bonobo–gorilla and bonobo–orangutan comparisons (Supplemental Table S4), the bonobo genic CNV being significantly enriched in high-frequency variants compared with the two other species.
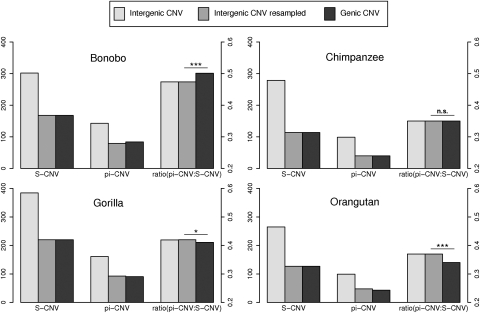
Afterward, we focused on within species variation of πCNV /SCNV. To see whether the frequency distribution of CNVRs was different for structural variants overlapping genes than for variants elsewhere in the genome, we resampled intergenic CNVRs to match the sample size of genic CNVs. We observed that for genic CNVRs the πCNV /SCNV ratio is significantly lower in gorilla (P = 0.029) and highly significantly lower in the orangutan (P < 10×10−5) just as expected if purifying selection was keeping structural variants at lower frequencies when they overlap genes (Fig. 5). In chimpanzee, there is no significant variation of the πCNV /SCNV ratio between in genic or intergenic regions. Interestingly, bonobo is the only species presenting a highly significant increase (P < 10×10−5) in the πCNV /SCNV ratio in genic CNVRs compared with intergenic CNVRs, which might be suggestive of either positive selection or relaxed purifying selection.
Figure 5.
Patterns of CNV diversity in the four species. The left axis gives the scale of the πCNV and SCNV values and the right axis scale shows the values of their ratio. In each case, intergenic CNVs are resampled because they are always more numerous than genic CNVs. Values of intergenic πCNV resampled are the mean πCNV obtained in 100,000 random resamplings of N intergenic CNV, N being the number of genic CNV (SCNV genic). We can observe that the resampling procedure maintains the ratio.
Relationship between structural and nucleotide diversity
Data on copy number variation can be related to the levels of nucleotide diversity in the same species. In the literature there are some instances of diversity measures obtained from putatively neutral genome regions in several primates (Yu et al. 2003, 2004; Fischer et al. 2006). These data have been gathered in Supplemental Table S5, where we can see that orangutan presents the highest level of nucleotide diversity, followed by either gorilla or chimpanzee (depending on the study), and finally bonobo and human. Since the five bonobos and the six gorillas used in the present study were analyzed for sequence diversity in Yu et al. (2003, 2004), we could compare nucleotide polymorphism data and CNV diversity for the very same individuals. We calculated the standard diversity statistics θw (theta Watterson, as estimated from the number of segregating sites) and π (or the mean number of pairwise differences) for each species. The values of the two statistics we obtained with five bonobos and six gorillas are nearly identical to those calculated by Yu et al. (2003, 2004) on nine bonobos and 15 gorillas (πbonobo = 0.067%, θw bonobo = 0.069%, πgorilla = 0.162%, θw gorilla = 0.159%) (see Supplemental Table S5), confirming the absence of selective pressures in the genomic regions examined by Yu et al. (2003, 2004) and indicating that our two subsets of individuals have no obvious genomic idiosyncrasy.
As explained above, πCNV varies with SCNV, which itself depends on sample size. To compare the relationship between structural and nucleotide diversity in gorilla and bonobo, we used resampling to create pseudosamples of gorilla CNVs down to the value of SCNV in bonobo (100,000 random resamplings). When doing so, we observed that the average πCNV in gorilla is lower than in bonobo (dashed bars on Supplemental Fig. S3). In contrast, gorillas present higher nucleotide diversity than bonobo (π in gorilla is higher than in bonobo), which suggests that structural variants may be under different selective pressures and mutational dynamics than single-nucleotide variants.
Measured in terms of the average number of pairwise differences that any given individual presents with the rest (i.e., the individual's πCNV and nucleotide π), there seems to be a correlation between levels of nucleotide and CNV diversity (bonobo: Pearson's r = 0.66; gorilla, r = 0.14) (see Supplemental Fig. S4). This trend is strikingly similar between genic and intergenic CNVs in bonobo, whereas in gorilla, genic and intergenic CNV present the opposite patterns (Supplemental Fig. S4). In gorilla, the average nucleotide differentiation of each individual increases with the average CNV differentiation for intergenic CNVs (r = 0.51) but decreases in genic CNVs (r = −0.09). However, due to our reduced sample size, neither of these correlations is significant. Only larger sample sizes will allow us to ascertain whether these are real trends.
The relationship between CNVs and SDs
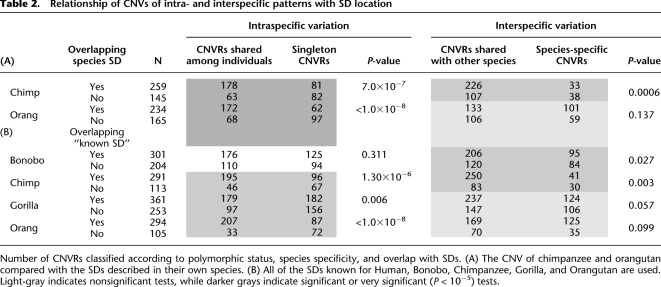
A strong relationship between segmental duplications (SDs) and CNVs has been widely documented in the literature. In particular, human CNVs and human SDs tend to overlap greatly (e.g., Marques-Bonet et al. 2009; Sudmant et al. 2010). In order to investigate this relationship in great apes, we compared locations of ape CNVs and SDs. A direct analysis was possible for chimpanzee and orangutan, for which SD data are available. CNVRs that do not overlap SDs are more likely to be species specific in chimpanzee, but not in orangutan. We also see that CNVRs overlapping SDs tend to be more frequent (i.e., present in more individuals) than CNVRs that do not overlap known SDs, in both species (Table 2A; Fig. 4B).
Table 2.
Relationship of CNVs of intra- and interspecific patterns with SD location
For bonobos and gorillas, SD coordinates remain partial, so for these species we used an indirect analysis. We built a set constituted of all of the SDs that are known to be present in any primate species studied so far (defined as “known SDs,” see Methods). The use of such a “known SD” list is justified by the high overlap described in primate SD (see Marques-Bonet et al. 2009; Supplemental Fig. S1), which makes it likely that most of these SDs are shared by bonobos and gorillas. Additionally, we checked the validity of this approach on chimpanzee and orangutan and obtained results that are consistent with the ones reported in the previous paragraph (Table 2, cf. A and B). Thus, the pool of “known SDs” can be considered a reasonable proxy of each species' SD complement, even if final results will only be obtained when refined data for SDs in the bonobo and gorilla genomes become available. Analysis shows that the CNVRs that do not overlap SDs are significantly more likely to be species-specific CNVRs in bonobo (Table 2B) and marginally nonsignificantly in gorilla (P = 0.057). As to intraspecific variation, CNVRs overlapping SDs appear to be significantly more frequent than CNVRs that do not overlap known SDs in gorilla, but not in bonobo.
Overall, we observed that around two-thirds of the CNVRs described in each species overlap at least partially with known SDs (ranging from 59% in gorilla to 72% in chimpanzee) (Table 2A). This overlap is higher than expected by chance, taking into account the regions tiled in our oligo array (in all species permutation test, P < 10−5). We could rule out that this observation is due a longer size of CNVRs overlapping SD (see Supplemental Information). Interestingly, when we restrict this comparison to the subset of CNVRs that are species-specific, we see that the proportion overlapping SD is relatively similar among species (53% in bonobo, 54% in gorilla, 58% in chimpanzee), except in orangutans, where it is more elevated (78% in orangutan). This higher figure in the latter species suggests that there are common CNV features in African apes, distinguishing them from orangutans. Examination of CNVRs that are strictly shared between the three African great apes showed that they present a stronger association with known SDs than the species-specific CNVRs of each of the same three species (χ2 = 0.75, P = 0.0018). In contrast, when orangutan is added to this test, that is, when we consider the CNVRs shared among the four species, the overlap with SDs is no longer significant (χ2 = 1.20, P = 0.27). This indicates that the common location of CNVRs shared among bonobo, chimpanzee, and gorilla is related to their overlap with SD, suggesting similar origins.
Positive selection upon fixed structural variation
We devised a McDonald-Kreitman-like test that considers CNVRs as polymorphic sites in each given species and species-specific SDs as fixed differences between species. The original McDonald-Kreitman test (McDonald and Kreitman 1991) was devised to compare protein-coding sequences between two Drosophila species. It checks fixed against polymorphic sequence changes in synonymous and nonsynonymous sites and leverages on the fact that most nonsynonymous changes will be under the effect of purifying selection, while the vast majority of synonymous changes are neutral. If, for example, the two compared species differ in more nonsynonymous positions than would be expected out of the proportion of nonsynonymous polymorphisms, one can infer that positive selection drove to fixation a burst of amino acid changes. Some basic assumptions underlie this test. The first and foremost being that SDs and CNVs are largely related, as reported in the literature (e.g., Marques-Bonet et al. 2009; Sudmant et al. 2010), and that, indeed, species-specific SDs are the result of fixed CNVs. Additionally, in our version of the test, fixed differences correspond to species-specific SDs not overlapping CNVs and polymorphic changes correspond to CNVs. Finally, in each category (fixed or polymorphic), we approximate functional changes by counting regions overlapping genes and neutral changes by counting intergenic regions. The McDonald-Kreitman test is known to be sensitive to homoplasy (i.e., parallel or repeated mutations at the same site) because it tends to blur any signal and may generate biases. To be conservative and avoid homoplasy as much as possible, we only applied this test to orangutans and chimpanzees, for which full SD information is currently available. Also, we focused on SDs that are species specific in each species relative also to human and macaque SDs; that is, we considered as SDs fixed in a given species only those that do not overlap SDs from any other known primate species. This strategy reduces drastically the number of putative homoplasic SDs, since they are known to be absent from all other species, and our McDonald-Kreitman-like analysis effectively became a test for each branch separately. In the orangutan, we could detect a significantly higher ratio of fixed over polymorphic changes for putatively functional (genic) sites than for putatively neutral (intergenic) sites (P = 0.00027) (Supplemental Table S6), which might be indicative of genic SD fixations having been driven by positive selection in the orangutan lineage. The same test was marginally nonsignificant for the chimpanzee (P = 0.087).
The genic content of CNVRs
We performed a functional characterization of CNVRs based on GO terms (Supplemental Table S7A). It is interesting to note the elevated number of significantly enriched GO categories in gorilla-specific CNVRs compared with other species. A comparison with bonobos is particularly striking, since they present a similar number of GO-annotated genes in species-specific CNVRs, but CNVRs are significantly enriched only for two GO categories. This suggests that in gorilla CNVRs are limited to some particular functional categories, while in bonobo, genic CNVRs are more widely distributed. For orangutans, given that the process of SD fixation seems to have been driven by positive selection, we compared the gene content of orangutan-specific SDs with the rest of orangutan SDs. Our results show that genes overlapping SDs fixed in the orangutan lineage are enriched for various biological processes, such as response to toxin and pheromone, and under-represented in various developmental and morphogenesis pathways (see Supplemental Table S7B).
Human-Specific NonPolymorphic Regions (HSNPs)
The data generated in this study allowed us to define genomic regions of particular interest that contain CNVs in the great apes, which either present no evidence of structural polymorphism in humans or very low-frequency polymorphism. This category is likely to represent structural variants that either became fixed in the human lineage or appeared recently and/or are maintained at low frequency by natural selection (e.g., pathogenic CNVs). We refer to these regions as Human-Specific NonPolymorphic regions (HSNPs). Given the huge amount of human CNV data, we entrust a high confidence in the fact that the regions without described CNV or with rare variant CNVs (frequencies below 5% or 1%) can be classified as nonpolymorphic human regions.
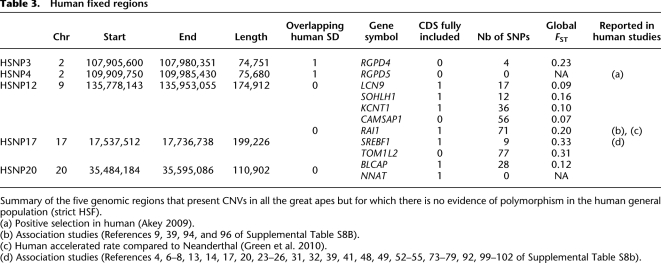
In order to construct a list of HSNP regions, we proceeded in two steps. First, we surveyed the database of genomic variants (DGV), excluding studies performed with technologies that are known to present increased uncertainty in CNV detection (such as BAC or SNP arrays) (see Winchester et al. 2009) and short CNVs (<5 kb) to make this initial survey consistent with the minimum CNV size that we can detect with our oligo array (see Methods). We identified 21 regions that present CNVRs in all African great apes, but do not harbor structural polymorphisms in humans (see Supplemental Table 8A). Out of these 21 regions, six presented CNVRs in all four great apes, including orangutans. To ensure a thorough analysis, we manually checked every single study referring to these regions, including CNVs shorter than 5 kb and irrespective of the technologies used in the study. Out of the 21 original regions, we found three regions with no current evidence for human CNVs, seven and four regions with low-frequency CNVs (frequency inferior to 1% or 5%, respectively), and seven with at least one study reporting a CNV with frequency higher than 5% (Supplemental Table S8A). These seven regions are represented in Supplemental Table S8A, but for an increased conservativeness, they were excluded from the following analyses. Out of the 14 remaining HSNP regions, the detail of the five that presented CNVRs shared by the four great apes is shown in Table 3. Some of these 14 HSNP regions do not overlap human or other great ape SDs and, thus, are very likely to be single copy in these species. A total of 58 genes are included in the HSNP loci, including genes that have been associated with disease (see list in Supplemental Table S8B), genes for which positive selection has been detected (e.g., RGPD5), or that have been shown to have derived amino acid changes in our species relative to Neanderthals (RAI1). It is also interesting to note that almost one-third of the pathologies reported in the HSNP list belong to disease classes that are related to immune function, brain function, and reproduction.
Table 3.
Human fixed regions
A complementary way to look at HSNPs is to study genome regions harboring known human pathogenic structural variants and check whether they present CNVs in the great apes. A list of genome regions containing pathogenic variants was obtained from the DECIPHERv5.1 database of submicroscopic chromosomal imbalance (see Methods). DECIPHER records all structural variants in patients with pathogenic phenotypes, but does not establish a direct one-to-one relationship between the phenotype and one or some of the various structural variants detected in each of the patients. To increase the probability to focus on pathogenic variants, we filtered out the DECIPHER coordinates that overlap CNVs in the DGV, which are less likely to be causal structural variants. The analysis shows that there are 285 regions that contain rare structural variants with strongly deleterious phenotypic effects in humans, but that present high-frequency CNVs in at least one great ape species (Supplemental Table S10A), while 534 do not show CNVs in any primates. From these 285 regions, 23 overlap CNVRs shared by the three African great apes and seven overlap CNVRs shared by all of the four great apes. Among these shared primate CNVRs overlapped by pathogenic loci, 13 are HSNP regions as defined above (Supplemental Table S10B).
HSNP regions can be the result of a combination of either of several possibilities: (1) independent acquisition of CNVRs in all great apes; (2) human-specific loss of structural polymorphism; (3) numerically congruent but independent loss of polymorphism in different human populations, such that most humans have a largely similar number of copies, but that different allelic copies were fixed in different populations. The first hypothesis seems to be the less parsimonious, especially because some CNVRs do not overlap SDs, which would be the clearest source of homoplasy. To try distinguishing between the other two, we studied the degree of differentiation between human populations in these regions by means of FST statistics (HapMap Phase 2 populations were used, see Methods). A total of 58 out of 62 genes included in HSNP regions harbor polymorphic SNPs. Taking all HapMap populations together, the average FST for these 58 genes is, on average, higher than the average FST for genes in the rest of the genome (FST = 0.161 vs. FST = 0.128, P = 0.0027). Since there is a weak but positive correlation between the number of SNPs in a gene and its average FST value (Spearman's rho = 0.045, P < 0.01), we compared the average number of SNPs in HSNP genes with the average number of SNPs in the genome. Although genes in HSNP regions have, on average, less SNPs than the rest of the genome, they still present significantly higher FST even after correcting for SNP density (P = 0.0021). When considering only the genes in the regions covered by the oligonucleotide array instead of the whole genome, the average FST for genes in the covered regions is still higher than in the rest of the genome (P = 0.0044). If we repeat the analysis for each pair of populations, the results are maintained for YRI vs. CEU and YRI vs. ASN, but not between CEU and ASN (Supplemental Table S9). Overall, these results suggest higher human differentiation in HSNPs and would favor either independent fixation of different paralogous copies of primate CNV regions in different human populations or common fixation in the human lineage, followed by local adaptation. The high FST between African and non-African populations, together with the low FST between European and Asian populations, rather supports the second hypothesis.
Discussion
CNVs, SDs, and the formation of CNVs
We produced the first study of structural variation in all of the great apes, in which we detected and studied hundreds of CNVs in each species. Our first observation is that the majority of CNVRs are not species specific, but shared among two, three, or four species. These structural variations are, in general, consistent with the known phylogenetic relationship between species (Fig. 3), with the exception of the unexpectedly large amount of CNVRs shared between gorilla on one side and chimpanzee and bonobo on the other. Shared CNVs could be the result of the conservation of ancient structural polymorphisms, but it is more likely that they are the result of high segmental duplication activity facilitating recurrent loss or creation of new copies by Non Allelic Homologous Recombination (NAHR). In full agreement with this idea, we observed that bonobos, chimpanzees, and gorillas present an enrichment of shared CNVRs in places of known SD (Table 2). Moreover, when we consider the CNV regions shared among these three species together, the association with SDs is very significant, which is not the not the case when orangutan is included. These results support the idea that the known burst of segmental duplication activity in the ancestor of the African Great Apes (Marques-Bonet et al. 2009) is the major cause of the pattern of sharedness of CNVRs between bonobo, chimpanzee, and gorilla, and suggest an important role of SDs as facilitators of CNVs in the three African great apes.
Comparing intraspecific CNV variability and SDs, we observed that in chimpanzee, gorilla, and orangutan, CNVs overlapping SDs are supported by more individuals, whereas CNVs not overlapping SDs are more likely to be singletons. Similar observations had already been made in humans (Wong et al. 2007) and chimpanzees (Perry et al. 2006). However, this is not the case in bonobo, where common CNVRs are not significantly associated with SD. This latter feature may be a specificity of this species, although it is more likely that it is due to lack of power caused by, first, the under-representation of bonobo SDs in our array; second, the fact that the SD analysis for bonobo and gorilla was performed with a proxy set of SDs (“known SDs”), since detailed information on SDs in not available for these species; and third, the relatively low sample size for bonobos in our study. More data, particularly larger sample sizes and SD maps, are required to settle this issue.
Thus, overall, rare CNVRs (within or among species) do not tend to overlap with SDs, while shared CNVRs and species-specific high-frequency CNVs do tend to overlap with SDs. Similar observations have been made with population-specific CNVs in humans (Itsara et al. 2009). Again, our results on primate CNVs are further evidence that common CNVs tend to be generated in places of shared SDs, probably via NAHR, and are shared between species and populations within a species because of their high rates of homoplasy. In contrast, rare CNVs may originate from other sequences with high similarity or via other mechanisms (e.g., Ranz et al. 2007) and/or may be affected by stronger selective pressures.
Patterns of variation and the signature of selection
Population genetics studies are increasingly possible in humans, for which abundant sequence data are available. In the rest of the great apes, data are less complete but do exist. For example, the effective population sizes (Ne) of several primate species have been estimated on the basis of nucleotide polymorphism (Yu et al. 2003, 2004). Because some bonobo and gorilla individuals from these studies were used in ours, we could compare the average differentiation of each individual in nucleotide sequence and CNV number. Overall, correlations between the two levels of polymorphism were positive, albeit nonsignificant, probably due to sample size. This is also what we observed when intergenic CNVs were considered alone. Given that the nucleotide sequences that are used for comparison are putatively neutral, this suggests that, in general terms, intergenic CNVs are neutral. In genic CNVs, two patterns stand out. First, the positive correlation between nucleotide and CNV diversity in bonobo suggests that genic CNV are also evolving neutrally in this species. Second, for gorilla CNVs containing genes, the average CNV differentiation between individuals decreases with increasing nucleotide differences, suggesting that, in gorilla, CNVs that overlap genes are under some selective pressure. Two further observations come in support of this idea. First, the number of CNVRs is higher in gorilla than in bonobo (Supplemental Fig. S3), which is consistent with Ne estimates from nucleotide polymorphism data (25,200 in gorilla and 12,300 in bonobo), but, secondly, at equal numbers of polymorphic sites (SCNV), πCNV is lower in gorilla. Therefore, the low frequency of CNVs in gorilla might be interpreted as either the result of stronger purifying selection against CNVs or higher efficiency of natural selection due to greater Ne.
For chimpanzees and orangutans, the available nucleotide polymorphism data do not come from the same individuals used in this study, but CNV polymorphism can be compared with independent measures of sequence diversity. The number of segregating CNVRs in these species, as measured by SCNV, is very similar (392 and 393), while estimates of Ne based on nucleotide diversity are much higher in orangutan than in chimpanzee. This is consistent with the recently unveiled structural stability of the orangutan lineage, which has less SDs, and is thus expected to have less CNV sites than the rest of the great apes (Locke et al. 2011). Given that orangutan has more species-specific CNVs than chimpanzee, relaxed purifying selection upon chimpanzee CNVs, together with the higher mutation rates induced by the segmental duplication burst in the ancestor of the African apes (Marques-Bonet et al. 2009), seems the most plausible explanation for this pattern. However, because the individuals in the present work have not been studied at the nucleotide-sequence level, their population structure is not fully known and the possibility remains that demography may help in explaining low-frequency CNVRs in these two species, particularly in the orangutan.
Comparing the levels of structural polymorphism in genic and nongenic CNVRs among and within species, orangutan and gorilla show a decrease of common variants in genic CNVs, which may be interpreted as genic CNVs being under stronger purifying selection within a species and/or having experienced accelerated evolution in the lineage of these species. This result would be expected if genic CNVs are a proxy for putatively functional CNVs. More surprisingly, bonobo presents an increase of common polymorphism in genic CNV compared with intergenic ones. The fact that bonobo was the only species for which the πCNV/SCNV ratio increased in genic CNVRs when compared with intergenic CNVRs (Fig. 5) may be related to the over-representation of defense and immunity genes in the CNVs of that species. Such genes may be under balancing selection and, thus, drive increased variability levels. Of course, the observation could also be caused by the poverty of significant GO terms in bonobo CNVs relative to gorilla. Smaller effective population size in bonobo would make selection less efficient and would be reflected in a random dispersion of CNVs across functional categories. This idea is supported by the strikingly similar increase of both genic and intergenic CNV diversity with neutral nucleotide diversity (Supplemental Fig. S4). Although it is difficult to draw final conclusions, the overall heterogeneity in diversity patterns suggests that CNVs are subject to different selective forces in different lineages, probably linked to their differential functional content.
Finally, we devised a McDonald-Kreitman approach to investigate the selective pressures acting on structural variants of chimpanzees and orangutans. Using a similar method Perry et al. (2008) had attempted to distinguish between the neutral accumulation of CNV by random drift versus their adaptive fixation by selection. Their results were alternatively compatible with purifying selection and with positive selection, depending on the GO categories that were represented in CNVs. For several reasons, our analysis could be more precise. Firstly, having more species allowed us to be far more conservative when counting species-specific fixed differences, and thus we were less affected by the usually high levels of homoplasy in structural variants (Marques-Bonet et al. 2009). Secondly, instead of analyzing separately structural variants that contain genes belonging to different GO categories, we performed a genome-wide test that would not incur in multiple testing issues and that would allow us to detect any overall selective force driving SD fixation in primates. We could detect the influence of positive selection in the fixation of SDs in the orangutan lineage. For orangutans, a higher ratio of fixed over polymorphic changes for putatively functional sites than for putatively neutral sites can be interpreted as the mark of adaptive structural variation that became fixed by positive selection during the divergence of orangutan from the rest of the great apes (see Supplemental Table S6). A similar trend is apparent in the chimpanzee lineage, even if it does not reach statistical significance. These results are quite suggestive, but they have to be interpreted with care because, first, intergenic CNVs are not necessarily a good proxy for neutral sites, as they may have a role in gene regulation (Stranger et al. 2007; Palacios et al. 2009; Conrad et al. 2010); second, the original McDonald-Kreitman test compares polymorphic versus fixed nucleotide changes, all arising by the same mutational process. In the case of structural changes, even if it is known that SDs and CNVs are very closely related, precise mutational models are still not available; and, third, related to the previous point, while we strived to remove homoplasy, it is not possible to measure how much of it remains and, in fact, only full sequence would help to solve the homoplasy issue. At any rate, and even taking all these caveats into consideration, it does make sense that some fixed SDs containing genes would have first appeared as CNVs and would have reached fixation under the influence of positive selection (e.g., Johnson et al. 2001). This pattern would be more obvious in the orangutan lineage than in that of the chimpanzee, because the latter species was affected by the SD outburst (Marques-Bonet et al. 2009), and the effect of recurrent segmental duplication would blur potential signals of natural selection acting upon structural variation.
So far, most genome-wide scans for positive selection in the primates (e.g., Kosiol et al. 2008) have focused on single-copy genes due to the difficulty of assigning orthologs. However, Han et al. (2009) showed that between 6% and 10% of recently duplicated genes present evidence for positive selection. Since all of these gene duplications have had an ancestral CNV and SD state, looking at more recent duplication events would allow us to know how many of these thousands of CNVs and SDs actually constitute an incipient gene duplication process. Our results support the pressing need to extend these studies to genes included in SDs and CNVs.
Human-specific nonpolymorphic regions
Finally, our study allowed us to identify, for the first time, genomic regions that have no or very low-frequency structural polymorphism in the general human population, while presenting large, high-frequency CNVRs in all four great apes or in all three African great apes. Some of these regions do not even overlap SDs in humans, so they are single-copy in our species. In addition, these regions present higher levels of differentiation in their world-wide diversity patterns, which is usually taken as an indication of the action of natural selection (Gardner et al. 2007; Itsara et al. 2009; Pickrell et al. 2009). The high differentiation between African and non-African samples in these HSNP regions, together with the low FST among non-African populations, suggests that human fixation, probably followed by adaptation, was an early event in the history of modern humans, at least predating the Out-of-Africa emergence. Furthermore, they contain genes that have been associated with disease or for which positive selection or differences between modern humans and Neanderthals have been reported (Table 3). Another interesting set of regions are 285 out of 819 locations harboring known human pathogenic structural variants that present high-frequency large CNVRs in some or all of the great apes. The absence of primate polymorphism in the remaining set of 534 out of 819 pathogenic regions may be due to our limited primate sample size. However, they are also interesting candidate regions that may contain genes or regulatory elements particularly sensitive to dosage variations, where gain or loss in copy number may result in strong pathogenic phenotypes that are not present in any of our healthy adult primate samples. Taking all of this into consideration, the more likely scenario is that adaptive importance of these regions drove their fixation in the human lineage, and that once fixed, they have still been important in human adaptation to local environments. To what extent these regions may have played a role in differentiating humans from the rest of primates is another open question that requires further research.
Methods
Sample collection
A total of 51 primate samples were analyzed (16 orangutans, 15 gorillas, 14 chimpanzees, and six bonobos). A list of individuals, origins, and hybridization arrays can be found in Supplemental Table S11. Detailed information on these individuals is provided as Supplemental Information. Different sets of samples were used in different parts of the study, as detailed in the Results section. A subset of individuals, five bonobos (KB1650, KB4229, KB7036, OR1166, OR310) and six of the gorillas (KB3456, KB3784, KB5712, KB5829, KB7973, OR934) had been previously used in measuring sequence diversity in these species (Yu et al. 2003, 2004).
CNV detection
For each set of great ape samples, array comparative genomic hybridization (aCGH) was performed against a reference of the same species. Hybridizations were performed as previously described (Marques-Bonet et al. 2009). We proceeded in two phases: discovery and validation/refinement.
Discovery phase
CNV polymorphism was examined at the genome-wide scale using a human 32K set v2.2 BAC array spanning the human genome (from Microarray Facility, Nijmegen, NL; described in http://bacpac.chori.org/pHumanMinSet.htm). The BAC array was used to discover intraspecific CNV polymorphism in a subset of samples. A total of 24 individuals were investigated: nine chimpanzees, eight gorillas, and seven orangutans were individually hybridized (one at a time) against a single reference of their own species (px14, pg20, and po14, respectively). Reversed-dye labeling of the samples was always used to minimize the effect of dye-specific biases. Hybridizations were performed as described by Wang et al. (2004). Details of the analyses that were performed on data obtained from these hybridizations can be found in the Supplemental Information.
Raw data were filtered and normalized (loess method) using the limma R package (Smyth and Speed 2003). Statistical analysis of the aCGH data obtained from the hybridizations with the BAC array was performed with R (Ihaka and Gentleman 1996; see Supplemental Information). Data of each species (chimpanzee, gorilla, orangutan) were analyzed separately and the same procedure was applied. Only autosomes were considered in the analysis.
Validation and refinement phase
Higher-resolution targeted hybridizations were performed to refine and validate the data of CNV polymorphism detected in the first phase and to discover new CNVs (see Supplemental Table S11). We designed a customized oligonucleotide microarray (NimbleGen, 385,000 isothermal probes) specifically designed to cover all of the regions that had at least one CNV in one of the primate samples hybridized in the discovery phase, plus the macaque CNV regions that are described in Lee et al. (2008), plus the positions of SD (regions >20 kb) that had been experimentally validated by Marques-Bonet et al. (2009) and their 5000-bp flanking regions. This covered 184 Mb of corresponding sequence from the human genome at an average density of 1 probe every 450 bp. In this phase, and depending on DNA availability, a total of 29 individuals was used. Five bonobos were individually hybridized against a single bonobo reference (LB502) and six gorillas against a single gorilla reference (pg20, same as the one used in the discovery phase). Of the nine chimpanzees used in this phase, three were hybridized against a single chimpanzee reference (Clint) and six against the chimpanzee reference used in the discovery phase (px14). Of the nine orangutans, seven were hybridized against a single orangutan reference (Susie) and two against the orangutan reference of the discovery phase (po14). Reversed-dye labeling of the samples was always used to minimize the effect of dye-specific biases. A total of 58 intraspecific hybridizations were performed and the log2 relative hybridization intensity was calculated for each probe.
CNV calling on the targeted NimbleGen array was performed using a modified version of the HMMseg algorithm (Day et al. 2007). HMMseg was originally designed for continuous genomic data, and our array is targeted to noncontiguous genomic regions of interest. We modified the program so that we were able to run the HMM algorithm independently on each of the regions covered by the array. A 3-state model was used (amplification, null, deletion), where transition probabilities among different states were set at 0.98 for remaining in the same state and 0.01 for changing from one state to another one. Since noise and average probe intensity may vary with experimental conditions, emission probabilities (mean and standard deviation of the states) were adapted to each individual hybridization (see Supplemental Information).
Then, pairs of dye combinations were compared and only calls that were consistent in both dye combinations were deemed correct and kept for analysis. In the case of partially overlapping (partially consistent) calls, only the fragments that strictly overlapped between dye combinations were kept. To further avoid false positives, all of the calls were visually revised, with a special care for CNV shorter than 10 kb, to make sure that they corresponded to largely overlapping small calls in both dye combinations and were not a by-product of poorly overlapping long calls that were not consistent between dye combinations. In addition, to further reduce the possibility of false positives, consistent calls that were made by less than five probes or shorter than 5 kb were eliminated. Finally, to ensure the quality of the calling, a further step of visual checking was applied to all of the CNVs, and minor modifications have been applied when necessary (see Supplemental Table S13B). The overall concordance in number of calls before and after manual checking is 99.0%, indicating a high quality in the initial calling strategy. The list of high-confidence CNVs detected in this analysis with their %GC and gene content is given in Supplemental Table S16. The numbers of calls obtained with the BAC array that were validated with the oligonucleotide array are shown in Supplemental Table S13C.
CNV calls, CNV regions, and shared CNV regions
We performed parts of the analyses directly upon individual CNV calls. For other aspects of our study we followed the practice of defining copy number variant regions (CNVRs) as genome regions that contain CNV calls that are fully or partially overlapping within each species (Redon et al. 2006; Perry et al. 2008). Coordinates of CNV calls or CNVRs are always referred to the human genome (Build 35). The edges of CNV regions are the most extreme coordinates of the set of CNV calls included in the region and, thus, they tend to be larger than any actual CNVs detected in individuals.
When comparing species, we defined shared CNV Regions as CNVRs that totally or partially overlap between species. Coordinates of shared CNVRs are, as before, the most extreme coordinates of the set of CNV regions included in the shared region. CNVRs have similar sizes to CNV (CNVRs are around 8% larger than CNV in bonobo and chimpanzee, and 13% and 19% larger in orangutan and gorilla, respectively), showing that CNVRs are formed from strongly overlapping CNVs. The minimum overlap between the CNVRs of two species is 400 bp, but the average overlap length is 61,987 kb, representing almost 73% of the length of the CNVRs considered. Overall, the use of CNVRs fairly represents the overlap CNVs within species, or CNVRs among species. This definition of CNVR is applied everywhere in the analysis, except for one case. Only in the analyses of CNV frequency (Fig. 4) and number of CNVRs shared among species (Fig. 2), did we count the number of strictly overlapping fragments of CNVs or CNVRs. This is because CNVs are differently fragmented among individuals, and one continuous CNV in one individual can be represented by two adjacent CNVs in another individual. Similarly, in interspecies comparisons, one larger CNVR in one species can overlap two smaller adjacent CNVRs in another species. For this reason, the exact individual frequency and numbers of species sharing a CNVR can only be established for fragments of fully overlapping CNVs or CNVRs.
Human CNV map
To compare the primate CNV information obtained in our study with human CNVs, we constructed a comparable human CNV map based on part of the data contained in the Database of Genomic Variants (DGV, available at http://projects.tcag.ca/variation/). We used the hg18 version of the database, since it contains more data; but, for consistency with the rest of available data, we converted the coordinates to hg17. From that data set, we limited our study to data coming from technologies that have proven to be more accurate: aCGH, pair-end mapping, sequencing, ROMA, Beadmicroarray, MLPA, and oligonucleotide arrays. In addition, we included calls from Conrad et al. (2010). From all of these studies we only kept the CNV calls that overlap the regions tiled in our oligonucleotide array. We additionally excluded CNVs with a length <5 kb to make human data readily comparable to our primate calls, which had been made using such criterion (see above). Altogether, this represents thousands of human individuals screened for CNVs. In the analysis part corresponding to 14 HSNP regions, six HSNP presented tiny variants (whose lengths are 576, 750, 1356, 1970, 2125, 2590, 3121, and 4295 bp). Given that the length of the primate CNVRs supporting HSNP regions is several orders of magnitude larger (ranging from 18,460 to 337,405 bp, with an average of 154,494 bp), consideration of these fragments would only fragment the large HSNP regions that we are reporting, but would not invalidate any of them.
Human pathogenic structural variation
The DECIPHERv5.1 database of submicroscopic chromosomal imbalance was used to gather the chromosomal location of pathogenic structural variants. All of the entries of the database were downloaded from http://decipher.sanger.ac.uk/. Only the portions of the DECIPHER loci that overlapped the genomic regions covered by the oligonucleotide array were considered in this analysis. Partially or fully overlapping DECIPHER loci were merged in 1695 pathogenic regions, following the same criteria used for CNVRs. Since many patients present copy numbers on different chromosomes simultaneously, it is not obvious which ones may be causative or not, so the analysis may account for some false positives. To increase our probabilities of focusing on really pathogenic copy number gains or losses, we filtered out pathogenic regions overlapping the CNVs reported in the DGV. The 819 remaining pathogenic regions were compared with the positions of the great ape CNVRs. In this analysis, the oligonucleotide array regions and the primate CNV positions were converted to hg19 to match the assembly of the coordinates of the DECIPHERv5.1 data. The DGV data were the same as described above, also converted to hg19.
Segmental duplication map
We used the SD map from Marques-Bonet et al. (2009) (available at http://humanparalogy.gs.washington.edu/primates2009/burst.htm). In this study, SDs were computationally determined for human, chimpanzee, and orangutan using whole genome shotgun sequence detection (WSSD). From this database, chimpanzee and orangutan SD covered 1004 (45.4 Mb) and 649 (27.7 Mb) of our tiling, respectively. SDs larger than 20 kb were experimentally validated with aCGH hybridizations using the same species, and species specificity was further refined with experimental aCGH hybridizations with bonobo and gorilla. Therefore, no direct SD data were available for bonobo and gorilla, except for those of experimental cross-species hybridizations mentioned above. For this reason, direct comparisons between CNVs detected in this study and for bonobo or gorilla SDs was not possible. However, given the high sharedness of primate SDs (Marques-Bonet et al. 2009) chimpanzee, orangutan, and human, we used a list of SD for all of these species together to study overlap between SD and CNV in all of the four species in our study. This list of SDs is referred to as “known SD” and it provides a reasonable proxy for bonobo and gorilla SD (see Results). To define coordinates of ”known SD” regions, we proceeded similarly to CNVRs, concatenating fully contiguous SD. After this process, the number of SDs and the length of the autosomal portion of the genome they covered was 1407 (66.9 Mb) for all species together.
The independence of localization of CNV regions with respect to SDs was tested by random permutations of all the CNVs within the regions tiled on the oligonucletide array used in the validation phase, while SDs were kept in their actual position. After each permutation, the number of CNVs overlapping SDs was counted. The P-values correspond to the number of times that the number of CNVRs overlapping SDs was equal to or greater than the actual number of CNVRs observed to be overlapping SDs.
Clustering analysis
To group individuals according to their CNV similarities, we summarized the CNVR data for each individual as a vector of 0s and 1s according to the absence or presence in the individual of any given CNVR. A hierarchical agglomerative clustering was applied on this matrix of individual vectors using the pvclust function from the pvclust R package (Suzuki and Shimodaira 2006). This method builds a hierarchy from the individual elements by successively merging clusters together, starting from the two closest elements, according to the chosen distance. The agglomerative method chosen was Unweighted Pair-Group Average, UPGMA. Cells in the matrix with a 0 (absence of CNVR) do contain information and should not be underweighted relative to cells with 1, so we used binary distances, which consider that 0 and 1 carry the same weight when a proximity measure is computed. We assessed the robustness of branches with 10,000 bootstraps.
Relationship between structural and nucleotide diversity
The six gorillas and the five bonobos used in this analysis are part of the set used in Yu et al. (2003, 2004), where nucleotide sequence diversity was estimated by the sequencing of 50 noncoding segments, totaling a final length of around 23.5 kb. Data from the individuals included in our study were downloaded from GenBank and standard estimates of nucleotide diversity (the number of pairwise differences, π, and Watterson's estimate of the neutral parameter of molecular evolution, θ, which is based on the number of segregating sites, S) were computed with DnaSPv5 (Librado and Rozas 2009).
To estimate the CNV diversity distribution of each species, for every individual we built a vector of 0s and 1s according to the absence or presence of each CNVR detected in this study. Using these vectors as sequences, we calculated πCNV, the average number of pairwise CNVR differences between individuals, and SCNV, the total number of segregating CNVRs in the species sample. From the vectors of all of the individuals, we built a polymorphism matrix for each species (with individuals in columns and CNVR information in rows). In this analysis, only segregating CNVRs were considered, meaning that we removed CNVRs differentiating all of the individuals of a given species from the reference of that species, implying that rows of 1s were removed from the matrices.
Measures of CNV polymorphism
The values of πCNV and SCNV could not be directly compared between pairs of species for two reasons. First, both πCNV and SCNV are dependent on sample size, which varies in the different species, and second, just as it happens with nucleotide variability, both variables are correlated and, thus, a difference in SCNV results in differences in πCNV. These problems were solved by using statistical tests based on the difference in the ratio between the two variables (πCNV/SCNV) and, in addition, by using a resampling strategy that was designed in such a way that every comparison was always done with equivalent sample size and equivalent number of CNVRs (SCNV). This strategy was as follows. First, for each comparison between pairs of species, individuals were resampled in the species with the largest sample size. On each resampling, nonsegregating sites (rows of 1s or 0s) which may have been generated by getting a subset of individuals were removed. Secondly, segregating sites were resampled in the species with the largest number of segregating sites. At this stage and by construction, the matrices of both species had an equal number of individuals and segregating sites. This resampling was repeated 10,000 times. On each of these 10,000 configurations, 10,000 permutations of polymorphic sites were performed among the two species. The P-value of the test is the number of times the difference πCNV/SCNV between two species computed after permutation was larger than the observed πCNV/SCNV computed after resampling. This two-phase procedure (resampling + permutation) was repeated for each pairwise comparison and values of πCNV/SCNV were computed for each pseudo-sample. To study the potential effects of the overlap of CNVs with genes, we considered CNVRs as genic when their overlap with any RefSeq gene was at least 100 nucleotides. The resampling + permutation procedure described above was also used to test, within each species, the difference in the πCNV/SCNV ratio between genic CNVs and non-genic CNVs.
Functional characterization of CNVs
To compare the gene content in different genome regions of interest, we compiled a list of Ensembl (http://www.ensembl.org) genes (release 58) with at least a 100-bp overlap with the feature under study. From such an original list, we removed any ENSG_IDs that matched only a clone-based sequence, with no corresponding RefSeq_ID or with no associated gene name. The largest transcript was kept for every gene. This final list was annotated for genetic and phenotypic information querying additional databases. Disease genes were obtained from OMIM (http://www.ncbi.nlm.nih.gov/omim), Genetic Association Database (http://geneticassociationdb.nih.gov/), HuGE Navigator (http://www.hugenavigator.net/), and the NIH catalog of GWASs (http://www.genome.gov/26525384). Information on positive selection was obtained from Akey (2009) and Kosiol et al. (2008), and information on Neanderthals from the Neanderthal genome study (Green et al. 2010). The over- or under-representation of functional terms in each given set of genes overlapping genomic features of interest were evaluated using Panther/tools (Thomas et al. 2003) and Babelomics/Fatigo web servers (Al-Shahrour et al. 2006).
McDonald-Kreitman-type test
In order to assess selection pressures exerted on CNVs, we adopted a MacDonald-Kreitman-like approach similar to that in Perry et al. (2008). For a given species, polymorphic sites were the CNVRs in this species. Fixed differences were species-specific SDs not overlapping CNVs. In each category (fixed or polymorphic), we approximated functional changes by counting regions overlapping with RefSeq genes and neutral changes by counting intergenic regions. In this test, we restricted SDs to the ones located in our tiling since they are the only ones for which we could be sure that they were not polymorphic.
Human population differentiation
To estimate genetic differentiation among human populations, data on Single Nucleotide Polymorphisms (SNPs) were obtained for the four populations from Phase 2 of the HapMap Project (release 22, April 2007). Following previous work (Frazer et al. 2007), JPT and CHB samples were pooled together. Only SNPs that match an Ensembl Gene ID were used in the analysis (n = 1,226,302 SNPs, from 18,883 genes). For each SNP, allele frequencies and measures of the FST statistic (used as a measure of genetic distance between populations, Weir and Cockerham 1984) were calculated with Arlequin v3.11 (Excoffier et al. 2005) as implemented in SNPator (Morcillo-Suarez et al. 2008). For each SNP, we calculated three pairwise FST values (European-Asian, European-African, and Asian-African) and a global FST value including the three HapMap populations.
To test for different FST values for genes within a given genomic region relative either to the rest of the genome or to the regions covered by our array, we used a resampling procedure that took into account that genes with different numbers of SNPs present different degrees of differentiation among human populations (data not shown). A genome-wide distribution of SNP densities in Ensembl genes was obtained according to the number of polymorphic SNPs harbored by each gene. This distribution was split in deciles, and genes within the genomic region of interest were assigned a score corresponding to the decile they occupied in the distribution of number of SNPs. We then resampled (either from the whole genome or from the regions covered in our array) a number of genes with a SNP density distribution that is equivalent to the number of genes and SNP density distribution in our regions of interest. This procedure was repeated 100,000 times, and each time, FST values were compared. The P-value is the number of times the average FST for genes in the resampled set was higher than the average FST in the observed gene list.
Data access
The aCGH data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE30559. Data are also accessible from http://biologiaevolutiva.org/anavarro/software-data/.
Acknowledgments
We are indebted to George Perry for valuable discussion. Financial support was provided by a Beatriu de Pinos postdoctoral Grant to E.G., the Spanish Ministry of Science and Innovation (Grant BFU2009-13409-C02-02 to A.N.), and the Spanish National Institute for Bioinformatics (INB, www.inab.org). E.E.E. is an investigator of the Howard Hughes Medical Institute. M.R. is grateful to CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare) and MIUR (Ministero Italiano della Universita' e della Ricerca; Cluster CO3, Prog. L.488/92).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.117242.110.
References
- Akey JM 2009. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Res 19: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J 2006. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res 34: W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol G, Knuutila S, Lozano JJ, Madrigal I, Caballin MR 2010. Identification of human specific gene duplications relative to other primates by array CGH and quantitative PCR. Genomics 95: 203–209 [DOI] [PubMed] [Google Scholar]
- Cahan P, Li Y, Izumi M, Graubert TA 2009. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nat Genet 41: 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WK, Swartz JD, Rush LJ, Alvarez CE 2009. Mapping DNA structural variation in dogs. Genome Res 19: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS 2007. Unsupervised segmentation of continuous genomic data. Bioinformatics 23: 1424–1426 [DOI] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci 104: 19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, Pollack JR, Sikela JM 2007. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res 17: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan CM, Sridhar S, Wigler M, Hall IM 2007. Recurrent DNA copy number variation in the laboratory mouse. Nat Genet 39: 1384–1389 [DOI] [PubMed] [Google Scholar]
- Eichler EE 2006. Widening the spectrum of human genetic variation. Nat Genet 38: 9–11 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50 [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S 2006. Demographic history and genetic differentiation in apes. Curr Biol 16: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Fontanesi L, Martelli PL, Beretti F, Riggio V, Dall'Olio S, Colombo M, Casadio R, Russo V, Portolano B 2010. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genomics 11: 639 doi: 10.1186/1471-2164-11-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol 2: E207 doi: 10.1371/journal.pbio.0020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M, Williamson S, Casals F, Bosch E, Navarro A, Calafell F, Bertranpetit J, Comas D 2007. Extreme individual marker F(ST)values do not imply population-specific selection in humans: the NRG1 example. Hum Genet 121: 759–762 [DOI] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, et al. 2007. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet 3: e3 doi: 10.1371/journal.pgen.0030003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, et al. 2008. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 40: 538–545 [DOI] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW 2009. Adaptive evolution of young gene duplicates in mammals. Genome Res 19: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C 2004. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951 [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R 1996. R: a language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, et al. 2009. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Lian EC, Foroud T, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514–519 [DOI] [PubMed] [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A 2008. Patterns of positive selection in six Mammalian genomes. PLoS Genet 4: e1000144 doi: 10.1371/journal.pgen.1000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Gutierrez-Arcelus M, Perry GH, Vallender EJ, Johnson WE, Miller GM, Korbel JO, Lee C 2008. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet 17: 1127–1136 [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654 [DOI] [PubMed] [Google Scholar]
- Morcillo-Suarez C, Alegre J, Sangros R, Gazave E, de Cid R, Milne R, Amigo J, Ferrer-Admetlla A, Moreno-Estrada A, Gardner M, et al. 2008. SNP analysis to results (SNPator): a web-based environment oriented to statistical genomics analyses upon SNP data. Bioinformatics 24: 1643–1644 [DOI] [PubMed] [Google Scholar]
- Newman TL, Tuzun E, Morrison VA, Hayden KE, Ventura M, McGrath SD, Rocchi M, Eichler EE 2005. A genome-wide survey of structural variation between human and chimpanzee. Genome Res 15: 1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Cokus SJ, Ghazalpour A, Ingram-Drake L, Wang S, van Nas A, Che N, Araujo JA, Pellegrini M, Lusis AJ 2009. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet 18: 4118–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios R, Gazave E, Goni J, Piedrafita G, Fernando O, Navarro A, Villoslada P 2009. Allele-specific gene expression is widespread across the genome and biological processes. PLoS ONE 4: e4150 doi: 10.1371/journal.pone,0004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Tchinda J, McGrath SD, Zhang J, Picker SR, Caceres AM, Iafrate AJ, Tyler-Smith C, Scherer SW, Eichler EE, et al. 2006. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci 103: 8006–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Yang F, Marques-Bonet T, Murphy C, Fitzgerald T, Lee AS, Hyland C, Stone AC, Hurles ME, Tyler-Smith C, et al. 2008. Copy number variation and evolution in humans and chimpanzees. Genome Res 18: 1698–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res 19: 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo Caldas Y, Castello A, Pena RN, Alves E, Mercade A, Souza CA, Fernandez AI, Perez-Enciso M, Folch JM 2010. Copy number variation in the porcine genome inferred from a 60k SNP BeadChip. BMC Genomics 11: 593 doi: 10.1186/1471-2164-11-593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, Ashburner M, Bergman CM 2007. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol 5: e152 doi: 10.1371/journal.pbio.0050152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528 [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, et al. 2005. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T 2003. Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. 2003. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Wang NJ, Liu D, Parokonny AS, Schanen NC 2004. High-resolution molecular characterization of 15q11-q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage. Am J Hum Genet 75: 267–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC 1984. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Wilson GM, Flibotte S, Missirlis PI, Marra MA, Jones S, Thornton K, Clark AG, Holt RA 2006. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res 16: 173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester L, Yau C, Ragoussis J 2009. Comparing CNV detection methods for SNP arrays. Brief Funct Genomics Proteomics 8: 353–366 [DOI] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, et al. 2007. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet 80: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Kidd JR, Deinard AS, Ryder O, Kidd KK, Li WH 2003. Low nucleotide diversity in chimpanzees and bonobos. Genetics 164: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li WH 2004. Nucleotide diversity in gorillas. Genetics 166: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]