Abstract
Structural variation has played an important role in the evolutionary restructuring of human and great ape genomes. Recent analyses have suggested that the genomes of chimpanzee and human have been particularly enriched for this form of genetic variation. Here, we set out to assess the extent of structural variation in the gorilla lineage by generating 10-fold genomic sequence coverage from a western lowland gorilla and integrating these data into a physical and cytogenetic framework of structural variation. We discovered and validated over 7665 structural changes within the gorilla lineage, including sequence resolution of inversions, deletions, duplications, and mobile element insertions. A comparison with human and other ape genomes shows that the gorilla genome has been subjected to the highest rate of segmental duplication. We show that both the gorilla and chimpanzee genomes have experienced independent yet convergent patterns of structural mutation that have not occurred in humans, including the formation of subtelomeric heterochromatic caps, the hyperexpansion of segmental duplications, and bursts of retroviral integrations. Our analysis suggests that the chimpanzee and gorilla genomes are structurally more derived than either orangutan or human genomes.
The nature of the genetic differences between humans and other great apes has fascinated scientists since the discovery of DNA in the 1950s (Sarich and Wilson 1973; Yunis and Prakash 1982; Goodman et al. 1989). The genetic relationship and phylogeny of humans and great apes is well established, based primarily on studies of single nucleotide variation (Koop et al. 1986; Enard and Paabo 2004). A surprising finding has been the extent of larger forms of structural variation among hominid genomes well below the limit of cytogenetic resolution (Locke et al. 2003; Fortna et al. 2004; Cheng et al. 2005; Bailey and Eichler 2006; Gibbs et al. 2007; Marques-Bonet et al. 2009a). Interestingly, the hominid genomes appear to be enriched with respect to structural variation, but the extent to which this has impacted each of the major lineages is not yet completely known. To date, three hominid genomes have been sequenced and assembled to the working draft stage using capillary-based approaches [human (The International Human Genome Sequencing Consortium 2001, 2004), chimpanzee (The Chimpanzee Sequencing and Analysis Consortium 2005), and orangutan (Locke et al. 2011)]. Projects are underway to sequence additional apes including the bonobo, gorilla, and gibbon. Many of these remaining ape genomes will be sequenced and assembled using a combination of next-generation sequencing and capillary whole-genome shotgun sequence data sets (Marques-Bonet et al. 2009b). Studies of structural variation, however, are complicated by difficulties in detecting and accurately resolving the sequence structure of these regions. In this study, we set out to systematically investigate the pattern of structural variation in the gorilla genome, combining capillary-based clone sequencing and next-generation genome sequencing in conjunction with detailed cytogenetic characterization and experimental validation. We present a comprehensive overview of inversions, deletions, segmental duplications, and retrotranspositions within the gorilla genome. Comparisons with humans and other apes reveal that parallel and independent mutational processes have more dramatically restructured chimpanzee and gorilla genomes when compared with other hominid genomes.
Results
In order to investigate the gorilla's pattern of genome structural variation, we undertook a three-pronged approach. First, we tested 788 human BAC clones by fluorescence in situ hybridization (FISH), comparing the probe order on human and gorilla chromosomal metaphases, thus providing a refined cytogenetic framework of large-scale and intermediate-sized rearrangement events (Supplemental Note). Next, we completely end sequenced 176,880 BAC clones (http://www.genome.gov/Pages/Research/Sequencing/BACLibrary/primateProposal.pdf) from a gorilla BAC library (CH277) and mapped them to the human reference genome [NCBI build 35 (NCBI35)] to generate a clone-based framework of the gorilla genome (Eichler and DeJong 2002). This approach defined potential rearrangements based on discordant end sequence placements. Last, we obtained blood DNA from Kwan, a middle-aged silverback gorilla, and generated 9.6-fold effective sequence coverage using massively parallel Illumina sequencing. While this sequence coverage means that each base is represented on average nine to 10 times, the paired-end sequences flanking a portion of the insert that is not sequenced means a larger fraction of the genome is spanned by anchored mate pairs (34-fold). These data were used to identify regions of copy number variation based on sequence read-depth and paired-end mapping revealing smaller forms of structural variation including mobile element insertions (>300 bp) using end sequence profiling approaches (Tuzun et al. 2005; Hormozdiari et al. 2009; Hormozdiari et al. 2010). The experimental and molecular data were integrated (Table 1; Supplemental Note), allowing us to correctly reclassify events that particularly distinguished translocations from duplicative transposition events and inversions from segmental duplications (SDs). For example, translocations could be distinguished from duplicative transpositions because read-depth and array comparative genomic hybridization (arrayCGH) predicted copy number changes of a segment of DNA but with no evidence of chromosomal rearrangement using cytogenetic markers. In those cases where we were able to completely sequence the corresponding BAC clone, the breakpoints could be resolved at the single base pair level. We summarize the pattern of gorilla genome structural variation from the perspective of size and class, and then compare our findings with human and other great ape genomes.
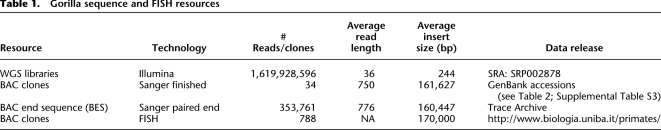
Table 1.
Gorilla sequence and FISH resources
Large-scale rearrangements and duplicative transpositions
Yunis and Prakash originally reported 11 large-scale cytogenetic differences between human and gorilla (eight pericentric inversions, one paracentric inversion, one translocation, and one fusion) (Yunis and Prakash 1982). Using FISH and data from other studies (Dutrillaux 1980; Yunis and Prakash 1982; Montefalcone et al. 1999; Muller et al. 2000, 2003; Carbone et al. 2002; Eder et al. 2003; Locke et al. 2003; Misceo et al. 2003, 2005; Ventura et al. 2003, 2004; Cardone et al. 2006, 2007; Stanyon et al. 2008), we refined all classical evolutionary breakpoints that distinguish the human and gorilla karyotypes (Supplemental Table S1). It should be noted that cytogenetics typically only resolves megabase pair-level variation, while FISH analyses utilizing overlapping probes can be used to resolve events ∼50 kb in size, depending on the complexity of the region. Using gorilla BAC end-sequence (BES) data mapped against the human genome (NCBI35), we identified 424 putative chromosomal rearrangements (Supplemental Table S2; Supplemental Note) including six of the 11 original classical rearrangements. The remaining five were confirmed by FISH but mapped to large and nearly identical SDs that could not be traversed by BES. We selected 14 representative BAC clones corresponding to the classical gorilla-human breakpoints and completely sequenced them using capillary-based sequencing methods (Table 2). Detailed breakpoint analyses (Fig. 1; Supplemental Figs. S1–S5) showed SDs (translocation 5;17 and inversion 7) or common repeat elements (Alus and LINEs) at or in close proximity to all breakpoints. Due to the abundance of these elements in the hominid genome and the small number of sequenced sites, this enrichment is not significant. In most cases, the repetitive sequences were not homologous, suggesting that mechanisms other than nonallelic homologous recombination were responsible for these evolutionary rearrangements. None of these rearrangements disrupted unique genes.
Table 2.
BAC sequenced chromosomal breakpoints
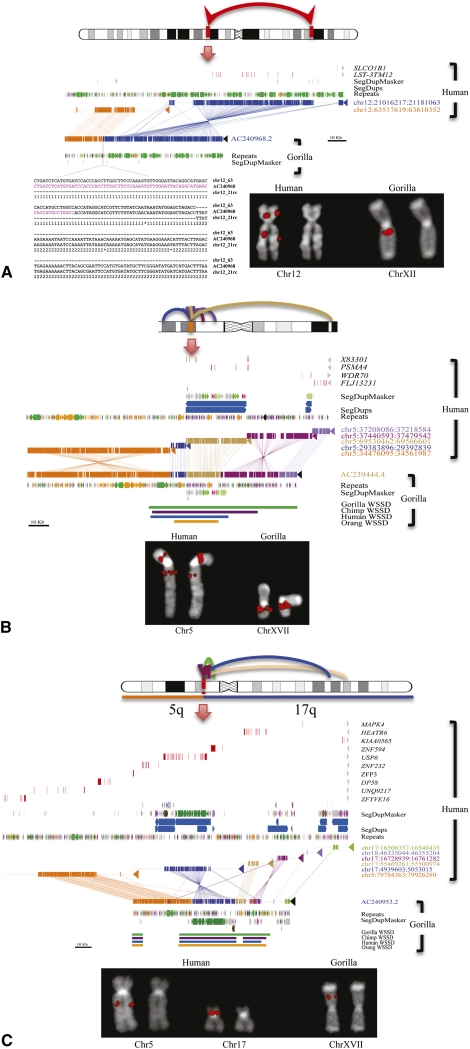
Figure 1.
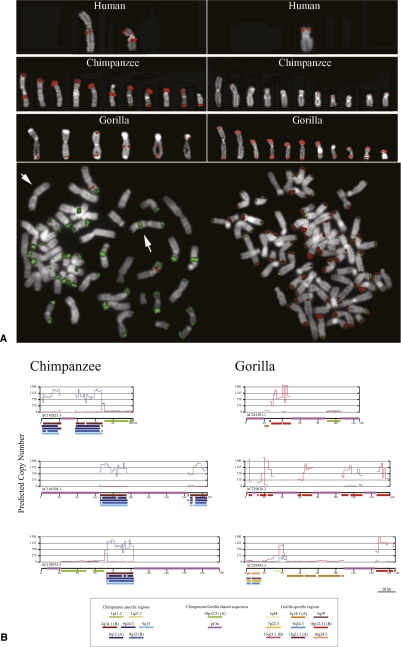
Sequencing of large-scale gorilla chromosome rearrangements. (A) Sequence characterization of a gorilla inversion on chromosome 12 (Egozcue and Chiarelli 1967; Miller et al. 1974). A schematic of the 42.5 Mb inversion (top panel ideogram) is refined by paired-end BES and confirmed by FISH (bottom right panel). A gorilla BAC clone (CH277-205P14) corresponding to the long-arm breakpoint is completely sequenced (AC240968). Miropeats analysis (Parsons 1995) compares two regions on human chromosome 12 (middle panel; blue and orange) with the gorilla sequence. (Repeat elements, LINEs, SINEs, LTRs, SDs, and genes are annotated based on the human genome.) (Bottom left panel) ClustalW alignment pinpoints an AluSx element (purple) at the precise breakpoint. (B) Sequence characterization of a duplicative transposition region in gorilla. (Top panel) A region on chromosome 5p13 near the retinoic acid-induced 14 gene (RAI14) has acquired at least four SDs in the gorilla lineage. (Middle panel) Sequencing of gorilla BAC clone CH277-50D8 (AC239444.4) and Miropeats analysis show SDs ranging in size from 10 to 28 kb along chromosome 5. Read-depth analyses (WSSD tracks) predict that these gorilla duplicative transpositions are focused on a more ancient duplication carrying the spinal muscular atrophy type 4 gene (PSMA4), with flanking duplications becoming increasingly lineage-specific. (Lower panel) FISH analysis with the clone as a probe shows multiple signals. Unique probes proximal and distal to the region indicate no change in the order between human and gorilla. Duplicative transpositions have occurred without a large-scale chromosomal rearrangement. (C) Duplicative transposition at translocation fusion point. (Top panel) Gorilla chromosome XVII arose as a lineage-specific fusion between chromosome 5 and 17 (Stankiewicz et al. 2001). Cloning and sequencing of the breakpoint (CH277-159N16, AC240953) show the presence of a >85 kb complex gorilla-specific duplication block at the breakpoint. The duplication block is a mosaic composed of at least five distinct SDs originating primarily from chromosome 5 (see Supplemental Note for more detail).
The BAC read-pair analysis predicted an unusually large number of putative inversions and translocations, which is inconsistent with previous chromosomal analyses and our own cytogenetic framework. We selected a subset of these events (six putative translocations and 14 inversions) (Supplemental Table S3) for further investigation. For each rearrangement, if the predicted translocations and inversions were bona fide, we would expect a change in the order of flanking probes (inversions) or a change in chromosomal location (translocation) when comparing human and gorilla. For each of these breakpoints, we selected gorilla BAC clones spanning the putative rearrangement breakpoints, as well as gorilla BAC clones located distally and proximally to each breakpoint, and tested their order between human and gorilla. In all cases, no change in the order of flanking unique sequences was observed. These FISH results suggested the presence of duplicated sequences at new locations in the gorilla genome.
We completely sequenced 20 of the corresponding BAC clones (3.2 Mb of finished capillary-based sequence) to resolve the structure of these selected loci (Supplemental Table S3). In each case, we confirmed duplicative transpositions and gorilla-specific juxtapositions of SDs as opposed to inversions and translocations. For example, BLAST sequence similarity searches of the sequenced gorilla BACs from chromosome 5 (AC23944) (Fig. 1B) indicate that this portion of the gorilla genome consists of a mosaic of four diverse SDs originating from different locations on human chromosome 5. The Miropeats analysis (Parsons 1995) shows the extent of each duplicated segment (ranging in length from 9 to 86 kb). Sequence read-depth analysis among the various species [whole-genome shotgun sequence detection (WSSD) tracks; see Supplemental Note] suggests that different segments have been duplicated at different time points during evolution. We conclude that this particular architecture is unique to gorilla originating from a series of duplicative transpositions to gorilla chromosome 5p13.2. We note that none of the 20 sequenced regions from BACs were collinear with the human genome. These regions carry, on average, three reconfigured or newly interspersed duplications with an average length of 29 kb. We estimate 79% map interstitially within gorilla chromosomes, with another 21% mapping to subtelomeric or pericentromeric regions. These data reveal extensive duplicative transposition in the gorilla genome creating a complex pattern of SDs unique to this lineage (see below).
Deletions
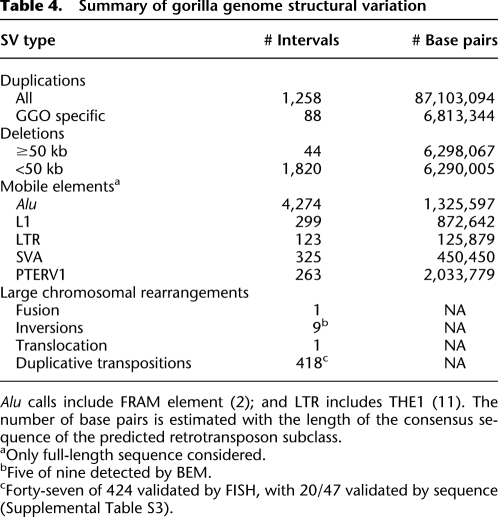
We initially detected 79 large deletions (>50 kb) compared with human using a combination of interspecies arrayCGH, BES mapping, and depletion in sequence read-depth. Of these events, 89% (70/79) were confirmed experimentally by arrayCGH and/or FISH (Supplemental Table S4). Based on human genome annotation, these regions contained 61 genes that were either completely (38) or partially (23) deleted in Kwan (Supplemental Table S4). We examined 52 of these regions by FISH and found that 62% (32/52) of these apparent deletions correspond to regions of duplication in the human where the gorilla simply showed reduced copy number. Only 16 of the regions contained unique hominid genomic regions that had been completely deleted in the gorilla lineage (Supplemental Fig. S6; Supplemental Table S4). In order to detect smaller deletions in the gorilla genome, we searched for clusters of discordant read pairs based on mapping gorilla sequence (Supplemental Note) against the human reference genome (Tuzun et al. 2005; Hormozdiari et al. 2009). We experimentally validated 1820 deletion intervals (6.7 Mb) using a customized microarray (Supplemental Note). This included 580 partial and 13 complete gene deletions (Supplemental Table S5; Supplemental Note). Many of the completely deleted genes belong to well-known gene families, including olfactory receptors (OR10K1, OR5L2, OR5D16, OR1M1, and OR7G2), keratin-associated proteins (KRTAP13-3 and KRTAP13-4) and HLA genes [HCP5 (HLA complex P5) and HCG26 (HLA complex group 26)]. One particularly intriguing deletion included SELV (selenoprotein V), thought to be important in the metabolism of dietary selenium, implicated in cancer prevention, immune function, aging, male reproduction, and other physiological processes (Kryukov et al. 2003). Among the partial gene deletions, 36 span more than a single exon—the largest being DNAH14 with 45 deleted exons. In total, we discovered and validated 1863 deletion intervals in the gorilla genome corresponding to 12.69 Mb.
Mobile elements
We initially excluded regions with >80% repeat content from our deletion analysis due to difficulties in validating these calls by arrayCGH. However, such deletions likely correspond to mobile element insertions in the human genome that occurred since the two lineages diverged. We specifically searched for the aforementioned events by identifying deletions where the corresponding locus in human was composed largely of a particular common repeat. We predicted 2481 Alu (887 kb), 1861 L1 (5.59 Mb), 663 SVA (1.17 Mb), 524 LTR (1.27 Mb), and 110 HERV (409 kb) insertions in human when compared with gorilla (Hormozdiari et al. 2010). A subset of these were full length, including 2372 Alu (≥275 bp) and 564 L1 (≥5.8 kb) elements. Of these mobile elements, 37% (2066/5639) mapped within 1650 genes (Supplemental Table S6). We performed the reciprocal analysis by searching for retrotranspositions specifically within the gorilla lineage. We detected the insertion breakpoints of various classes of active mobile elements (Alu, L1, SVA, and LTR) as well as nonhuman primate-specific endogenous retroviruses (PTERV1 and PTERV2) (Yohn et al. 2005) and predicted a total of 263 PTERV1, 4272 Alu, 325 SVA, 113 LTR, and 299 full-length L1 insertions (Supplemental Table S7). In order to estimate the rate of false positives, we tested 30 full-length gorilla Alu retrotransposons by PCR analysis of gorilla and human DNA (Supplemental Note). Of the events, 90% (27/30) validated as fixed insertions (Supplemental Fig. S7), with the remaining three being polymorphic. Consistent with recent analyses of human and other ape genomes, these results predict an acceleration of SVA retrotransposition in the chimpanzee-human ancestral lineage with a more recent surge of Alu retrotransposons in the human branch (Table 3). While we found no evidence of PTERV2 integrations in gorilla, we did identify 263 full-length integrations of PTERV1—an endogenous retrovirus initially discovered in chimpanzee (Yohn et al. 2005). A comparison with a previously developed integration map of chimpanzee revealed that 99.6% of these integrations are nonorthologous, mapping to different locations in the gorilla and chimpanzee genomes (Fig. 2). Both experimental and sequence analyses confirm that this mobile element is completely absent from human and orangutan genomes. This provides strong support that PTERV1 arose from an exogenous source that retrotransposed independently in both gorilla and chimpanzee lineages <6 million years ago.
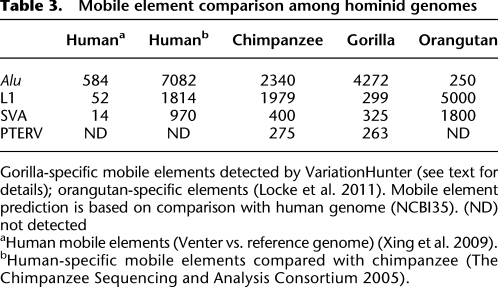
Table 3.
Mobile element comparison among hominid genomes
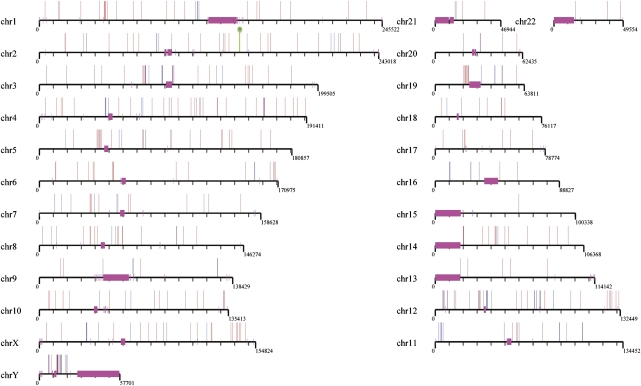
Figure 2.
Endogenous retroviral integration map. A comparison of chimpanzee (n = 275, blue) and gorilla (n = 265, red) PTERV1 sites of integration based on mapping to the human genome. None of the map positions in the two genomes are orthologous except one (indicated in green and corresponding to gorilla chr2: 143467521-143467682; chimpanzee chr2: 143467889-143468851). The endogenous retroviral element is absent in human and orangutan genomes and appears to have expanded largely independently in the two lineages after they diverged.
Segmental duplications
We developed an SD map of the gorilla genome based on detecting regions with excess sequence read-depth as described previously (Bailey et al. 2002; Alkan et al. 2009; Marques-Bonet et al. 2009a). We detected 99 Mb of SDs (>20 kb in length and >95% identity) (Cheng et al. 2005; Marques-Bonet et al. 2009a). We validated the duplications by interspecies arrayCGH, discovering 68 complete or partial gene duplications in the gorilla (Supplemental Note). Although most of the duplications are shared with other hominids (Fig. 3; Supplemental Note), we note an apparent excess of gorilla-specific duplications when compared with human, chimpanzee, or orangutan. Comparing the SD maps of five primate genomes, we assigned shared and lineage-specific SDs (Fig. 3B) and computed a genomic duplication rate along each branch under a maximum likelihood model, which assumes 20% homoplasy. The addition of gorilla duplication data into a maximum likelihood framework suggests an excess of SD in the human-African great ape ancestor that is larger than previously reported (Marques-Bonet et al. 2009a), with an estimated rate four- to fivefold higher when compared with the human or chimpanzee branches. Surprisingly, the gorilla-specific branch is also significantly accelerated compared with the human (approximately two to four times), but less so when compared with the common ancestral branch of humans and chimpanzees.
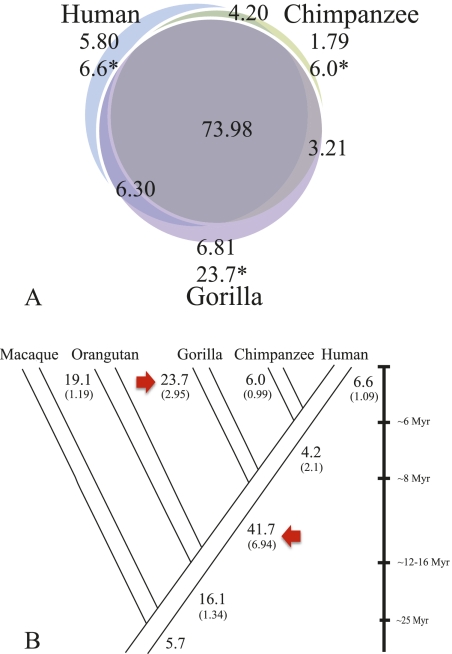
Figure 3.
Segmental duplication distributions. (A) SDs (>20 kb) were classified as lineage-specific or shared based on a three-way comparison of human, chimpanzee, and gorilla genomes. The inclusion of gorilla suggests that most SDs are shared among humans and African apes but not with Asian apes (Supplemental Note; Marques-Bonet et al. 2009a). Numbers are in megabase pairs; all SDs were validated by interspecies arrayCGH; (*) megabase pairs adjusted for copy number. (B) Using parsimony, we assigned the number of megabase pairs to different terminal and ancestral branches in the human-ape phylogeny. The copy-number-corrected megabase pairs are shown (bold type) and a calculated rate of megabase pairs/million years (in brackets) is estimated. A simple maximum-likelihood ratio test showed a dramatic SD burst in the African ape ancestor and in the common ancestor of humans and chimpanzees. The gorilla lineage-specific rate is greater than any other hominid.
This difference becomes more dramatic in the gorilla when adjusting for copy number (Fig. 3A), indicating that several sequences have expanded more prominently. We tested and validated by FISH (Supplemental Table S8) 11 gorilla-specific duplications showing the highest copy number increase (11–45 copies). Nine of these 11 regions contain genes completely and/or partially expanded specifically in the gorilla lineage (e.g., NDUFA8—unknown function, LRPAP1—low-density lipoprotein receptor-related protein, DOK7—downstream from tyrosine kinase 7, HGF—activator preproprotein, LETM1—leucine zipper-EF-hand containing transmembrane, and FGFR3—fibroblast growth factor receptor 3 isoform 2). Most of these expansions map to the termini of gorilla chromosomes indicating that the subtelomeric regions of gorilla have become increasingly complex as a result of duplicative transpositions (see above). In this regard, the most expanded gorilla-specific duplication (n = 45 copies) maps within 10 kb of the evolutionary fusion point that led to the formation of human chromosome 2 (chr2: 114145970-114215607). We also compared the copy number of shared duplications among humans, chimpanzees, and gorillas searching for regions of hyperexpansion (>500 copies) in one lineage when compared with the other two (Supplemental Table S9). Only in the gorilla and chimpanzee genomes were such hyperexpanded SDs identified with expansions of 1000–1500 copies mapping primarily to acrocentric, pericentromeric, subtelomeric, and subterminal cap regions of African ape chromosomes (Supplemental Table S9).
Subterminal caps
One of the most striking karyotypic differences between humans and African apes is the presence of subterminal heterochromatic caps at the ends of ape chromosomes (Fig. 4A). Evident by G banding (Yunis and Prakash 1982) and post-denaturation DAPI staining, these regions have been classified as subterminal heterochromatin found exclusively among gorillas (80/96 chromosomes), the common chimpanzee (Pan troglodytes) (42/96 chromosomes), and bonobo (Pan paniscus) (42/96 chromosomes). They are thought to be composed primarily of a 32-bp satellite repeat sequence (pCht7/13 sequence) arrayed in tandem (Royle et al. 1994). In addition, it is known that the formation of the subterminal cap in chimpanzee was accompanied by the hyperexpansion of SDs, which map near the human chromosome 2 fusion point (113997859-114024033, NCBI35) (Fan et al. 2002; Cheng et al. 2005). In gorilla, we find no evidence of an association of chromosome 2 sequences with the heterochromatic caps, but rather our copy number and FISH analysis suggests that a segment of chromosome 10 (19557646-19564636, NCBI35) is one of the primary components of the gorilla cap (Fig. 4A). To confirm these results, we selected three large-insert BAC clones corresponding to the cap regions of both gorilla and chimpanzee and subjected these to capillary-based sequence and assembly. The sequence analysis shows dramatic differences in the organization of heterochromatic caps between the two species. While both possess tracts of pCht satellite sequence ranging in size from 10 to 50 kb, chromosome 2 SDs are predominant in the chimpanzee cap, whereas chromosome 10 duplications define the cap organization in gorilla (Fig. 4B). These duplications appear to have expanded in concert with the satellite sequence creating a higher-order tandem array structure of several hundred copies in each species. Since subterminal heterochromatic blocks have also been reported among the lesser apes (Wijayanto et al. 2005), we tested pCht satellite probes on gibbon metaphase chromosomes and observed no hybridization signal above background (data not shown). Combined, these data strongly suggest that the heterochromatic caps have evolved independently in both chimpanzee and gorilla and possibly all ape species.
Figure 4.
Subterminal heterochromatic cap architecture in chimpanzee and gorilla. (A) Different FISH hybridization patterns using human fosmid probes (ABC8_40868200_C16 and ABC8_40925900_F12) corresponding to one hyperexpanded SD in gorilla (chr10: 19530349-19564732) and one in chimpanzee (chr2: 113978394-114020431). Extracted metaphase chromosomes (top panel) and cohybridization experiments (lower panel) in chimpanzee and gorilla reveal differences in the composition of the heterochromatic cap in each species. (B) Complete sequence analysis of three large-insert BAC clones sampled from gorilla and chimpanzee genomes confirm large-scale differences in the sequence organization. pCht satellite sequence (purple) interdigitates between different SDs (color bars) depending on the species. These SDs have expanded in copy from 500 to 1000 copies (y-axis represents copy number count based on read-depth) in chimpanzee (blue) and gorilla (red). This architecture has emerged in a species-specific fashion in conjunction with the evolution of the subterminal heterochromatic satellite.
Discussion
Structural variation has been extensive and episodic during human-great ape evolution. In this study, we identified and validated >7665 (Table 4) structural variant events in the gorilla when compared with human. It is important to note that our analysis is based primarily on a single gorilla genome (Kwan). Consistent with other studies, we expect 10%–30% of these variants to be polymorphic (i.e., not fixed within the gorilla lineage) (Chen and Li 2001; Ebersberger et al. 2002; Marques-Bonet et al. 2009a). Nevertheless, we find that the gorilla branch shows a significant increase in the rate of SD when compared with human or chimpanzee (p < 5.6 × 10−7), being more similar to the rate predicted in the human-African ape ancestor. Notably, our estimated Homo-Pan ancestral rate of duplication appears higher than rates estimated for the chimpanzee and human terminal branches. In general, our data support a model where SD activity slowed after hominid speciation events in all lineages, with this deceleration being the least evident for gorilla. This slowdown is supported by the observation that the sequence identity spectrum of SDs in humans peaks at 99.2% (Bailey and Eichler 2006) and the finding of few large-scale SD differences between human and Neandertal, which separated <1 million years ago (Green et al. 2010). The basis for deceleration is unknown, but it is possible that extensive differences in the SD architecture facilitated genetic isolation of emerging species during evolution (White 1978). We caution, however, that we cannot accurately estimate the time of such SDs with respect to hominid speciation events, so such correlations remain speculative. Thus, a reasonable line of inquiry going forward will be to compare the extent of genetic diversity in an unbiased fashion within each great ape lineage and compare these with divergence estimates between species.
Table 4.
Summary of gorilla genome structural variation
In this study, we document hundreds of duplicative transposition differences between human and gorilla that alter the structure of duplication blocks between the two lineages. Most of these structural differences are opaque to standard whole-genome shotgun sequence assembly methods or would be incorrectly classified without integration into a higher-level cytogenetic framework. Our structural variation analysis also suggests that the genomes of chimpanzee and gorilla have experienced several independent genomic rearrangements that did not occur during the evolution of the orangutan and human. The African ape genomes have been bombarded by retroviral integrations that entered the germline after the two lineages diverged. Neither orangutan nor human genomes carry these retroelements (Yohn et al. 2005) and the fact that fine mapping of the integration sites are largely nonorthologous argues for ancient parallel infections (Kaiser et al. 2007). Gorilla and chimpanzee have independently acquired subtelomeric heterochromatin caps, and this chromosome feature has been associated with the hyperexpansion of different SDs in the two lineages. Our molecular analyses suggest that these events occurred independently and in parallel early during the evolution of the Pan and Gorilla lineages, adding many new megabase pairs of DNA that altered the chromosome and chromatin architecture of these two species compared with all other primates. We propose that the orangutan and human genomes represent the hominid archetype, while the African ape genomes are more structurally derived with respect to these properties.
Methods
FISH
BAC and human fosmids clones (n = 1022) were used as probes to develop a comparative cytogenetic framework and to test rearrangements specific to the gorilla lineage. Ancestral state was determined based on comparison with other primate species. Metaphases from nonhuman primates were obtained from lymphoblastoid or fibroblast cell lines of the following species: common chimpanzee (Pan troglodytes, PTR); gorilla (Gorilla gorilla, GGO), and Borneo orangutan (Pongo pygmaeus pygmaeus, PPY) as representative of great apes; and rhesus monkey (Macaca mulatta, MMU, Cercopithecinae) as representative of Old World Monkeys. FISH experiments were essentially performed as previously described (Ventura et al. 2003).
Gorilla genome sequencing
Peripheral blood DNA was isolated from a male silverback gorilla, Kwan (Studbook #1107, b. 02/03/1989), housed at the Lincoln Park Zoo. Paired-end whole-genome sequence data were generated on an Illumina Genome Analyzer II using a modified protocol (see Supplemental Note). Sequence data have been deposited into the SRA under accession SRP002878.
BAC sequencing
End sequences from a gorilla BAC library (CH277) were retrieved from the NCBI Trace Archive and mapped to the human reference genome (NCBI35) to identify and clone rearrangement breakpoints as described previously (Newman et al. 2005). A subset of clones was selected for complete insert sequencing using capillary sequencing methods (McPherson et al. 2001) in order to obtain high quality finished sequence within duplicated regions. Rearrangements were visualized using Miropeats (Parsons 1995) and previously described in-house visualization tools (Kidd et al. 2010).
Structural variation discovery
Gorilla sequence reads were aligned to the human reference genome using the mrFAST and mrsFAST mapping algorithms (Alkan et al. 2009; Hach et al. 2010). Deletions and mobile element insertions were detected using VariationHunter (Hormozdiari et al. 2009; Hormozdiari et al. 2010), while SDs (>20 kb) were detected and copy number quantified using measures of read-depth (see Supplemental Note; Alkan et al. 2009).
ArrayCGH
We designed two oligonucleotide microarrays (n = 385,000) targeted to regions of gorilla deletions and duplications and performed cross-species arrayCGH as previously described (GEO accession numbers: GSE27072; samples: GSM665036, GSM665334, GSM665336, GSM665992, GSM665993, GSM667894, and GSM668114; and platforms GGO 2.1 custom: GPL11674 and Human 2.1 standard: GPL9684).
PCR
Thirty PCR assays were designed to test the specificity and polymorphism of predicted Alu insertions in the gorilla genome. We only tested loci not embedded within other repetitive elements or SDs to facilitate reliable primer design.
Data access
Gorilla sequence data (western lowland) have been deposited into the SRA under accession SRP002878.
We designed two oligonucleotide microarrays GEO accession number: GSE27072; samples: GSM665036, GSM665334, GSM665336, GSM665992, GSM665993, GSM667894, and GSM668114; and platforms GGO 2.1 custom: GPL11674 and Human 2.1 standard: GPL9684.
A subset of clones was selected for complete insert sequencing: AC243004.1, AC242656.3, AC242655.3, AC242627.3, AC242595.2, AC243002.1, AC241241.3, AC241522.2, AC240968.2, AC240954.2, AC239638.4, AC243003.2, AC243178.1, AC240953.2, AC239379.3, AC239356.2, AC239357.3, AC239280.2, AC239360.3, AC239282.3, AC239362.3, AC239380.3, AC239639.1, AC239363.3, AC239796.3, AC239381.3, AC239444.4, AC239382.3, AC239359.3, AC239358.3, AC239361.3 AC239281.3, AC239393.3, AC239640.2.
Acknowledgments
We thank Peter Sudmant and Tonia Brown for valuable comments in the preparation of this manuscript. We are grateful to Lisa Faust, Mollie Herget, and the staff of the Lincoln Park Zoo for the gorilla material used in this study. We are also indebted to the large genome sequencing centers for early access to BAC end sequence data for targeted analysis of structural rearrangements. This work was supported, in part, by an NIH grant HG002385 to E.E.E. and NIH grant U54 HG003079 to R.K.W. Financial support was provided by the Spanish Ministry of Science and Innovation (Grant BFU2009-13409-C02-02) and the Spanish National Institute for Bioinformatics (INB, www.inab.org) to A.N. T.M-B. is supported by a Ramon y Cajal program (MICINN-RYC) and the European FP7 (MC/StGr 2010). Resources for exploring the gorilla genome structural variation online are available at University of Washington Genome Sciences (http://gorillaparalogy.gs.washington.edu). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124461.111.
References
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 41: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE 2006. Primate segmental duplications: Crucibles of evolution, diversity, and disease. Natl Rev 7: 552–564 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Carbone L, Ventura M, Tempesta S, Rocchi M, Archidiacono N 2002. Evolutionary history of chromosome 10 in primates. Chromosoma 111: 267–272 [DOI] [PubMed] [Google Scholar]
- Cardone MF, Alonso A, Pazienza M, Ventura M, Montemurro G, Carbone L, de Jong PJ, Stanyon R, D'Addabbo P, Archidiacono N, et al. 2006. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol 7: R91 doi: 10.1186/gb-2006-7-10-r91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MF, Lomiento M, Teti MG, Misceo D, Roberto R, Capozzi O, D'Addabbo P, Ventura M, Rocchi M, Archidiacono N 2007. Evolutionary history of chromosome 11 featuring four distinct centromere repositioning events in Catarrhini. Genomics 90: 35–43 [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 68: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93 [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Dutrillaux B 1980. Chromosomal evolution of the great apes and man. J Reprod Fertil Suppl Suppl 28: 105–111 [PubMed] [Google Scholar]
- Ebersberger I, Metzler D, Schwarz C, Paabo S 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet 70: 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder V, Ventura M, Ianigro M, Teti M, Rocchi M, Archidiacono N 2003. Chromosome 6 phylogeny in primates and centromere repositioning. Mol Biol Evol 20: 1506–1512 [DOI] [PubMed] [Google Scholar]
- Egozcue J, Chiarelli B 1967. The idiogram of the lowland gorilla (Gorilla gorilla gorilla). Folia Primatol (Basel) 5: 237–240 [DOI] [PubMed] [Google Scholar]
- Eichler EE, DeJong PJ 2002. Biomedical applications and studies of molecular evolution: A proposal for a primate genomic library resource. Genome Res 12: 673–678 [DOI] [PubMed] [Google Scholar]
- Enard W, Paabo S 2004. Comparative primate genomics. Annu Rev Genomics Hum Genet 5: 351–378 [DOI] [PubMed] [Google Scholar]
- Fan Y, Linardopoulou E, Friedman C, Williams E, Trask BJ 2002. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Res 12: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol 2: E207 doi: 10.1371/journal.pbio.0020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234 [DOI] [PubMed] [Google Scholar]
- Goodman M, Koop BF, Czelusniak J, Fitch DH, Tagle DA, Slightom JL 1989. Molecular phylogeny of the family of apes and humans. Genome 31: 316–335 [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach F, Hormozdiari F, Alkan C, Birol I, Eichler EE, Sahinalp SC 2010. mrsFAST: A cache-oblivious algorithm for short-read mapping. Nat Methods 7: 576–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC 2009. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res 19: 1270–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Hajirasouliha I, Dao P, Hach F, Yorukoglu D, Alkan C, Eichler EE, Sahinalp SC 2010. Next-generation VariationHunter: Combinatorial algorithms for transposon insertion discovery. Bioinformatics 26: i350–1357 doi: 10.1093/bioinformatics/btq216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- The International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Kaiser SM, Malik HS, Emerman M 2007. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 316: 1756–1758 [DOI] [PubMed] [Google Scholar]
- Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, Hayden HS, Alkan C, Malig M, Ventura M, Giannuzzi G, et al. 2010. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat Methods 7: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop BF, Goodman M, Xu P, Chan K, Slightom JL 1986. Primate eta-globin DNA sequences and man's place among the great apes. Nature 319: 234–238 [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN 2003. Characterization of mammalian selenoproteomes. Science 30: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Locke DP, Archidiacono N, Misceo D, Cardone MF, Deschamps S, Roe B, Rocchi M, Eichler EE 2003. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol 4: R50 doi: 10.1186/gb-2003-4-8-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. 2011. Comparative and demographic analysis of orangutan genomes. Nature 469: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009a. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Ryder OA, Eichler EE 2009b. Sequencing primate genomes: What have we learned? Annu Rev Genomics Hum Genet 10: 355–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, Wallis J, Sekhon M, Wylie K, Mardis ER, Wilson RK, et al. 2001. A physical map of the human genome. Nature 409: 934–941 [DOI] [PubMed] [Google Scholar]
- Miller DA, Firschein IL, Dev VG, Tantravahi R, Miller OJ 1974. The gorilla karyotype: Chromosome lengths and polymorphisms. Cytogenet Cell Genet 13: 536–550 [DOI] [PubMed] [Google Scholar]
- Misceo D, Ventura M, Eder V, Rocchi M, Archidiacono N 2003. Human chromosome 16 conservation in primates. Chromosome Res 11: 323–326 [DOI] [PubMed] [Google Scholar]
- Misceo D, Cardone MF, Carbone L, D'Addabbo P, de Jong PJ, Rocchi M, Archidiacono N 2005. Evolutionary history of chromosome 20. Mol Biol Evol 22: 360–366 [DOI] [PubMed] [Google Scholar]
- Montefalcone G, Tempesta S, Rocchi M, Archidiacono N 1999. Centromere repositioning. Genome Res 9: 1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Stanyon R, Finelli P, Archidiacono N, Wienberg J 2000. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc Natl Acad Sci 97: 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Hollatz M, Wienberg J 2003. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum Genet 113: 493–501 [DOI] [PubMed] [Google Scholar]
- Newman TL, Tuzun E, Morrison VA, Hayden KE, Ventura M, McGrath SD, Rocchi M, Eichler EE 2005. A genome-wide survey of structural variation between human and chimpanzee. Genome Res 15: 1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JD 1995. Miropeats: Graphical DNA sequence comparisons. Comput Appl Biosci 11: 615–619 [DOI] [PubMed] [Google Scholar]
- Royle NJ, Baird DM, Jeffreys AJ 1994. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat Genet 6: 52–56 [DOI] [PubMed] [Google Scholar]
- Sarich VM, Wilson AC 1973. Generation time and genomic evolution in primates. Science 179: 1144–1147 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Park SS, Inoue K, Lupski JR 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res 11: 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone MF, Bigoni F, Archidiacono N 2008. Primate chromosome evolution: Ancestral karyotypes, marker order, and neocentromeres. Chromosome Res 16: 17–39 [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Ventura M, Mudge JM, Palumbo V, Burn S, Blennow E, Pierluigi M, Giorda R, Zuffardi O, Archidiacono N, Jackson MS, et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res 13: 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D'Addabbo P, Wandall A, Bjorck E, de Jong PJ, et al. 2004. Recurrent sites for new centromere seeding. Genome Res 14: 1696–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD 1978. Modes of speciation. W.H. Freeman, New York [Google Scholar]
- Wijayanto H, Hirai Y, Kamanaka Y, Katho A, Sajuthi D, Hirai H 2005. Patterns of C-heterochromatin and telomeric DNA in two representative groups of small apes, the genera Hylobates and Symphalangus. Chromosome Res 13: 717–724 [DOI] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. 2009. Mobile elements create structural variation: Analysis of a complete human genome. Genome Res 19: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CT, Jiang Z, McGrath SD, Hayden KE, Khaitovich P, Johnson ME, Eichler MY, McPherson JD, Zhao S, Paabo S, et al. 2005. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol 3: e110 doi: 10.1371/journal.pbio.0030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525–1530 [DOI] [PubMed] [Google Scholar]