Abstract
Independent determination of both haplotype sequences of an individual genome is essential to relate genetic variation to genome function, phenotype, and disease. To address the importance of phase, we have generated the most complete haplotype-resolved genome to date, “Max Planck One” (MP1), by fosmid pool-based next generation sequencing. Virtually all SNPs (>99%) and 80,000 indels were phased into haploid sequences of up to 6.3 Mb (N50 ∼1 Mb). The completeness of phasing allowed determination of the concrete molecular haplotype pairs for the vast majority of genes (81%) including potential regulatory sequences, of which >90% were found to be constituted by two different molecular forms. A subset of 159 genes with potentially severe mutations in either cis or trans configurations exemplified in particular the role of phase for gene function, disease, and clinical interpretation of personal genomes (e.g., BRCA1). Extended genomic regions harboring manifold combinations of physically and/or functionally related genes and regulatory elements were resolved into their underlying “haploid landscapes,” which may define the functional genome. Moreover, the majority of genes and functional sequences were found to contain individual or rare SNPs, which cannot be phased from population data alone, emphasizing the importance of molecular phasing for characterizing a genome in its molecular individuality. Our work provides the foundation to understand that the distinction of molecular haplotypes is essential to resolve the (inherently individual) biology of genes, genomes, and disease, establishing a reference point for “phase-sensitive” personal genomics. MP1's annotated haploid genomes are available as a public resource.
A central goal in biology and medicine is to understand individual genomes, their variation, and how it translates to organismal function, phenotype, and disease. Such knowledge will advance our insights into human individuality and prepare the ground for personalized medicine. Toward this goal, firstly, all existing variants in an individual must be catalogued, including particularly the rare and private ones (Durbin et al. 2010). Secondly, the phase of all variants must be known, that is, their organization into haplotypes, defined as the specific combinations of variants on each of the two chromosomes. Human individuals are diploid, and have about four million genetic variants on average (Durbin et al. 2010). Thus, any genes or noncoding functional sequences constituted by two homologous chromosomes can be genetically very different. Whether variant alleles reside on the same chromosome (in cis), or on opposite chromosomes (in trans), is key to understanding their impact on gene function and phenotype. Benzer (1957) demonstrated that different configurations of mutations result in different phenotypes: Two null mutations in cis left the second allele intact, but when in trans, no functional form of the gene was present. Cis versus trans configurations between mutations in cell essential genes and tumor suppressor genes, even megabases (Mb) apart, have been shown to result in profound alterations of cancer phenotype, spectrum, and progression (Biggs et al. 2003; Wang et al. 2010). Thus, identical genotypes may, depending on their phase, involve different clinical interpretations of potentially tremendous personal impact. Moreover, allele-specific expression (ASE) has been found common among autosomal genes (Knight 2004; Palacios et al. 2009) and related to a spectrum of diseases (de la Chapelle 2009) including cancer (Chen et al. 2008; Valle et al. 2008) and neurodevelopmental disorders (Chamberlain and Lalande 2010). This indicates the global importance of molecular diplotypes for the biology of genes and genomes, phenotype, and health and disease. A functional interpretation of phase information has been proposed earlier (Hoehe et al. 2000; Hoehe 2003) and recently been increasingly recognized (Levy et al. 2007; Tewhey et al. 2011). Ultimately, genetic variation can only be understood from phase.
However, human genome sequencing has for the most part been “phase-insensitive,” that is, generating “haploid composites” (Lander et al. 2001; Venter et al. 2001), partly because molecular genetic techniques to separate haplotypes have remained too cost and labor intensive, restricted in resolution, or not easily scalable to whole genome analysis (Zhang et al. 2006; Ma et al. 2010). Therefore, haplotypes are commonly inferred from population genotypic data by statistical methods (Stephens and Donnelly 2003; Scheet and Stephens 2006). Yet, even the presently most advanced resequencing-based population data source (Durbin et al. 2010) cannot predict the phase of rare or individual-specific variants, which potentially play an important role in complex human disease and individualized medicine (Cirulli and Goldstein 2010; McClellan and King 2010). Recently, whole genome sequencing-based approaches have been undertaken to haplotype-resolve individual genomes (Levy et al. 2007; Wang et al. 2008; McKernan et al. 2009; Kitzman et al. 2011) but remain restricted in extent and scope.
In this work, we aimed to perform a first systematic and comprehensive assessment of individual molecular haplotype architecture as it constitutes the biology of genes and the genome in a diploid human. To this end we haplotype-resolved a European individual, “Max Planck One” (MP1), to an extremely high degree of completeness, exceeding previous efforts in terms of both numbers of variants and length of contigs phased (Levy et al. 2007; Kitzman et al. 2011). We applied a fosmid pool-based next generation sequencing (NGS) approach developed in direct continuation of our previously described fosmid pool-based molecular haplotyping approach (Burgtorf et al. 2003); a similar method was described recently (Kitzman et al. 2011). The completeness of our phasing allowed determination of the molecular haplotype pairs for 81% of all autosomal protein-coding genes including upstream sequences of up to ∼5.7 Mb in length. It also allowed entire genomic regions to be separated into their underlying “haploid landscapes” extending up to ∼6.3 Mb. The diplotypic nature of genes, upstream and coding sequences, and extended genomic regions was seen to be both substantial and global. This highlights the importance of phase for genome biology in defining the functionally active transcriptome and ultimately proteome, and the indispensability of phase information for personal genome analysis. To gain first insights into the importance of phase for gene function, disease predisposition and clinical applications, we identified and annotated a set of 159 genes with two or more potentially significant protein-altering mutations in either cis or trans. To further advance the field, we provide the annotated molecular haploid genomes of MP1 as an easily browsable haplotype-resolved human genome reference sequence to the scientific community.
Results
Fosmid pool-based and whole genome NGS input data
Our fosmid pool-based NGS approach to haplotype-resolve the genome of MP1 is outlined in detail in Methods and Supplemental Figure S1. The principle of this method relies on the fact that sequencing of an individual fosmid produces a haploid 40-kb segment of the genome and that multiple fosmids can be pooled and sequenced together without significantly affecting the haploid nature of the output (Fig. 1A; Burgtorf et al. 2003). Sequencing multiple pools leads to increasing saturation of molecular haplotype sequences across the entire genome (Fig. 1B). Haplotype-informative, heterozygous positions within the genome are used to tile the haploid fosmids by allelic identity into long contiguous haploid sequences. Individual MP1 was from “PopGen” (http://www.popgen.de/), a population genetics research project to study the genetics of complex diseases (see Methods). He was recruited as part of the population representative control cohort, for which Affymetrix 1000K genotypic data were generated. Specifically, MP1 represents the first of 100 PopGen individuals from whom we established a “Haploid Reference Resource” of 100 fosmid libraries, formatted into 288 pools of ∼5000 fosmids each (Burgtorf et al. 2003). We have analyzed MP1's molecular haplotype architecture as a starting point and important reference for population level and disease studies.
Figure 1.
NGS of single and multiple fosmid pools: whole genome coverage. (A) Sequencing a pool of 15,000 fosmids covers ∼15% of the genome. The probability that complementary haplotypes may co-occur within a pool is P < 0.0112, resulting in only a small percentage (1%–2%) of variants likely to be covered by fosmids from both haplotypes. The insert shows a specific example of 19 fosmids detected in the MHC region, concordant with the expected number of fosmids. (B) Additional sequencing of fosmid pools (coverage shown for 32 pools) results in increasing fosmid clone coverage and saturation with molecular haplotype sequence coverage across the entire genome. As shown by simulation, low coverage regions are primarily explained by limitations inherent in short read mapping (Supplemental Fig. S2).
Based on estimated genome coverage (Supplemental Table S1), we sequenced 67 super-pools of ∼15,000 fosmids from MP1's fosmid library using the ABI (Life Technologies) SOLiD platform (see Methods); 142 Gb of uniquely mapped sequence data were produced (Supplemental Table S2) corresponding to a genome coverage of 47×, with 98.1% of autosomes covered ≥5×. The genomic positions of fosmids in each pool were detected by read coverage (see Methods). In total, 1.16 million phase-informative fosmids were detected and found distributed almost equally between the two haplotypes on each autosome (3.3% difference on average), ensuring equally robust phasing for each chromosomal haplotype. Each haplotype was physically covered by 6.38 fosmids on average; the corresponding read coverage was 25.1× (Supplemental Table S3). In addition to the fosmid pools, the “mixed diploid” DNA of MP1 was sequenced to 30× to control for heterozygote detection and cloning bias. Genetic variants were called and allele calls verified on haploid fosmid sequence data (see Methods). False positive and negative rates for SNP detection were 0.08% and 2.36%, respectively, as determined by cross-validation to Affymetrix 1000K genotyping data (Supplemental Table S4). Almost 90% of heterozygous positions were covered by fosmid sequences from both alleles. Only a small percentage of SNP calls (1.31% on average) were found to be heterozygous within each pool, confirming that fosmid pools contain very few complementary segments from both chromosome copies, as expected. For whole genome phasing, we used a specifically designed heuristic algorithm called RefHap (Duitama et al. 2010). Compared to other Single Individual Haplotyping algorithms, including those used by Kitzman et al. (2011) and Levy et al. (2007), ReFHap yields the best compromise between accuracy, completeness, and computational resources (Methods; Supplemental Fig. S3; Supplemental Table S5; J Duitama, GK McEwen, T Huebsch, S Palczewski, S Schulz, K Verstrepen, E-K Suk, MR Hoehe, in prep.). The power of fosmid-based phasing over haplotype assembly from paired-end “mixed diploid” NGS data (McKernan et al. 2009) was evident when directly comparing phase information obtained from MP1 for the same amount of read data (Methods; Supplemental Table S6).
A German genome and its genetic variation
The genome of MP1 comprised 3,258,774 SNPs, of which 258,195 (8%) were novel as compared to dbSNP129 and 1000 Genomes data (Supplemental Table S7). In addition, 221,984 structural variants (SVs) inserting or deleting up to ∼1-kb DNA sequence and 148 copy number variants (CNVs) were identified (Supplemental Table S7). MP1's variation profile is generally consistent with European ancestry including the genomes of Watson (Wheeler et al. 2008) and Venter (Levy et al. 2007) (Supplemental Table S8). More than one third of all SNPs (1,130,306) were located within genes, and of those, 21,966 SNPs were in coding exons. Approximately half (10,735) resulted in nonsynonymous amino acid (AA) exchanges including 154 stop mutations. Screening MP1's genome for potential disease risk variants identified 1677 nonsynonymous, potentially damaging SNPs (14% novel) as predicted by PolyPhen-2 (Adzhubei et al. 2010) and 691 known disease alleles, predominantly coding SNPs, uncovered by use of Trait-o-matic (Ashley et al. 2010) (http://snp.med.harvard.edu). In addition, 1429 GWA SNPs were detected. We found 178,616 SNPs in noncoding regions up to 10 kb upstream of genes and 6599 in conserved TFBS. Importantly, about two thirds of all variants, 2,050,124 SNPs (239,012 novel), 134,555 small indels, and 739 large indels were heterozygous and therefore informative for phasing.
A comprehensively molecular haplotype-resolved individual genome
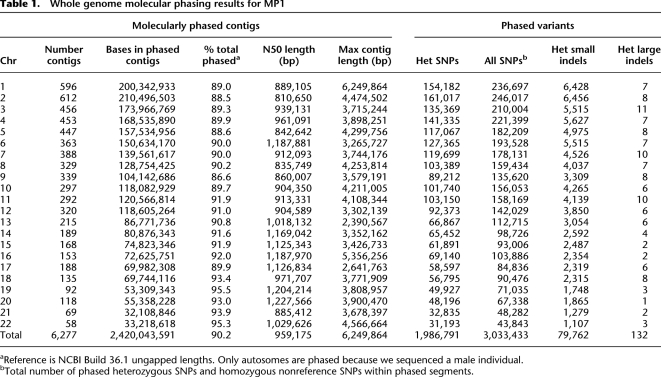
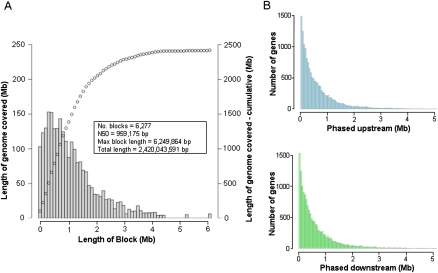
MP1 represents the most comprehensively haplotype-resolved human genome to date, with >90% (2.4 Gb) of the autosomal genome phased (Table 1). MP1's two haploid genomes were assembled into 6277 contiguous haploid sequences extending up to ∼6.25 Mb, with an N50 length close to 1 Mb (Fig. 2A). This is substantially (∼2.5-fold) longer than described in previous reports (Levy et al. 2007; Kidd et al. 2008; Kitzman et al. 2011) and exceeds the average length of common haplotype blocks in the HapMap-CEU panel (International HapMap Consortium 2007) by a factor of 50. We have anchored the phased haploid sequences within each chromosome (see Methods) to provide long range chromosomal haplotypes. Less than 5% of MP1's genome (134.4 Mb) could not be phased due to 2222 regions of homozygosity exceeding 40 kb, the size of a fosmid. The completeness of whole genome haplotype-resolution in MP1 is reflected by the fact that phase context was determined for virtually all SNPs (3,033,433) including >99% (1,986,791) of heterozygous ones (Table 1). Moreover, 132 large and ∼80,000 small indels were haplotype-resolved, making this the most complete report of integrating both SNPs and SVs into phase; these results are expected to improve with further development of the phasing algorithm. The maximum number of variants contained in a single haploid segment was 9179, and up to 113 genes were phased in a single segment (3.8 Mb). Half of the variants were in phased contigs containing ≥1045 variants.
Table 1.
Whole genome molecular phasing results for MP1
Figure 2.
Length of phased blocks. (A) Weighted histogram of genome coverage. Each gray bar shows summed length of genome covered per interval of contig size. Points show cumulative length of genome covered with increasing contig size. (B) Histogram of lengths of phased upstream/downstream regions from end of transcript to end of phased contigs, indicating the additional length of regions containing phased variants which can be analyzed in conjunction with variants within the gene in haploid context. Seventy-nine percent of genes had at least 10-kb phased upstream sequence.
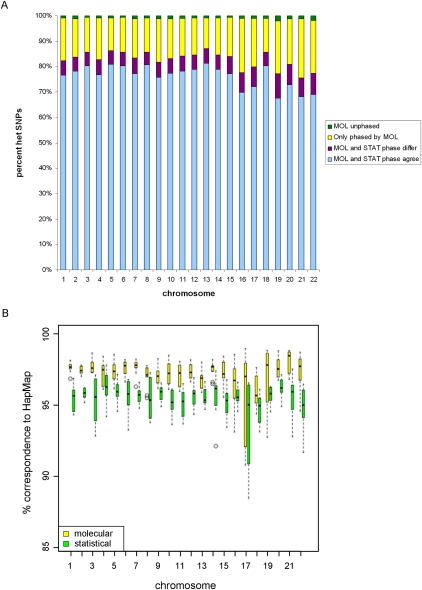
Importantly, we were able to phase >99% of the individual and rare SNPs of MP1, herein assessing an individual human genome in its molecular individuality. Novel variants were found in almost 60% of the genes, and in 75% of genes plus upstream regions, making the majority of individual molecular haplotypes unique. Over 50% of larger phased regions >1 Mb in length contained novel SNPs, and within such regions, a maximum of 26% of SNPs were novel. The ability of assigning private or novel SNPs to one of both haplotypes demonstrates the power of fosmid-based, molecular phasing over statistical approaches. For comparison, we statistically inferred MP1's haplotypes, using data from the 1000 Genomes Project as the required supplementary population data source (see Methods). Whereas the (mainly) common SNPs that were shared between MP1 and 1000 Genomes data (82.9%) could be phased statistically, 16.3% (323,180) of all heterozygous, novel and individual SNPs, remained unresolved (Fig. 3A; Supplemental Table S9). Of the statistically phased SNP positions, 6.4% were discordant to molecularly phased haplotypes. Comparison to HapMap-CEU trio-based haplotypes (see Methods) confirmed the accuracy of molecular phasing at these positions (Fig. 3B). Thus, fosmid-based phasing was superior to statistical phasing both in terms of completeness and accuracy. Discrepancies to statistical phase appeared to be most pronounced in regions harboring particularly high numbers of novel SNPs indicating that population-based methods cannot accurately resolve haplotypes of genomic regions where individual variation is high. For example, within a ∼2.5-Mb region of weak LD on chromosome 4q35.2 containing 2355 novel SNPs, 61% of all heterozygous SNPs remained statistically unphased. Where alleles were phased, a 25% difference was observed to molecular phase. Indeed, the emerging picture of molecular haplotype architecture being highly individual-specific underscores the importance of molecular phasing to prepare the ground for valid individual (diploid) functional genomics and medicine.
Figure 3.
Comparative evaluation of molecular vs. statistical phasing. (A) Stacked bar chart showing haplotype agreement between molecular (MOL) and statistical (STAT) phasing (blue) and disagreement (purple). A proportion of SNPs could only be phased molecularly (yellow) with <1% on average remaining unphased (green). Analysis includes all autosomal heterozygous SNPs in MP1. (B) Box plot showing concordance of MP1 molecular haplotypes (yellow) to the reference set of HapMap haplotypes derived by trio-based phasing (20 individuals). Statistically derived haplotypes for MP1 (green) show a lower concordance to HapMap haplotypes.
The completeness and extent of phasing allowed determination of the concrete molecular haplotype pairs for a total of 14,507 autosomal protein-coding genes (81%). These include long genes >400 kb up to ∼2.1 Mb (Supplemental Table S10), such as the pharmacogenetically relevant ABCA13 transporter gene (Supplemental Fig. S4A) or numerous disease-related genes, such as the schizophrenia-associated ALK gene, which spans >700 kb and harbors a wealth of SNPs, novel variants, and SVs (Supplemental Fig. S4B). Moreover, with up to 5.8-Mb up- and downstream phased (Fig. 2B), genes can be analyzed in their regulatory and larger functional contexts. This allows for instance examination of specific molecular gene forms in relation to their cis-regulatory elements or eQTLs across Mb distances (Stranger et al. 2007; Wray 2007; Epstein 2009; Visel et al. 2009; Teslovich et al. 2010). Approximately 90% of noncoding functional elements and >5000 variable transcription factor binding sites were contained in phased sequence (Supplemental Table S11). Moreover ∼40% of genes and noncoding functional elements were contained in long phased segments >1 Mb, allowing specific long range haplotypes in MP1 to be discerned from the multifold possible combinations of alleles. Both the critical mass of genes phased in their entirety including potential regulatory sequences and the feasibility of exploring larger functional entities represent a significant advance over previous efforts (Levy et al. 2007; Kidd et al. 2008; Kitzman et al. 2011). It prepared the ground to assess and characterize an individual's diplotype (Tewhey et al. 2011), both at the level of genes and the genome.
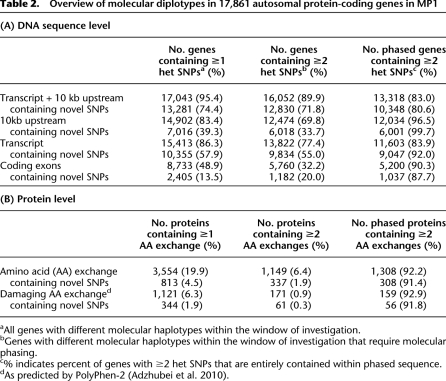
Molecular diplotypes constituting the majority of 17,861 autosomal protein-coding genes
At the DNA sequence level, the vast majority of genes and discrete functional units contain at least one heterozygous SNP and so have two different molecular forms (Table 2A). The small proportion of genes that were invariable between the two homologous chromosomes (13.7%) showed overrepresentation of GO ontologies for developmental processes (P < 1×10−100), which is in accordance with their conservation. Over 77% of all genes in MP1 had two or more SNPs that could exist in either cis or trans configurations and therefore required phasing to resolve their molecular diplotypes; we were able to phase ∼84% of these. This abundance of diplotypic gene regions increased when upstream regions were included, rising to almost 90% (Table 2A). We were able to determine the concrete molecular diplotypes for almost all upstream sequences that required phasing (∼70%) (Table 2A), providing indispensable information to be able to dissect the molecular basis of differential allelic expression, supposed to result from haplotypic combinations of multiple SNPs (Tao et al. 2006). Substantial fractions of diplotypes contained novel SNPs (Table 2A), underscoring the need for molecular phasing.
Table 2.
Overview of molecular diplotypes in 17,861 autosomal protein-coding genes in MP1
Approximately 20% of all genes (3554) encoded two different proteins, defined by presence of one nonsynonymous SNP causing an AA exchange. About one third of those (1149) contained two or more AA exchanges, which could exist in either cis or trans configurations; over 92% of these were phased (Table 2B). A subset of 182 genes had five or more AA exchanges; these were found to be overrepresented with GO ontologies for sensory perception (P < 2.7×10−13) and neurological system processes (P < 1.0×10−24). An additional 138 genes contained multiple SVs in their coding sequences and therefore could also encode two different proteins (Supplemental Table S12).
Given the ubiquity of regulatory, genic, and protein diplotypes, multifold combinatorial possibilities encoding the diploid state are conceivable (Supplemental Fig. S5). Where genes possess two different proteins, diploid gene function may be further diversified by the presence of variants in potential regulatory regions causing differential expression. Within the 10-kb upstream regions for genes that encode two different proteins, >90% contained at least one heterozygous SNP (maximum 182). This could affect expression of either an altered or intact protein, or modify gene dosage in case of a trans configuration. Examples of molecular diplotypic configurations observed in MP1 are shown (Supplemental Fig. S6). Thus, determination of phase is key to characterize and elucidate diploid gene function and dysfunction.
Importance of phase: cis and trans configurations in biology and disease
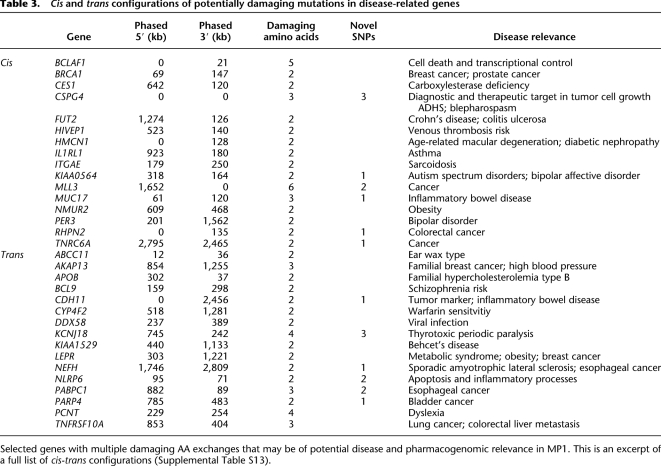
Phase is most likely to impact gene function and phenotype within genes that contain two or more potentially functionally significant mutations. We identified a subset of 171 genes that had two or more AA exchanges predicted to alter the expressed protein in MP1 (Table 2B). We were able to phase a total of 159 genes (92.9%) (Supplemental Table S13). The mutations were found to exist in cis in 86, and in trans in 73 genes. As outlined, cis configurations leave the second protein intact and trans configurations may affect structure and function of both proteins (Fig. 4). Protein-truncating stop mutations, multiples of which were observed in 15 genes, were found three times as often in cis as in trans, supporting this view.
Figure 4.
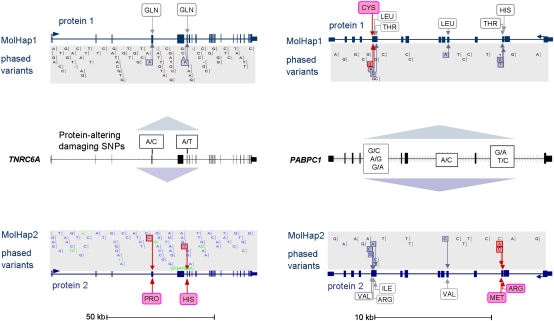
Examples of cis vs. trans configurations of potentially protein damaging mutations. Damaging mutations shown in pink. In cis (left), both mutations reside on the same chromosome, thus the second protein is left intact, shown for the TNRC6A gene. Mutations of TNRC6A may contribute to gastric and colorectal cancer development. In trans (right), multiple damaging mutations affect both haplotypes, shown for PABPC1 which has been associated with esophageal cancer progression and poor prognosis. Gene–gene interaction between TNRC6A and PABPC1 seems to play a role in miRNA silencing (Huntzinger et al. 2010), indicating global relevance of phase. All variants assigned to each of the two molecular haplotypes are shown, including nonsynonymous SNPs which have no predicted damaging effect on the protein (amino acids in gray boxes).
Among these 159 genes were many zinc finger (C2H2-ZNFs) and olfactory receptor (OR) genes. Members of some of the largest mammalian gene families, these genes were found to be significantly enriched even after correcting for the number of family members, e.g., by Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005): for genes involved in an “olfactory signaling pathway” P < 6.68×10−6; C2H2-ZNF genes (genes in cytogenetic band chr19q13), P < 2.39×10−4 (Supplemental Table S14). Thus, these genes could serve as models to explore the role of phase in exerting biological functions. Zinc finger proteins are transcription factors, key regulators of multiple cellular functions (Urrutia 2003). Mutations rendering the protein dysfunctional (e.g., in the KRAB domain and in residues, which are important for protein structure formation and DNA-binding) are exclusively found in cis, leaving one copy of the gene intact. On the other hand, mutations within the DNA-binding domain important for target recognition tend to reside in trans, potentially allowing diversification of function (Supplemental Fig. S7A). Similarly, mutations in OR genes were mainly located within the variable transmembrane domains that are important for ligand binding, a mechanism important in discrimination of different odors (Supplemental Fig. S7B). Potentially functionally significant mutations tended to occur more frequently in trans configurations (e.g., OR1A2). Since OR genes are monoallelicly expressed (Breer 2003), human individuals seem to increase the range of odors they can perceive by having two different forms of the protein (Breer 2003). Expression of two different molecular forms of the genes may enhance the repertoire of stimuli the receptors can detect. Such mechanisms of functional diversification could be advantageous from an evolutionary point of view.
Moreover, among the 159 genes with two or more potentially damaging mutations in either cis or trans configurations, more than half (89) have been documented to play a role in disease and pathophysiology; clinical relevance was collected from OMIM, GWAs, and the program Trait-o-matic (Methods; Ashley et al. 2010). Individual MP1 carried his mutations in cis in 49 of these disease-related genes, and in trans in 40 genes. The genes with mutations in cis have been associated with a broad spectrum of diseases including for instance breast and prostate cancer (BRCA1), other cancers (TNRC6A, MLL3), and asthma (IL1RL1) (Table 3; Fig. 4). Checking his record of ascertainment, at the age of 51, individual MP1 had no documented evidence of clinical disease or pathology. Therefore, we were able to exclude any cis configuration related to early onset diseases, particularly those with an autosomal recessive mode of inheritance. Moreover, no late onset Mendelian diseases were identified. The impact of mutations in cis on complex phenotypes, however, remains to be discussed. The potential clinical relevance of phase for prediction and prevention can be illustrated in MP1 by the cancer-related gene BRCA1. Male carriers of mutations are at increased risk of developing prostate cancer (Mitra et al. 2011). However, MP1 has all BRCA1 mutations in cis leaving one of the two gene forms intact, which could potentially attenuate or eliminate harmful effects. Thus, the same genotype may imply entirely different molecular and phenotypic conditions depending on haplotype. Nevertheless, medical decisions of vital importance are based on simple presence or absence of mutations. Today, BRCA1 female mutation carriers often face the stark prospect of preventive breast ablation. However, knowledge of the underlying molecular haplotypes could redefine diagnostic risk assessment and influence treatment decisions (Ashley et al. 2010). For families inflicted with hereditary cancer, testing a cis configuration of mutations in a gene could raise the hope of transmitting an intact allele to a child. Thus, knowledge of haplotype could change medical consequences entirely.
Table 3.
Cis and trans configurations of potentially damaging mutations in disease-related genes
Trans configurations are more likely to cause phenotypic effects, as has for instance been observed for a small number of autosomal recessive diseases known to be caused by compound heterozygote mutations (Tewhey et al. 2011). In trans, disease-related genes included a broad spectrum of (mostly complex) disease phenotypes (Supplemental Table S13), such as obesity (LEPR), familial hypobetalipoproteinemia (APOB), warfarin sensitivity (CYP4F2), and potential cancer protective genes (TNFRSF10A, CDH11). At his age, MP1 may still be at risk to develop any of the diseases listed in Table 3. We have extracted those genes that were related to complex phenotypes and may cause symptoms in MP1 with progressing age or under drug treatment and are therefore of higher relevance to preventive care strategies for MP1 (Table 3). One such example of potential clinical relevance in MP1 included two AA exchanges in trans in CYP4F2 encoding a drug metabolizing enzyme, in addition to a homozygous V433M mutation known to significantly influence warfarin metabolism (Caldwell et al. 2008). The mutations in trans may exert an additional effect on CYP4F2 function, with important implications for dosage requirement to prevent MP1 from severe side effects such as bleeding or thrombembolism in the case of warfarin treatment.
Taken together, we provide a first globally collected repertoire of potentially disease predisposing mutations as they exist in either cis or trans configurations in MP1. Our results exemplify that knowledge of phase may have far reaching clinical and personal consequences but also indicate the tremendous complexities of establishing genotype/haplotype-phenotype relationships. Ultimately analyses will need to consider phased variants within multiple genes and noncoding regions across large haploid landscapes to tackle the complex interactions that may drive the development and expression of the phenotype.
Extended phase in the MHC region resolves haplotypes of key clinical relevance
In this context, the ability to phase across the major-histocompatibility complex (MHC) extending over 4 Mb is of key clinical and medical relevance. MHC possibly harbors the highest density of disease genes in the human genome (Horton et al. 2008). In transplant medicine, being able to unambiguously determine the highly variable HLA alleles and, in addition, their combined phase across the MHC is of immediate medical relevance to a patient. As has been demonstrated, transplantation outcome in HLA-identical patients is critically impacted by the specific combination, or phase, of multiple HLA alleles; nonmatching long-range MHC haplotypes significantly increased the risk for severe graft-versus-host-disease (GvHD) (Petersdorf et al. 2007). Thus, accurate phase information allows identification of donor-recipient matches at high risk for life-threatening transplant complications and, in addition, the detection of novel genetic factors related to GvHD. However, long range phasing of the MHC presents a particular challenge due to abundant sequence variability, structural complexity, and large distances between the HLA genes (Guo et al. 2006). In a first step, we aimed to demonstrate validity of our extended MHC sequences phased in MP1. Alignment of our haplotypes to the IMGT/HLA database (http://www.ebi.ac.uk/imgt/hla) determined HLA alleles as A*0301 and A*2501/2601, B*0702 and B*5101, DRB1*15 and DRB1*11 (Methods; Supplemental Fig S8). These results were highly concordant with the genomic four-digit HLA typing results available for MP1. We then compared our long-range MHC haplotypes (the specific combinations of HLA alleles) to the known MHC haplotype sequences (http://www.ucl.ac.uk/cancer/research-groups/medical-genomics/past_projects/MHC.shtml) (see Methods). One of the two MHC haplotypes in MP1 carried HLA alleles A*0301, B*0702, and DRB1*15, a combination perfectly matching the homozygous PGF haplotype (Supplemental Fig. S8). PGF, the so-called “ancestral DR2 haplotype,” is one of the two most frequent long-range MHC haplotypes in Europeans (10% frequency) with typically high linkage of constituent HLA alleles, confirming our results. Substantial genetic differences in the MHC region between HLA-identical “tissue-matched” genomes have been found, pointing to the existence of significant, yet unexplored, transplantation determinants potentially linked to GvHD development (Proll et al. 2011) and a variety of immunogenetic diseases. In this context, our work has made a first important contribution by resolving the underlying haplotypes across the MHC region for an individual human. Notably, among MP1's two extended MHC haplotypes, 10,079 genetic differences (SNPs and indels) were found, including 221 novel heterozygous SNPs, of relevance in tissue matching or disease.
Haploid landscapes: Toward “phase-sensitive” functional genomics
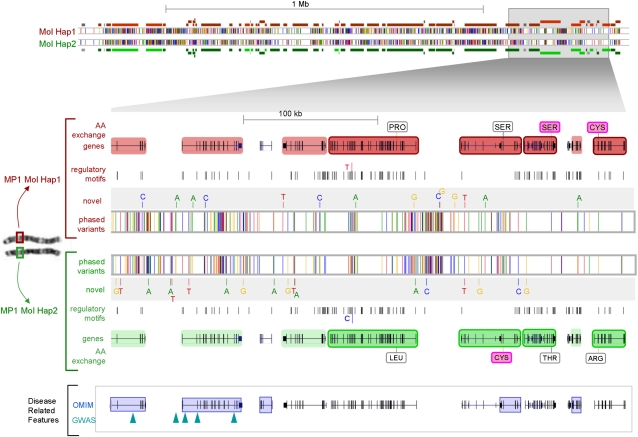
The determination of contiguous molecular haplotype sequences in the Mb range is crucial to translate individual genomic variation into the functionally active proteome. Physically and/or functionally related genes and regulatory elements, which can extend over large distances, may represent functional entities that affect gene expression in a coordinated way (Epstein 2009; Navratilova and Becker 2009). Given the global nature of differences within pairs of molecular haplotypes described above, it is therefore essential to determine the specific combinations of alleles that co-occur on each of the two chromosomes (“haploid landscapes”), as demonstrated in Figure 5. These constitute the concretely co-expressed and interacting forms of the genes. We have characterized a subset of over 700 haploid landscapes >1 Mb (examples are summarized in Supplemental Table S15). One such example containing strikingly diverse features is a >3.8-Mb haplotype on chr. 19q13 (Supplemental Fig. S9A), which spans one of the most gene dense regions in the human genome comprising the LRC/KIR (killer cell immunoglobulin-like receptor) and numerous zinc finger (ZNF) genes; 95.6% of these genes (108) displayed different molecular haplotypes, 22 of which had multiple protein-damaging mutations in either cis or trans configurations. Prototypic examples of functionally co-regulated genes to be examined as a haploid entity include the HOXA cluster, contained in an ∼2-Mb haplotype on chr. 7 (Supplemental Fig. S9B), and a zinc finger cluster on a 2.7-Mb haplotype on chr. 19. Co-regulation of numerous of these functional units arrayed on the same chromosome may influence downstream targets or functional cascades in a haplotype-specific manner. Corresponding landscapes with particularly high diplotypic diversity of regulatory sequences demonstrate the importance of phasing the entirety of functionally related alleles in conjunction with their cis-regulatory elements (Supplemental Fig. S9C,D; Epstein 2009). We provide the set of 700 long range haplotypes >1 Mb (www.molgen.mpg.de/∼genetic-variation/MaxPlanckOneLandscapes) to the scientific community to further study their particular regions of interest in phase context.
Figure 5.
Example of a 1.7-Mb haploid landscape of functional variation on chr. 19. Both molecular haplotypes are shown. Differences between the two are shown at the nucleotide level with bases in yellow = G, red = T, green = A, and blue = C, within framed bars (centered). Novel SNPs are assigned to each of the two haplotypes. Regulatory motifs (TFBSs track) differ in one SNP. At the level of genome organization, genes that have two different molecular haplotypes are shaded red or green per haplotype, and those encoding two differing proteins are highlighted and framed. Nonsynonymous mutations on each haplotype are annotated (damaging AA exchanges [Adzhubei et al. 2010] in pink). Disease-related features (GWAS and OMIM) are shown in lower track.
Further, with such extended phased sequences, the complete set of existing individual variants (genic and noncoding functional) that co-occur with a GWA marker on the same chromosome can be examined to track the candidate causative variant(s). These particularly also include the rare variants, which alternatively to a common variant could explain association signals in common disease (Dickson et al. 2010). We provide phase context in the Mb range for a total of 1218 identified GWA signals associated with a spectrum of important common diseases such as cancer and metabolic disorders. Interesting examples of haploid landscapes of genetic association, such as an ∼1.8-Mb gene rich haplotype on chr. 1 with a particularly high number of GWA signals, point to the highly variable architectures of such associated regions (Supplemental Fig. S9E). This example shows pronounced diplotypic diversity of contained features, with 63% of genes encoded by two different molecular forms. Others are characterized for instance by high proportions of novel SNPs (up to 26%) (Supplemental Table S15).
To provide first insights into importance of phase for translation of genetic variation into the functionally active proteome and functional genomics, we screened MP1's genes that encoded two different proteins, for potential interactions within the NCI cancer-related pathways. Preliminary analyses were carried out for 3554 genes with at least one AA exchange (Supplemental Table S16) and 1149 genes with AA mutations in either cis or trans configurations. One-hundred-seventy pathway interactions overlapped with genes, that had at least one AA exchange, and 70 pathways were affected by genes with multiple AA mutations (Supplemental Table S17); 202 of these genes were found to co-occur with multiple other phased cis or trans genes within the same pathway. Their functionality in such pathways and associated downstream interactions may critically depend on the specific forms of the proteins, or haplotypes involved (e.g., BRCA1) (Supplemental Fig. S10). The many combinatorial possibilities illustrate the potential importance of molecular diplotypes in diversification of gene and organismal function and phenotype. Taken together, coherent phasing of all potentially functionally active alleles and their regulatory environment is an indispensable prerequisite in functional genomics.
“Max Planck One”: A browsable phased genome reference
The haploid genomes of MP1 are available as browsable tracks available via the UCSC (http://www.molgen.mpg.de/∼genetic-variation/MaxPlanckOneUCSC) and Ensembl Genome Browsers (http://www.ebi.ac.uk/das-srv/easydas/MP1/das/sources). The phased variants have been annotated for their status of novelty (against dbSNP129) and potential for being damaging (Adzhubei et al. 2010), and can be examined for each of the two haplotypes separately. In addition, the input matrix used for fosmid-based phasing by ReFHap, which can be used to check the phasing quality at any particular locus, can also be downloaded (http://www.molgen.mpg.de/∼genetic-variation/MaxPlanckOneLandscapes/phasing_matrix.tar.gz). Providing a browsable phased genome resource, where haplotypic information can be viewed in context with additional whole genome data, e.g., eQTLs, noncoding functional elements or methylation, will allow the scientific community to further study their genes or particular regions of interest in phase context. This resource inherently includes a catalogue of the specific pairs of haplotypes, or molecular diplotypic configurations, of protein-coding genes in a human individual. This example of a reference phased genome will help to advance the diploid biology of genes and genomes and drive forward “phase-sensitive” approaches in many other areas of “omics.”
Discussion
The prospects of experimental whole genome haplotyping have recently captured increasing attention (Bansal et al. 2011; Fan et al. 2011; Kitzman et al. 2011). The need for phasing has been recognized for a broad range of applications in human genetics. Moreover, the importance of phase for genome biology as a whole has recently been increasingly emphasized (Hoehe 2003; Tewhey et al. 2011). Our work adds a major contribution by a first systematic and comprehensive characterization of the molecular haplotype architecture of an individual human genome. The diplotypic nature of functional regions was a ubiquitous occurrence throughout the genome, indicating indispensability of phase information to be able to unravel the complexities of genome function. Concrete and comprehensive knowledge of phase will advance new approaches, conceptually and experimentally, that will ultimately lead to an improved understanding of the biology of genes, genomes, and disease.
The observation that the vast majority of genes exist as two different molecular forms raises fundamental questions about the nature of the relationship between these two and their role in exerting “gene function.” Different scenarios of the nature and mechanisms of dual gene action are conceivable, including preferential allelic activity (Knight 2004; Palacios et al. 2009), synergism, fail-safe systems, counteraction, complementation or compensation. For example, differential expression of diplotypic genes encoding two protein isoforms may be detrimental if a disease protein is favored. When two different molecular forms of a receptor gene are co-expressed, this could broaden its response to drugs or immune tolerance and thus confer functional adaptability. Conventional single mutation testing models may no longer apply to establish relationships between variation, structure, and function of a diplotypic gene. It has been shown that functional or therapeutic phenotypes cannot be predicted from single variants, or the sum of single effects (Drysdale et al. 2000; Tao et al. 2006). Hence, the functional characterization and annotation of “genes” calls for novel experimental paradigms, using molecular haplotypes in their entirety, and pairs thereof (Drysdale et al. 2000; Hoehe et al. 2000), as the units of analysis. Our catalogue of molecular haplotypes, which includes many disease-related genes, provides the required templates to test and generate hypotheses on the dual nature of gene action.
From a broader perspective, the huge extent of molecular diplotypic differences across extended haploid sequences representing complex and larger functional entities translates into multilayered scenarios of “phase-driven” functional genomics. For example, in MP1, a notable number of genes with two different forms of coding haplotypes co-occurred in multiple disease-related pathways. Their interaction has the potential to disrupt pathways or impact entire downstream networks. In a positive sense they may create biological subtleties in unimagined ways. The ability to activate different haplotypic or diplotypic combinations of genes and regulatory elements points to the high versatility of a diploid genome, which may guarantee physiological resilience and is a rich playground for evolutionary forces to create phenotype diversity.
Multiple phenomena including ASE and methylation, imprinting effects, or dosage compensation testify to the “fundamental importance of diploidy in human biology” (Bansal et al. 2011). Overall, our results suggest that we must move from the notion of disperse “allelic” phenomena to the conception of the “molecular two-allele state” as a general foundational principle in approaches to explain the complexities of genome function. These inherently have a strong individual component. Sequencing-based phasing approaches generate a genome in its molecular individuality, through the phase of all individual variants. We have shown the importance of molecular phasing for the resolution of novel variants. Thus, our approach also delivers critical information on a pool of variants of importance for disease (Cirulli and Goldstein 2010; McClellan and King 2010) and actionable individually tailored medicine. However, to translate haplotype-resolved genomes into personalized genomics, phase-informed data from all conceivable “omics” sources, e.g., RNA-seq, ChIP-seq, or epigenomic methylation data, need to be integrated that describe genome function at increasing levels of complexity (Lunshof et al. 2010).
Knowledge of phase may be critical for clinical interpretation of genome data as has been illustrated using BRCA1 as an example. Nevertheless, medical measures seem yet “phase-insensitive” (Diamandis et al. 2010; van El and Cornel 2011) and based on genotypic data alone. To validate importance of phase information for genetic mutation testing, disease prediction, and prevention as a working hypothesis, targeted studies of molecular phase in large cohorts will be required. Furthermore, by determining and phasing classical HLA alleles across the MHC region in MP1, we demonstrate a key application of phase with immediate clinical relevance for transplantation-related GvHD (Petersdorf et al. 2007). With >10,000 genetic differences between MP1's two extended MHC haplotypes identified, exploration of novel genetic determinants, for GvHD and many other diseases, becomes now feasible. Although no obvious haplotypes-phenotype relationship could be established in MP1, this does not exclude the potential role of trans configurations in disease-predisposing genes causing complex disease phenotypes at a later age of onset. An emerging era at its earliest stage, the establishment of haplotype-phenotype relationships represents a daunting challenge. It will require the analyses of haplotype-resolved genomes in larger numbers of cases and controls. In this regard, our molecularly haplotype-resolved and well characterized individual European genome provides a starting point for future comparisons.
We are currently applying our approach to haplotype-resolve whole genomes of many more individuals, which will allow our findings to be consolidated and broaden the picture of the diplotypic state and its role in relation to phenotype and disease. Scalability and cost-effectiveness of our fosmid pool-based approach given (sequencing two haploid genomes per week, at costs of under 6000 €), we can now explore molecular diversity not only within, but also between individual genomes. Moreover, highly complex genomes, for instance those of admixed individuals or cancer genomes, can be resolved independently of any population or parental information. Considering that fosmid pool-based sequencing does not only phase rare or novel variants, but also allows phasing of larger SVs, complex rearrangements or novel sequences (Kidd et al. 2010), this will complement the comprehensive determination of an individual's molecular haplotype architecture. Taken together, our work sets a reference point for “phase-sensitive” personal genomics. It will advance our understanding of diploid genome function and prepare the ground for actionable personalized medicine to the benefit of each individual human.
Methods
MP1: Selection, characterization, research context
MP1 was ascertained as a proband, by random sampling, for a population genetics research project conducted by the University of Kiel (http://www.popgen.de/) in the area of Northern Schleswig-Holstein (Germany). Probands were subjected to a clinical check-up, and up to 250 different phenotypic items per sample including demographic data collected. MP1 was at the time of ascertainment 51 yr old, with the exception of medically treated arterial hypertension, in good health, without pathologic findings in the physical examination or routine laboratory check, and without a history of severe diseases. Ethical guidelines for the PopGen biobank do not permit a clinical follow-up of the proband. MP1 is part of a “Haploid Reference Resource” of 100 fosmid libraries from 100 PopGen probands (40 males, 60 females, 50–71 yr), created for the analysis of molecular haplotype architecture in a population as a reference for specific disease studies. Genotypic data generated by Affymetrix 1000K typing were available for these 100 PopGen probands and showed a strong correlation of >0.95 (LD) with the HapMap-CEU samples. In addition, four-digit HLA-typing was available for all 100 individuals.
A fosmid pool-based NGS approach to haplotype-resolve whole genomes
We have established a fosmid pool-based approach to haplotype-resolve whole genomes directly by NGS (Supplemental Fig. S1). A similar method was recently described by Kitzman et al. (2011). (1) We constructed a fosmid library containing ∼1.44 million fosmid clones from the genomic DNA of MP1, equivalent to 7× coverage of each haploid genome. The fosmid library was formatted into three 96-well plates, with each well comprising a “haploid clone pool” (Burgtorf et al. 2003) of ∼5000 fosmids (random mixtures of 40-kb haploid DNA segments), representing ∼5% of the genome. To increase throughput, three fosmid pools were combined into working unit “super-pools” of ∼15,000 fosmids. The probability that complementary haplotypes may co-occur within a pool of ∼15,000 fosmids is P < 0.0112. (2) We sequenced multiple super-pools (67 in total) from this library using the SOLiD platform to reach sufficient coverage of molecular haplotype sequences across the entire genome. (3) For each pool separately, fosmids were detected from read coverage. (4) Variants were called on combined fosmid pools and on 30× genomic DNA to identify heterozygous, haplotype-informative alleles for phasing. (5) Fosmids were then tiled into contiguous molecular haplotype sequences based on allelic identity at multiple heterozygous positions using ReFHap, a heuristic phasing-algorithm similar to HapCUT (Bansal and Bafna 2008), developed to specifically phase fosmid pool data (Duitama et al. 2010). ReFHap performs consistently as accurately as HapCUT but with a much faster running time (Supplemental Table S7; Supplemental Fig. S3). RefHap constructs a graph that enforces separation of fragments (fosmids) rather than variant loci; this use of multiple variant loci makes the procedure robust compared to single SNP-based phasing. Moreover, with each variant position being covered by multiple fosmids from the same haplotype, potential SNP calling errors can be identified and compensated for in subsequent phasing. Finally, we anchored the phased haploid contigs onto their homologous autosomes (see below).
Fosmid library construction
Twenty micrograms of genomic DNA of MP1 was sheared to select haploid DNA fragments of 40 kb on average. These fragments were used to prepare a complex fosmid library using the Epicentre EpiFOS Fosmid Library Production Kit as per standard protocol. Phage particles, absorbing haploid ligation products, were used to transform Epi100 cells, and the transfection titer determined. Mass transfected Escherichia coli cells were aliquoted into three 96-deep-well LB plates, such that each well contained a pool of ∼5000 fosmid clones. In total, 288 pools were generated and stored as glycerol-bacterial stocks to ensure long term availability. Complexity and evenness of genome representation have been checked by PCR of 10 different chromosomal loci in the entire fosmid library, which showed a good agreement to prior expectations. In addition, we could confirm the library quality by retrospective evaluation of our fosmid pool sequencing data. More detailed information is in Supplemental Methods.
SOLiD sequencing
Super-pools were barcoded and up to 16 multiplex sequenced in a single flow cell with the SOLiD system (Supplemental Methods). We prepared in total 158 sequencing libraries comprising 36 barcoded, 21 mate-paired, and 43 fragment libraries from the purified fosmid pool DNA and a genomic DNA sample according to standard protocols for SOLiD sequencing. For barcoded and fragment libraries, ∼3 μg purified fosmid DNA was sheared, and 150–200-bp size-selected DNA ligated to SOLiD adaptor sequences, containing unique barcode identifiers and universal SOLiD priming sites. The final sequencing library was subjected to emulsion PCR for clonal amplification of unique sequencing library molecules onto magnetic beads. Quality of generated templated beads (on-axis, P2:P1 ratio, satay plot) has been checked by WFA prior to full sequencing of at most 650 million templated beads per slide (SOLiD V3+/V4). For mate-pair libraries, DNA was sheared to targeted insert sizes (800 bp, 1.5 kb, 2.5 kb, 3.5 kb, 8 kb, and 10 kb) followed by a circularization step, read tag generation by enzymatic cutting, and adaptor ligation as described above. Sequencing was carried out on 67 unique fosmid pools (∼15,000 fosmids per pool) and, of these, mate-pair sequencing was performed on 20 pools. Paired-end sequencing (Life Technologies) has been performed for a subset of 16 fosmid pools. For the genomic sample, we sequenced two mate-pair libraries with an insert size of 1.5 kb and 2.0 kb, and performed paired-end sequencing of the genomic sample to achieve a total 30× genome coverage. More detailed information is in Supplemental Methods.
Fosmid detection
To detect the genomic positions of fosmids sequenced within a pool, we mapped reads to the reference genome. SOLiD reads were aligned to the reference genome (Hg18) with Bioscope 1.2 (www.solidsoftwaretools.com) using default parameters. The alignment algorithm mapreads uses a seed and extend approach similar to BLAST. The reference (NCBI 36.6) was translated into color space and reads aligned to this reference starting with short matches (seed of 25 bases with two mismatches for 50-bp reads). The alignment is extended in both directions with a mismatch penalty of –2 and a matching score of +1 and the alignment with the highest score is reported. PCR duplicates (read pairs that share outer mapping coordinates) were marked.
To detect fosmids we used a sliding window approach to locate suitable length regions above a coverage threshold defined dynamically based on the total number of mapped bases. Fosmids were detected as ungapped contigs ranging from 3 to 45 kb. Due to low complexity elements in the genome, some fosmids are composed of two or more contigs. We finally ensured that predicted fosmids do not span heterozygous SNP calls. More detailed information is in Supplemental Methods.
Variant detection
SNPs were identified with the diBayes SNP caller (http://solidsoftwaretools.com/gf/project/dibayes) which is a Bayesian algorithm that includes color space error detection. The algorithm incorporates prior probabilities of heterozygosity, errors, and GC content. Heterozygous SNPs are called from 30× genomic DNA, and within each fosmid pool we verify that only one of the two alleles is present at 5× average coverage (see Fosmid Detection). DiBayes was used with a medium call stringency using default parameters (McKernan et al. 2009). SNPs were further filtered by eliminating calls based on reads with indels. Affymetrix 1000K SNP genotyping data were used to validate the accuracy of the called genotypes. SVs (small indels, large indels, and copy numbers) were called with Bioscope 1.2 using mate-pair and paired-end reads as described (McKernan et al. 2009). Candidate lists were combined for each fosmid pool to generate a final evidence list for the subject. Heterozygous indels were then phased using the information from each fosmid pool and the genomic DNA.
Fosmid pool-based phasing
All haplotype-informative alleles were consolidated into an input matrix, the basis for a heuristic phasing algorithm, RefHap, which we specifically developed for fosmid pool data (Duitama et al. 2010). This algorithm builds a graph with a vertex for each fosmid and an edge for each pair of fosmids sharing at least one SNP call, and assigns weights to each edge consistent with the agreement between the corresponding fosmids. This construction allows the Single Individual Haplotyping problem to be reduced to the well known Max-Cut problem allowing the haplotypes most consistent with the cut obtained in the previous step to be detected. Fosmids are thus separated and tiled into contiguous molecular haplotype sequences based on their allelic identity at heterozygous positions. Finally, we anchored the phased haploid contigs onto their homologous autosomes. For this, we used a genotypic data set obtained by whole genome Affymetrix 1000K chip-based genotyping of a total of 97 individuals from the same German reference cohort (PopGen) to infer the common haplotypes (Stephens and Donnelly 2003; Scheet and Stephens 2006) which then served as anchor. More detailed information on ReFHap is in Supplemental Methods.
Comparative evaluation molecular vs. statistical phasing
The program fastPHASE (Scheet and Stephens 2006) was used to generate a statistical haplotype for our individual based on population data from the 1000 Genomes Project for 57 CEU samples (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/2009_04/). In order to assess which phasing method produced the most accurate haplotypes for MP1 we used publicly available haplotypes from HapMap-CEU trios (International HapMap Consortium 2007) containing ∼380,000 SNPs as a reference for comparative evaluation. For each pair of SNPs in high LD (D′ = 1), the phase of the alleles between the two samples was compared and the number of discordant positions counted. More detailed information is in Supplemental Methods.
Comparison of fosmid pool-based vs. “mixed-diploid” phasing
The “mixed-diploid” phasing of MP1 was carried out on 30× genomic DNA as described in McKernan et al. (2009). The algorithm constructs haplotype blocks from overlapping paired reads spanning at least two heterozygous SNPs. In order to make a fair comparison, the same amount of sequence data (30×) was used for fosmid pool-based phasing. More detailed information is in Supplemental Methods.
Biological analysis methods
RefSeq (Pruitt et al. 2005) genes were downloaded from the UCSC table browser (Hg18). Only genes on chromosomes 1–22 were considered. All alternative transcripts belonging to a gene were merged and the coordinates defining this entire region used for subsequent analysis, resulting in a final set of 17,861 genes. The UCSC table browser was also used to download the following tracks: OMIM, GWAS, Vista Enhancers, and conserved TFBSs. CNEs were downloaded from the CONDOR Database (Woolfe et al. 2007). SNPs were annotated using SeattleSeq Annotation (Build 5.02, http://gvs.gs.washington.edu/SeattleSeqAnnotation). Polymorphism Phenotyping version 2 (PolyPhen-2) (Adzhubei et al. 2010) was used to predict possible impact of an amino acid substitution on the structure and function of a human protein. This program uses sequence-based and structure-based predictive features to determine whether AA exchanges are likely to be damaging or not. We use the HumVar trained version of PolyPhen-2 which includes disease-causing mutations from UniProt. SNPs were filtered to retain only nonsynonymous SNPs that occur within exons of the RefSeq gene set. BEDTools (v2.7.1) (Quinlan and Hall 2010) was used to calculate intersections between gene and SNP sets. Gene Ontologies were checked using GOstat (Beissbarth and Speed 2004). Pathway analysis was done using the NCI Nature Pathway Interaction Database (Schaefer et al. 2009). We intersected 1149 genes with two or more heterozygous coding SNPs and the NCI database, encompassing cancer-related pathways, by using the batch query tool (http://pid.nci.nih.gov/search/batch_query.shtml). Resulting pathways and interactions, which contained two or more of the cis-trans genes, were used for illustration.
Data access
Short-read sequence data have been deposited at the European Nucleotide Archive (ENA) under accession no. ERP000494. The SNP genotype data for MP1 have been submitted to NCBI dbSNP and will be available in dbSNP version B135. Data files can be downloaded from http://www.molgen.mpg.de/∼genetic-variation/MaxPlanckOneData. A DAS source with annotation of the haplotypes and phased variants is available under http://www.ebi.ac.uk/das-srv/easydas/MP1/das/sources and can be integrated into Ensembl. Haplotypes and haploid genomes are available in a UCSC session at http://www.molgen.mpg.de/∼genetic-variation/MaxPlanckOneUCSC, and megabase-sized haploid landscapes can be downloaded from http://www.molgen.mpg.de/∼genetic-variation/MaxPlanckOneLandscapes. The RefHap algorithm is available to download from http://www.molgen.mpg.de/∼genetic-variation/SIH/Data/algorithms.
Acknowledgments
We thank T. Kroslak, C. Burgtorf, B. Mentrup, S. Thottakatara, and A. Neubert for advice and/or production of human fosmid libraries; T. Borodina, A. Soldatov, D. Parkhomchuk, M. Schilhabel, and A. Franke for performing early sequencing tests; and H.v. Eberstein for providing access to the German PopGen cohort. We thank C. Schönemann for HLA testing of “Haploid Reference Resource” samples. We thank R. Horton for the original design and implementation of the fosmid detection algorithm. We thank K. Rohde and A. Bauerfeind for discussion on haplotypes. We thank R. Shamir for cooperation. We acknowledge S. Guenther, A. Sartori, and the German Lifetech team for providing support in initial setup. We particularly thank T. Harkins and K. McKernan, Life Technologies, for general advice and discussion. We thank R. Herwig for support of J.D. We particularly thank H. Lehrach and G.M. Church for helpful discussion. We thank two anonymous reviewers for their constructive suggestions. This work was supported by the Federal Ministry of Science and Education (BMBF), Germany, by the NGFN-2 program grant 201GR0414, and the NGFN-Plus program grant 01GS0863 (to M.R.H.), and in part by the German Israeli Foundation (GIF) grant I-837-156.9/2004 (to M.R.H. and Ron Shamir). Ethical consent for human subjects' research has been obtained from the Ethics Committee, University of Kiel (AZ: A156/03 POPGEN); informed consent has been obtained from the study participant.
Authors' contributions: E.-K.S., S.S., and S.P. established next generation sequencing platform and experimental methods, carried out wet lab production, and performed data analysis. T.H., G.K.M., and J.D. designed and implemented the main bioinformatic pipeline and performed data analysis. D.T.H., S.M., H.P., and C.L. carried out additional sequencing work and/or data analysis. K.N. contributed to writing and data analysis. St.S. provided DNA and Affymetrix 1000K genotypic data. M.R.H., E.-K.S., and G.K.M. wrote the manuscript. M.R.H. and E.-K.S. jointly designed and supervised the work. M.R.H. conceived the study based on longstanding research, development, and production on the theme.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.125047.111. Freely available online through the Genome Research Open Access option.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, et al. 2010. Clinical assessment incorporating a personal genome. Lancet 375: 1525–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Bafna V 2008. HapCUT: An efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 24: i153–i159 [DOI] [PubMed] [Google Scholar]
- Bansal V, Tewhey R, Topol EJ, Schork NJ 2011. The next phase in human genetics. Nat Biotechnol 29: 38–39 [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP 2004. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465 [DOI] [PubMed] [Google Scholar]
- Benzer S 1957. The elementary units of heredity. In The chemical basis of heredity (ed. McElroy WD, Glass B), pp. 70–93 Johns Hopkins University Press, Baltimore [Google Scholar]
- Biggs PJ, Vogel H, Sage M, Martin LA, Donehower LA, Bradley A 2003. Allelic phasing of a mouse chromosome 11 deficiency influences p53 tumorigenicity. Oncogene 22: 3288–3296 [DOI] [PubMed] [Google Scholar]
- Breer H 2003. Olfactory receptors: Molecular basis for recognition and discrimination of odors. Anal Bioanal Chem 377: 427–433 [DOI] [PubMed] [Google Scholar]
- Burgtorf C, Kepper P, Hoehe M, Schmitt C, Reinhardt R, Lehrach H, Sauer S 2003. Clone-based systematic haplotyping (CSH): A procedure for physical haplotyping of whole genomes. Genome Res 13: 2717–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, et al. 2008. CYP4F2 genetic variant alters required warfarin dose. Blood 111: 4106–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Lalande M 2010. Neurodevelopmental disorders involving genomic imprinting at human chromosome 15q11-q13. Neurobiol Dis 39: 13–20 [DOI] [PubMed] [Google Scholar]
- Chen X, Weaver J, Bove BA, Vanderveer LA, Weil SC, Miron A, Daly MB, Godwin AK 2008. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum Mol Genet 17: 1336–1348 [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 11: 415–425 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A 2009. Genetic predisposition to human disease: Allele-specific expression and low-penetrance regulatory loci. Oncogene 28: 3345–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamandis M, White NM, Yousef GM 2010. Personalized medicine: Marking a new epoch in cancer patient management. Mol Cancer Res 8: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB 2010. Rare variants create synthetic genome-wide associations. PLoS Biol 8: e1000294 doi: 10.1371/journal.pbio.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, Arnold K, Ruano G, Liggett SB 2000. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci 97: 10483–10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duitama J, Huebsch T, McEwen G, Suk E-K, Hoehe MR 2010. ReFHap: A reliable and fast algorithm for single individual haplotyping. In Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology, pp. 160–169 ACM, Niagara Falls, New York [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein DJ 2009. Cis-regulatory mutations in human disease. Brief Funct Genomics Proteomics 8: 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Wang J, Potanina A, Quake SR 2011. Whole-genome molecular haplotyping of single cells. Nat Biotechnol 29: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Hood L, Malkki M, Petersdorf EW 2006. Long-range multilocus haplotype phasing of the MHC. Proc Natl Acad Sci 103: 6964–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehe MR 2003. Haplotypes and the systematic analysis of genetic variation in genes and genomes. Pharmacogenomics 4: 547–570 [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Kopke K, Wendel B, Rohde K, Flachmeier C, Kidd KK, Berrettini WH, Church GM 2000. Sequence variability and candidate gene analysis in complex disease: Association of μ opioid receptor gene variation with substance dependence. Hum Mol Genet 9: 2895–2908 [DOI] [PubMed] [Google Scholar]
- Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, et al. 2008. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics 60: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29: 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cheng Z, Graves T, Fulton B, Wilson RK, Eichler EE 2008. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res 18: 2016–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, Hayden HS, Alkan C, Malig M, Ventura M, Giannuzzi G, et al. 2010. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat Methods 7: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, Mackenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, et al. 2011. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol 29: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC 2004. Allele-specific gene expression uncovered. Trends Genet 20: 113–116 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. 2007. The diploid genome sequence of an individual human. PLoS Biol 5: e254 doi: 10.1371/journal.pbio.0050254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunshof JE, Bobe J, Aach J, Angrist M, Thakuria JV, Vorhaus DB, Hoehe MR, Church GM 2010. Personal genomes in progress: From the human genome project to the personal genome project. Dialogues Clin Neurosci 12: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Xiao Y, Huang H, Wang Q, Rao W, Feng Y, Zhang K, Song Q 2010. Direct determination of molecular haplotypes by chromosome microdissection. Nat Methods 7: 299–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC 2010. Genetic heterogeneity in human disease. Cell 141: 210–217 [DOI] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, et al. 2009. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res 19: 1527–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra AV, Bancroft EK, Barbachano Y, Page EC, Foster CS, Jameson C, Mitchell G, Lindeman GJ, Stapleton A, Suthers G, et al. 2011. Targeted prostate cancer screening in men with mutations in BRCA1 and BRCA2 detects aggressive prostate cancer: Preliminary analysis of the results of the IMPACT study. BJU Int 107: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova P, Becker TS 2009. Genomic regulatory blocks in vertebrates and implications in human disease. Brief Funct Genomics Proteomics 8: 333–342 [DOI] [PubMed] [Google Scholar]
- Palacios R, Gazave E, Goñi J, Piedrafita G, Fernando O, Navarro A, Villoslada P 2009. Allele-specific gene expression is widespread across the genome and biological processes. PLoS ONE 4: e4150 doi: 10.1371/journal.pone.0004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z 2007. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med 4: e8 doi: 10.1371/journal.pmed.0040008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proll J, Danzer M, Stabentheiner S, Niklas N, Hackl C, Hofer K, Atzmuller S, Hufnagl P, Gully C, Hauser H, et al. 2011. Sequence capture and next generation resequencing of the MHC region highlights potential transplantation determinants in HLA identical haematopoietic stem cell transplantation. DNA Res. doi: 10.1093/dnares/dsr008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2005. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33: D501–D504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH 2009. PID: The Pathway Interaction Database. Nucleic Acids Res 37: D674–D679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M 2006. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P 2003. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. 2007. Population genomics of human gene expression. Nat Genet 39: 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Cox DR, Frazer KA 2006. Allele-specific KRT1 expression is a complex trait. PLoS Genet 2: e93 doi: 10.1371/journal.pgen.0020093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ 2011. The importance of phase information for human genomics. Nat Rev Genet 12: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol 4: 231 doi: 10.1186/gb-2003-4-10-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle L, Serena-Acedo T, Liyanarachchi S, Hampel H, Comeras I, Li Z, Zeng Q, Zhang HT, Pennison MJ, Sadim M, et al. 2008. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science 321: 1361–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van El CG, Cornel MC 2011. Genetic testing and common disorders in a public health framework. Eur J Hum Genet 19: 377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA 2009. Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. 2008. The diploid genome sequence of an Asian individual. Nature 456: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Edelmann L, Kolodner RD, Kucherlapati R, Edelmann W 2010. Cis lethal genetic interactions attenuate and alter p53 tumorigenesis. Proc Natl Acad Sci 107: 5511–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goode DK, Cooke J, Callaway H, Smith S, Snell P, McEwen GK, Elgar G 2007. CONDOR: A database resource of developmentally associated conserved non-coding elements. BMC Dev Biol 7: 100 doi: 10.1186/1471-213X-7-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhu J, Shendure J, Porreca GJ, Aach JD, Mitra RD, Church GM 2006. Long-range polony haplotyping of individual human chromosome molecules. Nat Genet 38: 382–387 [DOI] [PubMed] [Google Scholar]