Abstract
Comparison of protein-coding DNA sequences from diverse primates can provide insight into these species' evolutionary history and uncover the molecular basis for their phenotypic differences. Currently, the number of available primate reference genomes limits these genome-wide comparisons. Here we use targeted capture methods designed for human to sequence the protein-coding regions, or exomes, of four non-human primate species (three Old World monkeys and one New World monkey). Despite average sequence divergence of up to 4% from the human sequence probes, we are able to capture ∼96% of coding sequences. Using a combination of mapping and assembly techniques, we generated high-quality full-length coding sequences for each species. Both the number of nucleotide differences and the distribution of insertion and deletion (indel) lengths indicate that the quality of the assembled sequences is very high and exceeds that of most reference genomes. Using this expanded set of primate coding sequences, we performed a genome-wide scan for genes experiencing positive selection and identified a novel class of adaptively evolving genes involved in the conversion of epithelial cells in skin, hair, and nails to keratin. Interestingly, the genes we identify under positive selection also exhibit significantly increased allele frequency differences among human populations, suggesting that they play a role in both recent and long-term adaptation. We also identify several genes that have been lost on specific primate lineages, which illustrate the broad utility of this data set for other evolutionary analyses. These results demonstrate the power of second-generation sequencing in comparative genomics and greatly expand the repertoire of available primate coding sequences.
Comparative genomics is invaluable for the study of evolutionary processes such as mutation, selection, and speciation (Thomas et al. 2003). In many cases, our power to detect evolutionary events is limited by the number of species with high-quality genome sequences (Anisimova et al. 2001; Eddy 2005). For example, power to detect positive selection depends on the total sequence divergence of the species being studied (Anisimova et al. 2001). Additionally, for many evolutionary analyses, the sequences need to be of very high quality to limit the rate of false positives (Mallick et al. 2009; Fletcher and Yang 2010). Second-generation sequencing technologies have made sequencing new primate genomes more feasible, but it is still challenging to assemble these short reads into complete genomes.
New methods for targeted enrichment of the human exome allow high-coverage sequence to be generated for the coding fraction of the genome (Albert et al. 2007; Gnirke et al. 2009; Tewhey et al. 2009; Turner et al. 2009). These methods are currently used in human medical resequencing studies to identify causal genes in Mendelian disorders (Choi et al. 2009; Ng et al. 2009, 2010) but, in principle, can be extended to targeted sequencing of human orthologs in closely related species. Such an approach has the advantage that sequenced reads are limited to non-repetitive coding regions that are more easily assembled from short reads.
Here we use solution-based targeted capture (Bainbridge et al. 2010) designed to human exons to sequence the exomes of three Old World monkeys and one New World monkey. We combine our high-quality sequences with available primate reference genomes and conduct a genome-wide scan for genes experiencing positive selection in primates. Our analysis has greater statistical power than previous scans for positive selection in primates (Clark et al. 2003; Chimpanzee Sequencing and Analysis Consortium 2005; Nielsen et al. 2005; Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), which were limited by a low number of species and low total sequence divergence (Anisimova et al. 2001). Other studies, which used diverse mammals to identify targets of positive selection (Kosiol et al. 2008), are more powerful but provide little information on more recent adaptation in primates.
We identify more than 150 genes that show strong evidence of positive selection on the primate lineage, at least twice as many as previous studies with fewer species (Nielsen et al. 2005; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Many of the genes and gene classes we identify are in accordance with these previous scans (e.g., genes involved in defense and immunity); however, we also find several novel adaptively evolving genes, most notably several genes involved in keratinization.
Results and Discussion
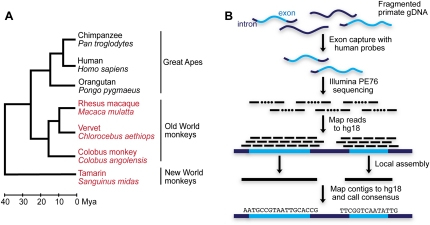
In total, we sequenced the exomes of three Old World monkeys (rhesus macaque, colobus monkey, and vervet) and one New World monkey (tamarin) (Fig. 1A). For each species, we targeted 25.3 Mb of unique protein-coding sequence from the Consensus Coding Sequence (CCDS) database (Pruitt et al. 2009) and generated, on average, 7.1 Gb of sequence per species with paired-end 76-bp reads. We aligned reads to the human reference genome and performed a local assembly of each target region (Fig. 1B; Supplemental Tables S1, S2). Our approach differs from de novo assembly in that it retains information from the human reference genome while still allowing for more diverged sequences than typical short read mapping techniques (for more details, see Methods).
Figure 1.
Sequence capture and assembly of non-human primates. (A) Phylogeny of primate species used in all analyses. Sequences from the species in red and black are from our exome assemblies and the publicly available primate reference genomes, respectively. For rhesus macaque, we generated an exome assembly and compared it to the macaque reference genome to assess the accuracy of our capture and assembly method. The phylogenetic relationship of the species and estimates of their divergence dates are from Goodman (1999). (B) Overview of sequencing, mapping, and assembly pipeline. Primate genomic DNA is fragmented, and protein-coding regions are captured using a solution-based hybridization method and sequenced as 76-bp paired-end reads. Reads are mapped to the repeat masked human reference genome using cross_match (v1.090518, http://www.phrap.org). Reads with overlapping mapped chromosomal coordinates are partitioned into groups and assembled independently using phrap (v1.090518, http://www.phrap.org). The resulting contigs are mapped back to the repeat masked human reference genome, and consensus bases are called from the highest-scoring mapped contigs.
To assess capture, sequencing, and assembly quality, we compared our rhesus macaque exome to the macaque reference genome (rheMac2). We captured and mapped macaque sequences for >96% of the target (Fig. 2; Table 1) and surprisingly found only a low association between our ability to capture macaque sequences and the number of nucleotide differences (R2 = 0.0087) or the number of indels (R2 = 0.0047) in targeted regions. In fact, human capture efficiency is the most informative predictor of macaque capture efficiency, suggesting that unknown conserved sequence features predominate in determining capture efficiency (Supplemental Table S3). We also compared targets that were successfully captured to those that failed to capture (<50% of bases covered by a read) despite having clear orthologs in the macaque genome (Supplemental Table S4). The failed targets have comparable GC content (51.6% vs. 49.9%), slightly higher divergence (3.9% vs. 3.1%), and a substantially greater proportion of bases that were inserted or deleted (0.50% vs. 0.17%). In total, only a small number of targets failed to capture by these criteria (1111 out of 155,707 targets with orthologs), and even targets up to 7% diverged from human are captured efficiently with a mean read depth >60× (Supplemental Fig. S1). Our ability to capture even the most divergent exons in macaque suggests that it will be possible to perform targeted capture of even more distantly related species.
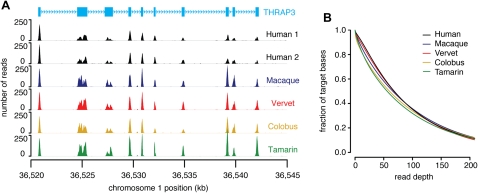
Figure 2.
Read depth of targeted regions. (A) An example of sequence capture. Read depth for a region on chromosome 1 encompassing the gene THRAP3 (CCDS405.1) from two human HapMap samples (Human 1: NA12878 and Human 2: NA18967) and four non-human primate samples (macaque, vervet, colobus, and tamarin). (B) Cumulative coverage of all targeted bases from one lane of paired-end 76-bp reads mapped to the human reference genome using cross_match for human (NA12878), macaque, vervet, colobus, and tamarin.
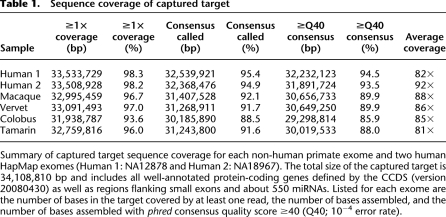
Table 1.
Sequence coverage of captured target
We assembled high-quality (≥Q40; error rate of <10−4) macaque sequences for ∼90% of our target (Table 1). These sequences are 2.24% different from the human reference genome, which corresponds almost precisely with sequence divergence in coding regions calculated from the macaque reference genome (Fig. 3A; Supplemental Table S5). We estimate the pairwise differences of our assembly relative to the macaque reference genome to be ∼0.10%, which agrees with a previous estimate of nucleotide diversity in Indian macaques (0.12%) (Hernandez et al. 2007). These data indicate that the quality of our macaque exome is at least that of the macaque reference genome and demonstrate that we can generate high-quality and accurate exome assemblies from short read data.
Figure 3.
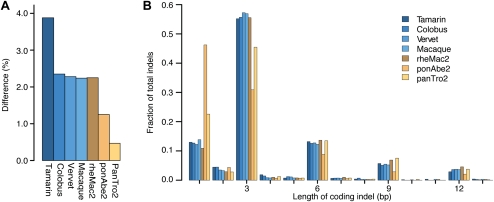
Sequence differences and indel lengths in protein-coding regions. (A) Coding sequence differences relative to the human reference genome for each assembled exome and non-human primate reference genome, calculated from the 9,106,235 sites that are high-quality in all species. (B) Distribution of coding indel lengths from the 4637 gene alignments where at least 75% of sites have high-quality sequence in all species. All indels are relative to the human reference genome. Low-quality indels are not included unless their read depth is ≥4 or they are confirmed by a high-quality indel in another species. Lengths from indels <15 bp apart are combined to account for uncertainty in the alignments. Indel lengths from the exome assemblies of macaque, vervet, colobus, and tamarin (blue); indel lengths from the reference genomes of chimpanzee (panTro2), orangutan (ponAbe2), and macaque (rheMac2) (yellow).
In the three other primates that we sequenced, between 95% and 97% of the targeted bases are covered by at least one read, and we generated high-quality consensus sequence for between 86% and 90% of the target (Table 1). Once again, divergence had very little impact on our ability to capture exome sequences from non-human primates (Fig. 2). Even for the most divergent species, the tamarin, we captured >96% and assembled >88% of the target at high quality (Table 1).
We performed extensive filtering of our exome assemblies because errors in sequencing, alignment, assembly, or ortholog assignment may introduce false positives in comparative genomic analyses (Mallick et al. 2009; Fletcher and Yang 2010). We removed sequences overlapping known segmental duplications in human (Cheng et al. 2005; Alkan et al. 2009), chimpanzee (Cheng et al. 2005), and macaque (Marques-Bonet et al. 2009), removed sequences with low read depth (<16×), and removed exons with very high levels of heterozygosity (which may reflect mis-assembly of paralogous sequences). We then removed exons and genes that had less than half of their sequence remaining after filtering. These filtering criteria exclude sequences that are more likely to be mis-assembled due to paralogous sequences (Supplemental Text S1) and do not appear to be biased toward removing known rapidly evolving genes, such as those involved in reproduction or immunity (Supplemental Text S2). For the exome assemblies, 61%–72% of each species' targeted coding sequence was retained for comparative analysis. This is comparable to the reference genome sequences, where we used less conservative filtering and retained 80%–89% of the targeted coding sequences post-filtering.
We combined our four assembled exomes with coding sequences from the reference genomes of human, chimpanzee, orangutan, and macaque and generated multiple sequence alignments for 16,707 genes. After filtering for high-quality regions in common to all species, we calculated the average nucleotide difference to the human reference genome to be 2.3% for Old World monkeys and 3.9% for the New World monkey (Fig. 3A; Supplemental Table S5).
As a test of assembly quality, we examined the distribution of indel lengths with respect to the human reference genome (Fig. 3B). On average, 80% of indels from our exome assemblies have lengths that are multiples of 3, an enrichment that is consistent with selection to preserve reading frame and remarkably similar to that of the macaque reference genome (Supplemental Table S6). This enrichment is substantially higher than that seen in other human exome studies (Ng et al. 2009; Pelak et al. 2010) or in the chimpanzee and orangutan reference genomes (Fig. 3B), which suggests that our exome assemblies are of higher quality. The increased rate of indel errors in the orangutan reference genome has also been previously noted (Meader et al. 2010).
From our coding sequence alignments, we filtered out sequences with too little sequence data, frameshifts, or internal stop codons (Supplemental Fig. S2) to obtain a highly confident set of 15,027 orthologs with sequence from at least three species. We then tested each of these orthologs for evidence of positive selection acting at any point during primate evolution using likelihood models that allow dN/dS to vary across codons (Nielsen and Yang 1998; Yang et al. 2000). The addition of our exome sequences increased the total branch length by threefold relative to analyses using just human, chimpanzee, and macaque (median S = 0.30 vs. S = 0.080 nucleotide substitutions per codon), and should substantially increase our power to detect positive selection (Anisimova et al. 2001). We find evidence of positive selection for 930 genes (nominal P-value < 0.05) without correcting for multiple testing, or a total of 157 at a false discovery rate (FDR) of 10% (Supplemental Tables S7, S8).
We compared these 157 genes to a previous scan for positive selection in primates that identified 67 positively selected genes (at 10% FDR) using coding sequences from the human, chimpanzee, and macaque genomes (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Of these 67 genes, we omitted 22 from our analysis because they were either not targeted or we could not confidently obtain sequences from at least three species, including the available primate reference genomes (Supplemental Table S9). The remaining 45 genes rank significantly higher than other genes in our scan for positive selection (p < 2.2 × 10−16; two-sided Mann-Whitney U-test) with 15 genes showing strong evidence for positive selection at an FDR of 10% and an additional 19 genes with a nominal P-value < 0.05. We thus identify 142 new candidates in our analysis. We find no evidence of positive selection for 11 of the genes identified by the previous analysis, which is likely due to differences in methodology, such as ortholog filtering, low-quality sequence filtering, or multiple sequence alignment. When we perform the same analysis using only human, chimpanzee, and macaque sequences, we find evidence of positive selection for only 25 genes (at 10% FDR). Our approach is more conservative and should have fewer false positives due to low quality or misaligned sequences.
We identified several biological processes enriched for genes under positive selection (Supplemental Table S10) using the Gene Ontology classification system (Ashburner et al. 2000). In agreement with previous scans for positive selection in primates (Clark et al. 2003; Nielsen et al. 2005; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), several of the top categories are involved in immunity (“defense response” and “antigen processing and presentation”), sensory perception (“sensory perception of a chemical stimulus”), and reproduction (“spermatogenesis” and “fertilization”). Among the genes that show the strongest evidence of positive selection are several that are known to be rapidly evolving (e.g., PTPRC [Filip and Mundy 2004], PRM1 [Wyckoff et al. 2000], and APOBEC3G [Zhang and Webb 2004]), and several with no previous evidence of positive selection in primates (e.g., TF, an iron transporter previously known to be under positive selection only in salmonids) (Ford 2001).
Interestingly, we also found an excess of positively selected genes involved in the process of keratinization (Supplemental Table S11). To our knowledge, none of these genes have been previously identified as targets of positive selection in primates, except for IVL, which was recently shown to be subject to positive selection in human populations (Tennessen et al. 2010). As keratinization is the process of converting outer epidermal cells in skin, hair, and nails to keratin, these genes may be important for setting up physical barriers between the body and the outside world and could evolve rapidly in response to changing environments.
We also tested whether the genes that we find under positive selection in primates also show evidence for recent selection in human populations. The 157 genes with strong evidence for positive selection in primates have increased allele frequency differences between Europeans and Africans (mean FST = 0.0928) compared to the remaining genes (mean FST = 0.0710; p < 2.2 × 10−16; two-sided Mann-Whitney U-test) (Tennessen et al. 2010). We also tested the 10 GO categories with the most significant enrichments for genes under positive selection and found that three of the categories (“sensory perception of chemical stimulus,” “oxidation reduction,” and “sensory perception”) have significantly higher levels of population differentiation (Supplemental Table S12). Both observations are consistent with the idea that many of the genes under long-term positive selection in primates are also important in recent human adaptation.
In addition to the 157 positively selected genes identified across all branches of the primate phylogeny, 142 genes show evidence for positive selection on specific lineages at an FDR of 10% (Supplemental Tables S13–S15). Of these, 28 overlap with the original set of 157 genes, bringing the total number of identified genes to 271. Two genes, KRTAP4-5 and CASP10, have evidence for positive selection on more than one lineage. KRTAP4-5 encodes a keratin-associated protein involved in the structure of hair fibers and shows evidence for positive selection on both the chimpanzee and the hominid branches. CASP10 encodes an apoptosis-related caspase that appears to be adaptively evolving on both the Old World monkey and tamarin lineages. The number of genes identified on each branch ranges from zero on the human branch to 84 on the tamarin branch. The longer branches (e.g., tamarin) probably have more significant genes because they provide more power to detect positive selection (Zhang et al. 2005).
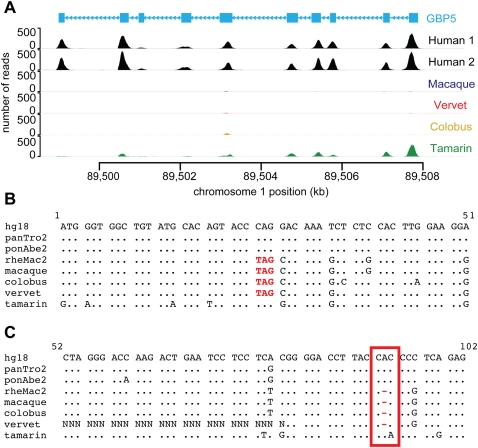
To demonstrate how this data set can be used for other types of evolutionary analyses, we identified genes that have been lost by either gene deletion or pseudogenization. For example, all or nearly all exons of the gene GBP5 are missing in the three Old World monkeys but are present in the New World monkey, the tamarin (Fig. 4A). Similarly, SNTN and CCL14 contain premature stop codons or frameshifts in all of the Old World monkeys, yet not in tamarin (Fig. 4B,C). Sequences from the chimpanzee, orangutan, and macaque reference genomes confirm the loss of these genes in the common ancestor of Old World monkeys.
Figure 4.
Examples of gene loss in Old World monkeys. (A) An example of a gene deletion detected by read depth differences between species. The read depth for GBP5 (CCDS722.1) is high in human (Human 1: NA12878 and Human 2: NA18967) and tamarin, but absent in macaque, vervet, and colobus. The absence of GBP5 in the macaque exome sequences is supported by the macaque reference genome. (B) The beginning of the multiple sequence alignment for the gene SNTN (CCDS33779.1) containing a premature stop codon in Old World monkeys. The substitution causing this premature stop codon is high quality (≥Q40) in macaque, vervet, and colobus and confirmed by the macaque reference genome. (C) A portion of the multiple sequence alignment for the gene CCL14 (CCDS32624.1) containing a frameshift in Old World monkeys. (Red box) A one-base gap in macaque, vervet and colobus, which disrupts the reading frame of the latter half of the gene. This gap is surrounded by high-quality (≥Q40) bases and also supported by the macaque reference genome.
We have demonstrated that solution-based hybrid capture is an efficient method for the sequencing of orthologs in other species. This method can be applied not just to non-human primate species, but to any species for which a closely related reference genome is available and works well for coding sequences up to an average divergence of 7% (and possibly greater). In principle, this method is not limited to coding sequences but can be extended by designing capture probes to other unique regions of the genome. We have used this method to identify 157 candidates for positive selection in primates and envision that this method can be applied to many other problems in molecular evolution and population genomics. By obtaining sequences from multiple individuals, it will be possible to characterize patterns of genetic variation in numerous species. Such data will help answer many questions about selection, demography, mutation, gene loss, and gene duplication.
Methods
Genomic DNA samples
Genomic DNA samples were obtained from Coriell Cell Repositories for a rhesus macaque (Macaca mulatta; NG07107), a colobus monkey (Colobus angolensis; PR00099), a vervet (Chlorocebus aethiops; PR00990), a tamarin (Saguinus midas; PR00550), and two HapMap human individuals (European-American NA12878 and East Asian NA18967).
Library oligonucleotides and adapters
Oligonucleotides used in the library construction were SLXA_Pair_For_Amp (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T), SLXA_Pair_Rev_Amp (CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T), Adapter_PE_Hi (ACACTCTTTCCCTACACGACGCTCTTCCGATC*T) and Adapter_PE_Lo (/5Phos/GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG), where “*” refers to a phosphorothioate bond.
Adapter_PE_Hi and Adapter_PE_Lo were annealed to form Y-adapters by incubating equimolar amounts at 95°C and then allowing them to cool to room temperature in a heat block.
Library construction
Genomic DNA from each sample (3 μg) was sheared (Covaris AFA) in 85 μL of elution buffer (Buffer EB, 10 mM Tris-Cl at pH 8.5; QIAGEN) using the settings: duty cycle 10%, intensity 5, and cycle/burst 200 for 600 sec. Fragmented DNA ends were repaired for 30 min at 20°C with 5 μL of End Repair Enzyme Mix and 1× End Repair Reaction Buffer in a total volume of 100 μL (NEBNext End Repair Module; New England Biolabs) and eluted in 45 μL of water after cleanup. A-tails were then added to the end-repaired DNA for 20 min at 70°C in a total volume of 100 μL (1× PCR buffer, 1.5 mM MgCl2, 1 mM dATP, and 5 units AmpliTaq DNA polymerase) and eluted in 38 μL of water after cleanup. Y-adapters were ligated to the A-tailed fragments for 20 min at 16°C in a total volume of 50 μL (1× T4 DNA Ligase Buffer [Enzymatics], 240 units of T4 DNA Ligase [Enzymatics], and 5 μL of Y-adapters [50 μM]) and eluted in 50 μL of water after cleanup. All cleanup steps were performed with 1.8× AmpureXP beads as directed by Agencourt.
The adapter-ligated fragments were PCR-amplified in four reactions per sample, each in a total volume of 40 μL (10 μL of adapter-ligated fragments, 1× iProof High Fidelity Master Mix [Bio-Rad], and 0.625 μM both SLXA_Pair_For_Amp and SLXA_Pair_Rev_Amp). The PCR conditions were 2 min at 96°C, 16 cycles of 20 sec at 96°C, 30 sec at 65°C, and 45 sec at 72°C, followed by a final 5 min at 72°C. The four reactions for each sample were then pooled, cleaned up (PCR Purification Kit; QIAGEN), and quantified (Nanodrop 8000 Spectrophotometer).
Library capture and sequencing
Each library (1 μg) was hybridized to SeqCap EZ Exome probes (v1.0, Nimblegen) according to manufacturer's protocols and blocked with 100 μL of 1 mg/mL human Cot-1 DNA (Invitrogen) and 10 μL of both SLXA_Pair_For_Amp (100 μM) and SLXA_Pair_Rev_Amp (100 μM). The hybridized library was captured and washed as directed by Nimblegen and eluted in 50 μL of water. The enriched library was PCR-amplified in 10 reactions per sample, each in a total volume of 50 μL (4 μL of library, 1× iProof High Fidelity Master Mix, and 0.625 μM both SLXA_Pair_For_Amp and SLXA_Pair_Rev_Amp). The PCR conditions were 30 sec at 98°C, 20 cycles of 10 sec at 98°C, 30 sec at 60°C, and 30 sec at 72°C, followed by a final 5 min at 72°C. The 10 reactions for each sample were then pooled and column-purified (PCR Purification Kit; QIAGEN).
One lane of 76-bp paired-end reads was generated for each sample on an Illumina Genome Analyzer IIx according to the manufacturer's instructions.
Target description
All unique, well-annotated protein-coding regions (including flanking regions for exons smaller than 200 bp) from the CCDS database (version 20080430) (Pruitt et al. 2009) and about 550 miRNAs were targeted by SeqCap EZ Exome probes. This resulted in 176,817 continuous captured genomic regions totaling to 34,108,810 bp. The 20080430 version of the CCDS database contains 164,367 protein-coding genomic regions spanning 28,000,325 bp after merging regions with overlapping coordinates. Repetitive regions are excluded from the tiling probes, reducing the final protein-coding target to 148,667 genomic regions for a total of 25,299,356 bp.
Merging overlapping paired-end reads
Although our genomic DNA was fragmented to an average size of 200 bp, a substantial fraction of our 76-bp paired-end reads overlapped their mate and could be merged into longer single reads. We aligned each pair of reads using a semi-global version of the Needleman-Wunsch (Needleman and Wunsch 1970) algorithm that constrained the alignment to the end of the left read and the start of the right read and used the following score scheme: match +1, mismatch −3, gap −5. If the alignment score was ≥10, the reads were merged, with any mismatching positions masked to “N.” When high-scoring alignments contained gaps, both reads were discarded. Quality scores for the overlapping portion of the merged reads were calculated by summing the two independent quality scores, capping the maximum value at Q40. When alignments did not meet the score threshold, both reads were kept individually. If the semi-global alignment spanned the entire length of either read, a local Smith-Waterman alignment (Smith and Waterman 1981) was performed, and only the aligned portion of both reads was kept. This prevented adapter sequences from being included in the merged reads.
Mapping reads to the human genome
We used the human genome to guide local exome assemblies for each of the primate species previously listed. Each merged read or pair of unmerged reads were mapped independently to the repeat masked human reference genome (hg18) using cross_match (v1.090518, http://www.phrap.org) with the parameters –minscore 25 -minmatch 12 -maxmatch 20. If a read mapped to more than one location, only the highest scoring match was kept, and if there was no single highest-scoring match, the read was discarded. Duplicate reads, which may result from PCR amplification or optical artifacts, were identified as those that mapped to exactly the same chromosomal location in the same orientation. For these reads, only the one with the highest mean quality score was kept.
Assessment of capture efficiency
We assessed the capture efficiency of our method by comparing the sequenced macaque exome with its reference genome (rheMac2). We looked at the correlation between read depth and the number of nucleotide differences or indels and also built a linear model (Supplemental Table S3) to identify sources of variability in captured target read depth. We used macaque read depth as our response variable and human read depth, number of nucleotide differences, number of indels, GC content, and mappability as predictors. As data points we used 155,707 capture targets, for which we could identify orthologs in rheMac2 by best reciprocal BLAST hits (requiring scores to be at least 1.2× greater than the next best alignment). To separate capture and mapping efficiencies, we gave each base in our capture target a “mappability” score determined from the depth of simulated 76-bp rheMac2 reads, which were aligned to human. Targets were discarded if they were <100 bp in length or if they contained a base with a rheMac2 quality score <40 (because this could affect the accuracy of our nucleotide difference and indel estimates). In total, 128,914 orthologous capture targets were used to fit the linear model.
Local assembly of mapped reads
Based on their mapped chromosomal locations, overlapping reads were partitioned into groups, which could be assembled independently. Overlap groups that contained more than 500 reads were split into equally sized subgroups to reduce computational time. Overlap groups and subgroups were assembled using phrap (v1.090518; http://www.phrap.org) with parameters that were previously optimized for short read assembly (Hiatt et al. 2010): –vector_bound 0 -forcelevel 1 -minscore 12 -minmatch 10 -indexwordsize 8. Contigs from split overlap groups were further assembled into longer contigs with a second round of phrap using the same parameters. The final contigs were then mapped back to the repeat masked human reference genome using cross_match with the same parameters as above. We discarded contigs that mapped to a different chromosomal location than the individual reads and discarded contigs that did not map to one location uniquely (requiring the score of the best alignment to be at least 1.2× greater than the next best alignment). These filters helped us reduce mis-assemblies caused by paralogous sequences.
Consensus calls and quality scores were determined from the contigs that overlapped the target sequences. When phrap created two or more contigs that mapped uniquely to the same chromosomal location, the contig with the highest cross_match score was used as the consensus. Target regions with no mapped contigs were assigned a base “N” with quality Q0, as were non-targeted regions that were present in the CCDS database. A fasta file containing these consensus contig sequences was then generated for each species. A summary of the sequence coverage and assembly of captured regions is found in Table 1 (whole captured target) and Supplemental Table S16 (captured miRNAs).
Mapping unique reads to the assembled consensus
To identify heterozygous sites and assess the quality of the phrap assemblies, we remapped the paired-end reads to the assembled contigs. From our cross_match output, we identified uniquely mapping read pairs that aligned to one location with a score at least 1.2× higher than at any other location. We used only read pairs where both individual reads mapped uniquely, and replaced merged unique reads with the individual reads from which they came. We then mapped read pairs to the phrap-assembled consensus using BWA 0.5.6 with default parameters for paired-end reads (Li and Durbin 2009). The alignments were sorted and filtered for duplicates using Picard 1.15 (http://picard.sourceforge.net), and a pileup file was generated with SAMtools 0.1.7a (Li et al. 2009), which lists the bases of all the aligned reads for each position in the assembled consensus.
Identification of heterozygous sites
We called genotypes at all consensus sites using the observed bases and quality scores from the pileup file described above. We assigned a genotype quality score to each base using the independent error model described by Li et al. (2008). For these calculations, we used a prior probability of 0.001 of a site being heterozygous and capped the base quality of individual reads at 30.
For comparison with our own data, we estimated nucleotide diversity in macaques from counts of previously identified segregating sites (Hernandez et al. 2007). In this study, a total of 1476 SNPs were identified in 150,372 bp, which were sequenced in 38 Indian macaques and nine Chinese macaques (94 chromosomes total). Of these SNPs, 486 were observed in both Indian and Chinese macaques, and 386 were observed only in the Indian sample. From these numbers, we estimated nucleotide diversity by calculating Watterson's population mutation rate estimator θ (Watterson 1975) and dividing by the number of sequenced bases. Nucleotide diversity for the Chinese and Indian macaques was estimated to be 0.22% and 0.12%, respectively.
Target masking
To avoid mis-assembly of paralogous sequences, we masked regions that we could not uniquely map human reads to. We simulated all possible human 76-bp reads (in one orientation), which overlapped the captured target and mapped them to the repeat masked human reference genome using cross_match (v1.090518, http://www.phrap.org) with parameters -minscore 68 -minmatch 12 -maxmatch 20. These parameters allowed reads to be mapped to all locations in the reference genome with one or two mismatches. We then tabulated the number of correctly and incorrectly mapped reads, for each base in the target. Target bases with less than 38 correctly mapped reads (half the expected 76) or more than 10 incorrectly mapped reads were considered “unmappable.” We also masked coding sequences overlapping segmentally duplicated regions of the human (Cheng et al. 2005; Alkan et al. 2009), chimpanzee (Cheng et al. 2005), or orangutan (Marques-Bonet et al. 2009) genomes. This resulted in 1,400,787 bp (4.1%) masked due to potential segmental duplications and 1,711,106 bp (5.0%) masked due to low mappability, for a total of 2,938,059 bp (8.6%) masked for downstream analyses.
Assembly quality filtering
From the pileup, we identified and filtered inconsistencies between the assembled consensus sequence and the individual reads. If the pileup consensus base disagreed with the phrap-assembled base, we masked that base to an “N” with quality Q0 unless the pileup contained eight or more reads, in which case we changed that base to the pileup consensus and flagged it with quality Q1. If the pileup indicated a non-polymorphic insertion or deletion, suggesting an incorrectly placed indel in the phrap assembly, that region and the two flanking bases were masked to an “N” with quality Q0. For heterozygous sites, the pileup base with the majority of reads was used as the consensus, regardless of whether or not it matched the phrap-assembled base, and given quality Q1. Exons with excess heterozygosity (≥3 heterozygous sites in any 20-bp window), which suggest paralogous assemblies, were removed completely. Exons with <16× read depth for more than half of their sequence were also removed completely.
CDS from exome assemblies
Coding sequences and quality scores were extracted from the quality-filtered consensus sequence for each CCDS entry using human coordinates. Gaps were removed from the coding sequences so that the multiple sequence alignment program could place them. Exons or genes missing more than half of their sequence were completely masked or removed to avoid alignment errors. In total, there are 20,091 entries in the CCDS (version 20080430). When coding sequences for multiple CCDS entries overlapped, only the entry with the longest sequence was kept, resulting in 16,707 unique coding sequences (27,492,897 bp).
CDS from reference genomes
Coding sequences were obtained from the publicly available reference assemblies of human (hg18), chimpanzee (panTro2), orangutan (ponAbe2), and macaque (rheMac2). Pairwise whole-genome alignments were downloaded from UCSC (Chiaromonte et al. 2002; Kent et al. 2003; Schwartz et al. 2003) for each of these species and filtered to be best-reciprocal and syntenic as described by McVicker et al. (2009). Bases overlapping segmentally duplicated regions of the human (Cheng et al. 2005; Alkan et al. 2009), chimpanzee (Cheng et al. 2005), or orangutan (Marques-Bonet et al. 2009) genomes were removed. Target sequences and quality scores were then extracted from the filtered alignments based on the human CCDS coordinates.
Multiple sequence alignments
Coding sequences for the combined set of species were aligned using PRANK (v0.100311) (Löytynoja and Goldman 2005) with parameters -t -F -twice -a -gapext=0.8 -kappa=2.0 -gaprate=0.05 and a species tree representing the standard primate phylogeny (Goodman 1999). Our assembled macaque sequences were included in addition to sequences from the macaque reference genome so that the quality of the two assemblies could be compared. This resulted in a total of eight sequences in the multiple alignments: hg18, panTro2, ponAbe2, rheMac2, macaque, vervet, colobus monkey, and tamarin.
Multiple sequence alignment quality filtering
We extensively filtered low-quality single-nucleotide differences and indels to produce a set of high-quality multiple sequence alignments. For each coding sequence alignment, we compared each non-human primate sequence to the human sequence and masked differences with quality scores less than Q40 in the non-human sequence to an “N.” This includes heterozygous sites in both the non-human reference genome sequences and the exome sequences that have quality Q0 and Q1, respectively.
For simplicity, we refer to alignment gaps in the human sequence as “insertions” and alignment gaps in the non-human sequences as “deletions,” even though it is unclear what the exact events were or on which lineage they occurred. Indels were retained if they met any one of the following criteria: (1) a minimum quality score ≥Q40 within (insertions) or surrounding (deletions) the indel; (2) a minimum read depth ≥4 within or surrounding the indel; or (3) the presence of a high-quality or high-read-depth indel in another non-human sequence (species confirmation). Indels not meeting one of these criteria were completely removed from all sequences of the alignment if they were insertions or masked if they were deletions.
Quality assessment of assemblies
We assessed the quality of our assemblies using the number of single-nucleotide differences and distribution of indel lengths relative to the human reference genome. To directly compare the number of nucleotide differences between each assembly, we limited our analysis to coding sites that were high quality (≥Q40) in all three of our non-human reference genomes and all four of our exome assemblies, resulting in a total of 9,106,235 sites (36.0% of the coding target). We then calculated the proportion of high-quality differences between each human reference base and its corresponding base in each non-human sequence. We note that these common high-quality sites may be biased toward more conserved regions and, thus, are likely to underestimate the average proportion of nucleotide differences between human and non-human coding sequences.
For calculating the proportion of high-quality indels, we restricted our set of alignments to 4637 genes containing >75% high-quality sequence in all species. To compute the length of each indel, we combined indels that were <15 bp apart in order to account for uncertainty in the alignment. For example, a 2-bp deletion followed closely by a 5-bp insertion would be considered a 3-bp insertion.
Additionally, we calculated the number of high-quality nucleotide differences between our macaque exome assembly and the macaque reference genome from 14,924,161 coding sites (59.0%) to get an estimate of pairwise polymorphism (π) in Indian rhesus macaques. This number is likely to be an underestimate of π in Indian rhesus macaques because heterozygous sites are masked in both sequences.
Ortholog filtering for evolutionary analysis
For each orthologous gene set, we filtered out sequences from species that suggested sequencing/assembly errors, alignment errors, or gene loss of function. Four hundred and one genes were completely removed because their CCDS status is currently listed as “withdrawn” (CCDS version 20100829), resulting in 16,303 genes before ortholog filtering. Of these genes, a species' sequence was removed if it: (1) contained <25% high-quality sequence; (2) contained a premature stop codon >25 bp from the end of the gene sequence; or (3) contained a frameshift disrupting >15 bp of sequence. Stop codons within 25 bp of the end of the coding sequence and any sequence following them were masked, as were frameshifted regions of <15 bp in length. Fourteen human coding sequences contained SECIS elements that direct internal UGA stop codons to be translated as selenocysteines. In these cases, the internal UGA was masked in each sequence of the alignment rather than throwing the gene out completely. Following this filtering, 15,037 remaining genes contained sequence from three or more species and were used in downstream analyses of positive selection (Supplemental Fig. S2).
Evolutionary analysis
We obtained likelihoods and dN/dS estimates for each orthologous set of genes using CODEML from the PAML 4.4 package (Yang 2007). Heterozygous and all missing or low-quality sites were masked to “N” and treated as missing data with the cleandata=0 option. An unrooted phylogeny corresponding to the accepted relationships between the studied primates (Goodman 1999) was used in each analysis.
To test for selection acting at any point on the phylogeny, we compared a neutral model of 0 ≤ dN/dS ≤ 1 (M7; model=1, NSsites=7) to a model of selection where an additional class of codons is allowed to have dN/dS > 1 (M8; model=1, NSsites=8) and performed a likelihood ratio test with a χ2 approximation to calculate P-values. q-values (Storey and Tibshirani 2003) were used to set a significance threshold corresponding to an FDR of 10%. CODEML was run with multiple dN/dS starting points to ensure convergence of model parameter estimates. Ten genes failed to converge and were removed from the analysis, resulting in 15,027 genes tested.
For tests of selection acting on individual lineages, we used CODEML's branch-site models (model=2, NSsites=2), which allow dN/dS to vary among codon sites and across branches of the phylogeny. For each branch, we compared a selection model, which allows a class of codons on that branch to have dN/dS > 1, to a neutral model, which constrains this additional class of sites to have dN/dS = 1 (fix_omega = 1, omega = 1). A likelihood ratio test was performed and P-values were computed from a 50:50 mixture of a χ2 distribution with 1 degree of freedom and a point mass at 0 (Zhang et al. 2005). The false discovery rate was estimated with q-values. Details on the number of genes tested and significant for each branch are provided in Supplemental Table S13.
Gene Ontology analysis
We used the following procedure to identify GO terms enriched for genes under positive selection. We first assigned each gene a UniProt identifier, using a list of CCDS to UniProt associations downloaded from BioMart (Smedley et al. 2009). We then assigned genes to GO terms using the UniProt identifiers and the human gene association file downloaded from http://geneontology.org (submission date 09/06/2010). Genes were also assigned to parent GO terms by propagating them up GO's hierarchy of “ISA” relationships.
We next ranked genes by their log-likelihood difference (between CODEML's M7 and M8 models) and performed one-sided Mann-Whitney U-tests to determine whether genes in a given GO term ranked significantly higher than genes in a null distribution. The null distribution consisted of all genes assigned to GO terms, excluding those assigned to the term being tested. Following this procedure, we reported the most significant GO term and removed its associated genes (to avoid reporting redundant or overlapping terms). This process was repeated iteratively until there remained no significant GO terms at the p < 0.05 level. In each iteration, only GO terms with at least 20 remaining genes were tested.
We used the same iterative procedure to identify GO terms enriched for “absent” genes (see Supplemental Text S2). In this case, we compared counts of present and absent genes for the GO term being tested to those in all other GO terms and assessed significance using a one-sided Fisher's exact test.
Human population differentiation
We examined whether genes with evidence for positive selection in primates also have increased human population differentiation, which may result from recent positive selection. To estimate differentiation between European and African populations, we assigned each gene in our filtered data set an average FST value. These values were calculated using exome sequences from four African individuals and six European individuals in a study by Tennessen et al. (2010). FST was calculated for 100-kb genomic regions that contained >500 bp of exonic sequence by averaging FST (Weir and Cockerham 1984) across all exonic polymorphic sites. If a gene fell into multiple 100-kb regions, the lowest FST value was used. We tested whether the mean FST for genes with evidence of positive selection in primates (at 10% FDR) was different from the mean FST among the remaining genes using a two-sided Mann-Whitney U-test. Additionally, we tested the top 10 GO categories enriched for genes under positive selection in primates by comparing the mean FST of genes within the category to the mean FST of remaining genes with two-sided Mann-Whitney U-tests.
Example of gene loss
To find an example of a deleted gene in our non-human primate samples, we considered exons with an average of at least 15 reads per base in human, but fewer than 5 reads per base in a non-human species as candidates for exon loss. Where possible, we confirmed these events in Old World monkeys using the macaque (rheMac2) reference genome. We are currently developing statistical methods for the detection of gene loss events genome-wide.
Data access
All raw sequencing reads can be retrieved from the NCBI Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRP005434. Raw sequencing reads, assembled coding sequences, multiple sequence alignments, and CODEML output are available from http://depts.washington.edu/swansonw/Swanson_Lab/Data.html.
Acknowledgments
We thank Debbie Nickerson, Josh Akey, Jacob Tennessen, Geoff Findlay, and members of the Swanson, Thomas and Shendure labs for helpful discussions and comments on the manuscript. We also thank Choli Lee for sequencing assistance. This work was supported in part by grants from the National Institutes of Health/National Heart Lung and Blood Institute (RO1 HL094967 to J.S.) and the National Institutes of Health/National Human Genome Research Institute (R21 HG004749 to J.S.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121327.111.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905 [DOI] [PubMed] [Google Scholar]
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 41: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol 18: 1585–1592 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Burgess DL, Kovar C, Rodesch MJ, D'Ascenzo M, Kitzman J, Wu YQ, Newsham I, Richmond TA, et al. 2010. Whole exome capture in solution with 3 Gbp of data. Genome Biol 11: R62 doi: 10.1186/gb-2010-11-6-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakewell MA, Shi P, Zhang J 2007. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc Natl Acad Sci 104: 7489–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson R, Pääbo S 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93 [DOI] [PubMed] [Google Scholar]
- Chiaromonte F, Yap VB, Miller W 2002. Scoring pairwise genomic sequence alignments. Pac Symp Biocomput 2002: 115–126 [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, et al. 2009. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci 106: 19096–19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. 2003. Inferring nonneutral evolution from human–chimp–mouse orthologous gene trios. Science 302: 1960–1963 [DOI] [PubMed] [Google Scholar]
- Eddy SR 2005. A model of the statistical power of comparative genome sequence analysis. PLoS Biol 3: e10 doi: 10.1371/journal.pbio.0030010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filip LC, Mundy NI 2004. Rapid evolution by positive Darwinian selection in the extracellular domain of the abundant lymphocyte protein CD45 in primates. Mol Biol Evol 21: 1504–1511 [DOI] [PubMed] [Google Scholar]
- Fletcher W, Yang Z 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol 27: 2257–2267 [DOI] [PubMed] [Google Scholar]
- Ford MJ 2001. Molecular evolution of transferrin: Evidence for positive selection in salmonids. Mol Biol Evol 18: 639–647 [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M 1999. The genomic record of humankind's evolutionary roots. Am J Hum Genet 64: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Hubisz M, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, et al. 2007. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science 316: 240–243 [DOI] [PubMed] [Google Scholar]
- Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J 2010. Parallel, tag-directed assembly of locally derived short sequence reads. Nat Methods 7: 119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D 2003. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci 100: 11484–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, Vinař T, Da Fonseca R, Hubisz M, Bustamante C, Nielsen R, Siepel A, Schierup M 2008. Patterns of positive selection in six mammalian genomes. PLoS Genet 4: e1000144 doi: 10.1371/journal.pgen.1000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci 102: 10557–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Gnerre S, Muller P, Reich D 2009. The difficulty of avoiding false positives in genome scans for natural selection. Genome Res 19: 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet 5: e1000471 doi: 10.1371/journal.pgen.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meader S, Hillier LW, Locke D, Ponting CP, Lunter G 2010. Genome assembly quality: Assessment and improvement using the neutral indel model. Genome Res 20: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48: 443–453 [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. 2009. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. 2010. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet 42: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz M, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol 3: e170 doi: 10.1371/journal.pbio.0030170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, Cirulli ET, Fellay J, Dickson SP, Gumbs CE, et al. 2010. The characterization of twenty sequenced human genomes. PLoS Genet 6: e1001111 doi: 10.1371/journal.pgen.1001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. 2009. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res 19: 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W 2003. Human–mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A 2009. BioMart—biological queries made easy. BMC Genomics 10: 22 doi: 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Waterman M 1981. Identification of common molecular subsequences. J Mol Biol 147: 195–197 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Madeoy J, Akey JM 2010. Signatures of positive selection apparent in a small sample of human exomes. Genome Res 20: 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB, Larson JW, et al. 2009. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol 27: 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788–793 [DOI] [PubMed] [Google Scholar]
- Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J 2009. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods 6: 315–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Weir B, Cockerham C 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu CI 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304–309 [DOI] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum Mol Genet 13: 1785–1791 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479 [DOI] [PubMed] [Google Scholar]