Abstract
The ability to uncover the phylogenetic history of recently extinct species and other species known only from archived museum material has rapidly improved due to the reduced cost and increased sequence capacity of next-generation sequencing technologies. One limitation of these approaches is the difficulty of isolating and sequencing large, orthologous DNA regions across multiple divergent species, which is exacerbated for museum specimens, where DNA quality varies greatly between samples and contamination levels are often high. Here we describe the use of cross-species DNA capture hybridization techniques and next-generation sequencing to selectively isolate and sequence partial to full-length mitochondrial DNA genomes from the degraded DNA of museum specimens, using probes generated from the DNA of a single extant species. We demonstrate our approach on specimens from an enigmatic gliding mammal, the Sunda colugo, which is widely distributed throughout Southeast Asia. We isolated DNA from 13 colugo specimens collected 47–170 years ago, and successfully captured and sequenced mitochondrial DNA from every specimen, frequently recovering fragments with 10%–13% sequence divergence from the capture probe sequence. Phylogenetic results reveal deep genetic divergence among colugos, both within and between the islands of Borneo and Java, as well as between the Malay Peninsula and different Sundaic islands. Our method is based on noninvasive sampling of minute amounts of soft tissue material from museum specimens, leaving the original specimen essentially undamaged. This approach represents a paradigm shift away from standard PCR-based approaches for accessing population genetic and phylogenomic information from poorly known and difficult-to-study species.
The advent of next-generation sequencing technologies (NGSTs) has transformed the way in which scientists approach a myriad of biological questions (Hawkins et al. 2010). Even with NGSTs' growing familiarity and broad range of applications such as ChIP-seq, RNA-seq, and genome-wide association studies, NGSTs still have the potential to influence the field of population genetics and phylogenetics with new methods to obtain genomic sequences of rare, difficult to sample, or extinct species (Millar et al. 2008). The ability to uncover the phylogenetic history of recently extinct species has rapidly improved due to the reduced cost and increased sequence capacity of NGSTs (Gilbert et al. 2007, 2008; Miller et al. 2008, 2009); however, obstacles do remain. The difficulties with applying NGSTs to phylogenetic problems do not lie with the sequencing technology itself, but with the preparative procedures for isolation and sequencing of large, orthologous DNA regions across multiple divergent species (Summerer 2009). This problem is exacerbated for museum specimens, where DNA quality varies greatly between samples and contamination levels are often high (Millar et al. 2008). Generation of whole genome sequences for museum specimens, or even complete mitochondrial DNA (mtDNA) genome sequences, is not cost-effective for most laboratories due to the large amount of sequencing required for adequate genome coverage of a single individual.
Capture hybridization methods are routinely used for genomic-scale enrichments of modern target DNA from the same species (Summerer 2009; Mamanova et al. 2010) and also for recovery of DNA from museum or fossil specimens by largely removing contaminants from the final product (Krause et al. 2010). However, capture hybridization techniques have not been applied to assembling phylogenetic data sets across divergent sets of taxa (e.g., millions of years of genetic divergence), largely due to lack of appropriate probes and lack of exploration of hybridization conditions to allow for heterologous sequence capture. Enrichment for target sequences by PCR (which has been the standard for most previous museum DNA studies) requires closely related reference sequences and painstaking efforts to design many oligonucleotide primers to amplify very short regions of the DNA of interest. Capture hybridization and sequencing of targeted loci from museum specimens promises to be a more flexible, cost-effective, and efficient approach than other enrichment procedures for degraded samples.
Here we describe the application of capture hybridization and selection techniques to recover mitochondrial DNA from 13 Sunda colugo (Galeopterus variegatus) museum specimens of varying ages (47–170 yr old) that represent major geographical locations throughout the Southeast Asian mainland and archipelago (Supplemental Fig. S1; Table 1). Colugos are arboreal mammals that are widely distributed throughout Southeast Asia and have the most extensive gliding membrane (patagium) of any known mammal. This allows them to glide for very large distances, the longest recorded being 136 m (Lim 2007). Colugos are rarely kept in captivity and are elusive in the wild (Lim 2007), factors that have obscured their evolutionary history for decades. Under current taxonomy, colugos comprise a unique mammalian order (Dermoptera) and are classified as two species: the Sunda colugo (Galeopterus variegatus) and the Philippine colugo (Cynocephalus volans) (Wilson and Reeder 2005). However, recent mtDNA and nuclear DNA data provide compelling evidence that the geographically widespread Sunda colugo in fact represents multiple species distributed throughout Southeast Asia (Janečka et al. 2008) and suggest that further genetic sampling may identify many additional divergent populations and/or species. Because of the extreme difficulty obtaining fresh tissue or DNA samples from colugos, we further explored this question using collections of museum specimens and devised a comprehensive method for capture, selection, and recovery of divergent mtDNA fragments using NGST.
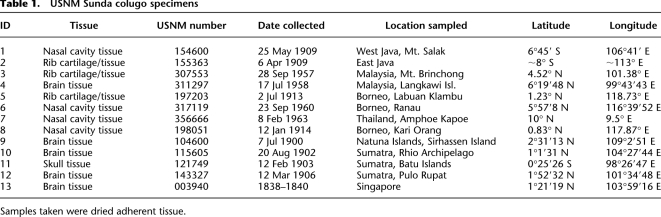
Table 1.
USNM Sunda colugo specimens
Results
Amplified adapter-ligated museum DNA
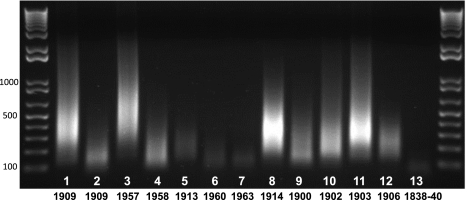
Extraction of DNA from tissues of 13 colugo museum specimens (Table 1) yielded varying amounts and qualities of DNA. DNA was recovered from every specimen, including the oldest, which was collected ∼170 yr ago (Supplemental Table 1). Due to variation in the initial quantity between samples, we measured the degree of degradation from the PCR-amplified adapter-ligated DNA (Fig. 1). The age of museum specimens showed little correlation with quality of DNA recovered (Fig. 1). Quality of DNA was measured based on the size and intensity of the amplified, adapter-ligated-DNA smear on an agarose gel. Several specimens that were nearly 100 yr old had higher-molecular-weight DNA when compared to specimens collected more recently. For example, specimen USNM 121749 (#11), collected in 1903, showed comparably high-quality DNA, while specimen USNM 356666 (#7), collected in 1963, yielded among the poorest DNA qualities. Furthermore, DNA quality did not appear to be influenced by the source tissue and was highly variable within and between tissue types, even when considering collection age. Thus, the quality of DNA recovered from each specimen seems to be largely influenced by how the specimen was treated and stored during and after collection rather than simply its age or tissue source.
Figure 1.
PCR-amplified, adapter-ligated museum DNA extracts (5 μL) for 13 colugo specimens, resolved on a 1% agarose gel, with year of collection indicated below. Note that DNA quality does not always correlate with the age of each specimen.
Hybrid DNA capture
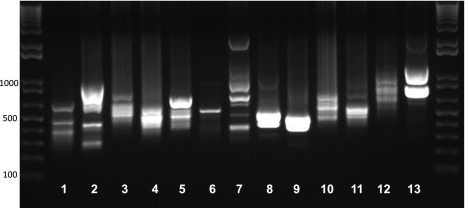
The distribution and banding patterns of selected mtDNA fragments after two rounds of hybrid capture and amplification (2°-selected, amplified museum products) are shown in Figure 2. There is a strong correlation between size and uniform distribution of the capture DNA smear with the percent genome coverage eventually obtained by Illumina DNA sequencing (Fig. 2; Table 2). Specimens yielding uniform smears yielded the highest overall genome coverage percentages because they have a more even distribution and concentration of products. Specimens that yielded more banded 2°-selected amplified products yielded more biased mtDNA genome sequence coverage, with a larger number of gaps.
Figure 2.
2° selected and amplified mtDNA from the 13 museum specimens (5 μL) resolved on a 1% agarose gel.
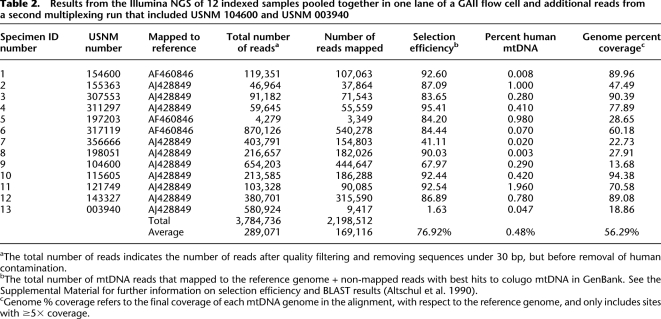
Table 2.
Results from the Illumina NGS of 12 indexed samples pooled together in one lane of a GAII flow cell and additional reads from a second multiplexing run that included USNM 104600 and USNM 003940
Illumina sequencing
To make a preliminary evaluation of the efficiency of our selection procedure, we incorporated Illumina index sequences into the 2°-selected-amplified museum products and created sequence libraries for two specimens: USNM 143327 (#12) and USNM 317119 (#6). These libraries were cloned, and 96 colonies were sequenced per library using standard Sanger capillary techniques and aligned to a published colugo reference mtDNA genome (Supplemental Fig. 2). This pilot study showed that between 70% and 90% of fragments recovered were of colugo mitochondrial origin, confirming the high selection efficiency of the capture procedure.
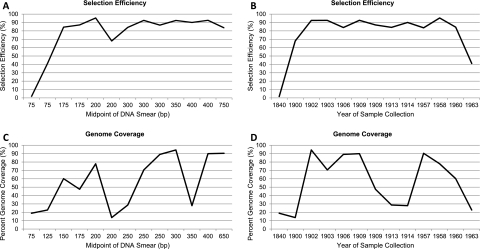
In light of these results, we indexed the remaining selection libraries and pooled 12 of the 13 specimens in a single lane of an Illumina GAII flowcell. A single-end read, 84-cycle run returned an average of 24.4 Mbp of sequence per individual (Table 2). After quality filtering and removing sequences under 30 bp, on average 76.92% of captured sequences showed strong similarity to the reference colugo mtDNA molecules (referred to hereafter as “selection efficiency”), 0.48% were of human mtDNA origin, and the remaining 22.60% represent other exogenous DNA (i.e., bacterial), nuclear DNA, or colugo sequence that was too divergent to map to the reference genome (Supplemental Table 2). When the oldest specimen, #13, is excluded, the average selection efficiency increases to 82.96%. Overall selection efficiency appeared to be influenced by starting DNA quality; however, this was only pronounced in highly degraded samples, <150–200 bp (Fig. 3A,B), as well as samples more than 110 yr old. In contrast, age of samples was not a good predictor of genome coverage (Fig. 3D).
Figure 3.
Plots of selection efficiency and mtDNA genome coverage relative to the original sample DNA quality (average size in base pairs of DNA fragments on 1% agarose gel) (see Fig. 1) and age of sample. (A) The effect of DNA quality on selection efficiency. (B) The effect of specimen age on selection efficiency. (C) The effect of DNA quality on mitochondrial genome coverage. (D) The effect of specimen age on mitochondrial genome coverage.
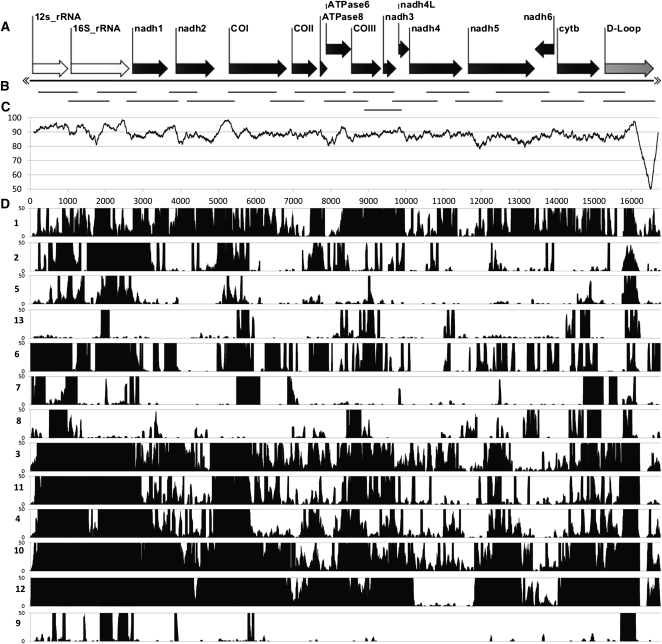
Hybrid capture yielded an average depth/site of ∼979× coverage (Supplemental Table 3); this extreme depth allowed for accurate calling of 99.99% of bases used in our analysis. Captured sequence fragments were not evenly distributed across the genome in every individual: Some mitochondrial genomes were nearly complete with >90% genome coverage, while other genomes had as little as 20% genome coverage (Fig. 4; Table 2). Across samples, conserved gene regions, such as the 12S and 16S rRNA genes and the conserved portion of the control region, possess higher depth of coverage than other areas of the mtDNA genome. On an individual basis, there was no obvious correlation between gaps in genome coverage and regions of low overall conservation across the genome, as assessed by the divergence plot between the Bornean and Javan reference mtDNA genomes (Fig. 4C). This would suggest that probe bias was minimal.
Figure 4.
Distribution and depth of coverage of captured mtDNA fragments from each Sunda colugo museum specimen, displayed relative to the reference colugo mtDNA genome. (A) Reference colugo genome, AJ428849, depicted horizontally with gene annotations displayed except tRNA genes. (B) Distribution of mtDNA probe fragments produced by PCR. (C) Plot of DNA sequence identity between two full-length mitochondrial reference genomes (AJ428849, Borneo; AF460846, West Java) calculated in overlapping, 200-bp sliding windows. (D) Histogram showing distribution of captured mtDNA sequence fragments for the 13 museum specimens (labeled on left side by ID number in Table 1), relative to the reference genome. Only coverage from 0× to 50× is shown for clarity, although most individuals have substantially higher coverage across the genome (Supplemental Fig. 6).
Assessment of DNA damage and numts
Both ancient and historical samples are known to contain nucleotides that are damaged, the most common form being due to deamination of cytosine to uracil, which will lead to an excess of C-to-T and G-to-A substitutions when compared to a modern reference genome (Millar et al. 2008; Briggs et al. 2009; Krause et al. 2010). Our assessment of complementary C-to-T and G-to-A transitions versus T-to-C and A-to-G transitions from each individual sequence compared to a reference sequence revealed no significant bias (α = 0.05) in favor of transitions typical of chemical damage (Supplemental Table 4B). This indicates that chemical damage has had little influence on our consensus sequences.
Numts (mtDNA that has been transposed into the nuclear genome) (Triant and DeWoody 2007; Hazkani-Covo et al. 2010) can potentially be isolated by hybrid capture when they are similar to the probe sequence itself. The results of our initial Sanger sequencing identified three putative numts out of 183 high-quality Sanger reads, based on the presence of stop codons, immediately flanking nuclear sequence, or repetitive elements in the same sequence read. Although this suggests that numts likely represent a very small fraction of captured sequences, we evaluated the 13 putative protein-coding regions of the colugo mitochondrial genome in all final consensus sequences for stop codons or indels that might indicate capture of predominantly numt rather than cymt (cytoplasmic, or “true” mtDNA) sequences. We identified four nonsense mutations in four different individuals and removed these sequence fragments as putative numts (Supplemental Table 5). Not all numts are characterized by stop codons or indels. Specifically, the recent transposition of numts into the nuclear genome could avoid detection by lacking indels or nonsense mutations (Hazkani-Covo et al. 2010). Therefore, we examined SNP frequencies and distributions within the read profiles of each individual, reasoning that high-frequency SNPs that are clustered in specific regions of the mtDNA genome may be evidence of capture of recent numts (Supplemental Fig. 5). Alternatively, SNPs that are shared in similar regions across individuals may indicate capture of ancestral numts. Although we did find evidence for high-frequency SNPs across most individuals, the average frequency of these sites/individual was 0.42% (Supplemental Fig. 5). We did not observe any significant stretches of SNPs of similar frequency that were clearly identified as numts. Furthermore, because of the multiple rounds of PCR amplification during the selection procedure, it is difficult to exclude any SNP as a PCR-induced error incorporated during early rounds of the amplification process.
Phylogeny
The genetic divergences between the published reference genomes (AJ428849 and AF460846) and the final mtDNA consensus sequences of the study specimens (and GVA5, which served as our probe) were substantial, averaging ∼9.0% for both AJ428849 comparisons (range: 5.0%–13.7%) and AF460846 comparisons (range: 0.3%–13.2%) (Supplemental Table 6). Notably, populations from Borneo, the Natuna Islands, and East Java were more divergent from the Bornean AJ428849 reference mtDNA genome than either the West Javan or Peninsular Malaysian populations (GVA1–6), which were considered divergent enough to warrant species-level distinction (Janečka et al. 2008). Average within-island genetic distances were also very large: 8.1% between Bornean populations, 3.7% between Javan populations, 1.4% between Northeastern Sumatran islands, and 6.9% between both Peninsular Malaysia and Thailand populations (Supplemental Table 6).
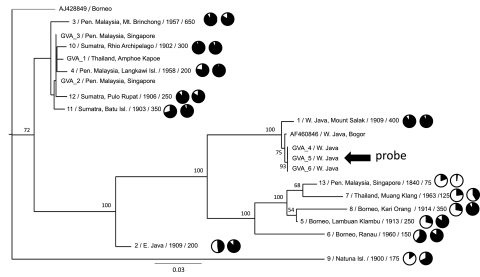
We constructed maximum likelihood (ML) trees for the complete mtDNA alignment of the 13 museum mtDNA sequences, two reference colugo mtDNA genomes and six published partial mitochondrial rRNA sequences from individuals from Peninsular Malaysia (GVA 1–3) and West Java (GVA 4–6) (Janečka et al. 2008). Enforcing different read depth thresholds for inclusion of a site in the alignment (e.g., read depth 5×–25×) had little effect on phylogenetic stability; all trees showed consistent clustering of the same major geographic lineages (Supplemental Fig. S3). In light of these data, we performed subsequent analyses on our minimum 5× depth data set. The ML tree based on all sites (minus ambiguous regions in the alignment of hypervariable regions of the rRNA genes and control region) is shown in Figure 5. To minimize any effect of missing data on phylogenetic accuracy, we performed an analysis of different alignments where we varied the threshold for the number of individuals in the alignment that had sequence present at a given site (Supplemental Fig. 4). Altering this parameter from 30% to 90% had little effect on well-supported nodes in Figure 5. Specimens from Peninsular Malaysia, Thailand, and several NE Sumatran islands formed a well-supported clade. This clade also consistently grouped with the Bornean reference genome (AJ428849) when the Natuna Island sequence (#9) was excluded from analyses. The remaining Bornean populations formed a divergent clade that also includes a separate Thailand/Peninsular Malaysia group. A third divergent clade includes the West Java populations. Two other populations, East Java and the Natuna Islands, were not consistently positioned within the phylogeny of colugos, possibly due to larger amounts of missing data and/or long branch attraction, and tended to lower bootstrap support for major geographic clusters (Supplemental Fig. 4). Indeed, analyses that removed the Natuna specimen (#9) showed increased bootstrap support for each of the major clusters shown in Supplemental Figure 4. An analysis that only included sites present in the Natuna specimen (∼2250 aligned base pairs) and only individuals where >50% of these sites were present revealed the Natuna colugo (#9) to be a deeply divergent lineage with no close affinity to remaining colugo populations (Supplemental Fig. 4E). In summary, our results identify divergent phylogenetic groups of individuals from geographically distinct populations, confirming previous observations (Janečka et al. 2008).
Figure 5.
Maximum likelihood phylogenetic tree of Sunda colugo mtDNA sequences and the effect of phylogenetic divergence on capture efficiency and genome coverage. Specimens are labeled with geographic location of specimen collection and their USNM catalog number, Texas A&M sample number (prefix “GVA”), or GenBank accession number. Additional sample information following the taxon ID includes (left to right): average starting DNA fragment size distribution (in base pairs), a pie chart showing percent genome coverage, and a pie chart showing selection efficiency. Bootstrap values displayed at each node are based on 1000 replicates. The tree shown is based on those sites where 50% of the individuals possess a base (14,008 bp, excluding hypervariable regions) (see Methods) at a minimum depth of 5×. The overall relationships were supported in other analyses that minimized missing data and maximized data overlap across individuals (Supplemental Fig. 4). The tree is displayed with midpoint rooting. The specimen (GVA4) from whom the mtDNA probe was derived is indicated with an arrow.
Sequence divergence and capture efficiency
Specimen #1 shows ∼99% sequence identity to the full-length mtDNA genome of specimen GVA4, a mtDNA sequence obtained from preliminary assembly of the Galeopterus genome, as well as specimen GVA5, from which our biotin-labeled mtDNA probe was amplified. All three individuals represent populations within close proximity in West Java. Therefore specimen #1 serves as a good reference for estimating the maximum selection efficiency and genome coverage that might be obtained with our probe and hybridization procedure (assuming that the DNA quality of this specimen is typical for what one might obtain from other museum specimens) (Fig. 1). Selection efficiency for individual #1 was ∼93%, and genome coverage ∼90%. Although these values are among the highest obtained for all of the museum specimens we analyzed, we obtained comparable selection efficiencies and genome coverage for individuals that were phylogenetically divergent from the probe sequence (Fig. 5; Table 2). Specifically, hybrid capture from Sumatran and Peninsular Malaysian specimens (#4, #10, and #11) achieved 92%–95% selection efficiency, and similar levels of genome coverage (∼90%) relative to specimen #1. Therefore, our method can efficiently capture mtDNA sequences with as great as 10%–13% sequence divergence from the probe sequence (Supplemental Table 7), with minimal levels of mtDNA genome coverage bias due to probe divergence.
Discussion
Here we were able to demonstrate efficient cross-species capture of orthologous mitochondrial DNA sequence fragments, and in many cases nearly complete mtDNA genomes, from the degraded DNA of museum specimens collected >100 yr ago. Below we discuss various experimental considerations for further improvement of capture hybridization across heterogeneous target loci and divergent species with unknown phylogenetic affinity. We then examine the contribution of these data to addressing the phylogeny, taxonomy, and biogeography of a poorly known group of mammals, Sundaic colugos.
Cross-species probe design
Probe construction is inherently important for successful recovery of target DNA, particularly when attempting cross-species hybridization with probes containing sequences with different levels of evolutionary conservation. While high sequence homology between the probe and target DNA allows for efficient and unbiased recovery of target fragments among closely related populations of the same species (Mamanova et al. 2010), the greatest potential for capture hybridization lies with extinct species of uncertain phylogenetic affinity, or when there are no modern specimens available from an isolated, and possibly divergent, geographic population/species. However, as sequence divergence increases between the probe and the target individual, the potential to hybridize to exogenous DNA (e.g., numts, human contaminants) also increases. However, our hybridization conditions seem to be adequately relaxed to retrieve divergent (10%–13%) mtDNA fragments, while maintaining high selection efficiency.
In the present study, we generated a probe from a single individual, although a pooled probe from multiple species or individuals from various locales might be more effective. This latter approach would lower annealing specificity but increase the probe's annealing potential (equivalent to a degenerate probe), which would be advantageous for probing taxa of uncertain phylogenetic affinity. By gradually relaxing the hybridization conditions, the touchdown approach (analogous to touchdown PCR) used here provides more stringent and accurate hybridization conditions for conserved orthologous fragments of DNA to anneal prior to subjecting the probe to less-specific hybridization conditions. Occupation of probe fragments under stringent hybridization conditions removes or reduces the possibility for later mis-pairing during less accurate hybridization to paralogous DNA sequences, such as numts or human contamination. The touchdown hybridization approach appears to be extremely efficient as our levels of human and numt capture were considerably reduced relative to colugo mtDNA, while allowing for capture (albeit unevenly) of sequences across the majority of the mtDNA genome from most specimens.
The objective of capture hybridization experiments is to obtain equal concentrations of every target base pair for equal sequencing depth coverage; hence, another facet of cross-species probe production is controlling for varying levels of genome evolution across target regions. Although the high sequence coverage made it possible to recover orthologous fragments across the mtDNA genome of each specimen, as expected there was clearly a significant bias of sequence depth toward more conserved regions of the mtDNA genome (e.g., the 12S and 16S rRNA genes). This bias is particularly notable for the Natuna Islands specimen, which appears to be the most divergent Sunda colugo we sampled, and explains the more limited recovery of mtDNA fragments in more rapidly evolving parts of this specimen's mtDNA genome. To address high coverage bias, future attempts might use multiple hybridization experiments, one probing for more conserved regions and the other for more divergent sections (based on pairwise sequence divergence), followed by equimolar pooling during NGS library production. In principle, this strategy would obviate the need for touchdown hybridization, enabling different probe sets to hybridize longer at either stringent or relaxed conditions, thus providing greater opportunity to hybridize under optimal conditions. Alternatively, one could adjust the proportions of different probes/oligos in the pool, with more conserved regions represented in lower concentrations and less conserved regions in higher molar concentrations, in proportion to genetic divergence observed across related groups of taxa. This would allow more accurate hybridization conditions for corresponding levels of divergence and more standardized representation of each base pair in the probe. Clearly, many variables need to be considered before attempting hybridization, depending on the extent of divergence anticipated between the probe and target individuals, allowing flexibility and customization in the hybridization capture experiment for successful recovery of a majority of the target DNA fragments.
Exogenous DNA
The initial quality of DNA derived from each museum specimen was evaluated based on the smear of DNA produced following amplification with adapter-ligated primers and provided an estimate of the extent of degradation and the potential for efficient hybridization. This initial DNA distribution could largely represent bacterial, or even human, contamination depending on how the specimen was handled and stored. Indeed, published reports of next-generation sequencing data from total DNA extracts of ancient mammalian hair or bone specimens indicate that a substantial proportion of reads may correspond to exogenous DNA (Miller et al. 2008; Briggs et al. 2009). However, capture hybridization has the benefit of enriching for only target DNA, hence requiring fewer sequencing reads to obtain sufficient depth of coverage for accurate base-calling. Overall, our data showed very low levels of human as well as exogenous (bacterial) contamination following selection, although this did vary across specimens. Nonetheless, the levels of target colugo DNA sequenced were an order of magnitude greater than human contaminating DNA, and we achieved more than sufficient depth of coverage (>1000-fold in most cases) such that additional pooling of samples/flowcell lane is feasible. In this study, only two museum specimens (#7 and #13) yielded <50% captured colugo mtDNA, while the remainder show a majority of reads to be of colugo origin. Even in those cases in which colugo DNA did not represent the majority of captured reads, we could easily distinguish true colugo mitochondrial DNA through comparison of de novo contig assemblies with BLAST (Altschul et al. 1990) and alignment to the reference mtDNA genome sequences.
Colugo phylogenetics and Southeast Asian biogeography
Under current taxonomy, the Sunda colugo individuals sampled here from geographically widespread populations are classified as one species, Galeopterus variegatus. A previous genetic study that compared mtDNA and nuclear DNA fragments from specimens from Peninsular Malaysia, Borneo, and West Java revealed a high degree of genetic divergence across colugo populations that exceeded levels observed for other pairs of well-established mammalian sister species (Janečka et al. 2008). Our expanded analysis of genetic divergence between new and published mitochondrial sequences further indicates very large genetic divergence between specimens from geographically widespread localities such as Borneo, Peninsular Malaysia, Thailand, East and West Java, and the Natuna Islands. Although molecular divergence dates were not estimated for the current data set (due to the absence of internal calibrations and lack of a full-length mtDNA genome from the closest outgroup, Cynocephalus volans), a previous study revealed potential species-level distinctions between populations from West Java (GVA4–6), Borneo, and the Malay Peninsula (GVA1–3), with estimated divergence times between these populations as great as 5 million years (Janečka et al. 2008). Our results indicate similar or greater amounts of genetic divergence within and between island populations (e.g., Borneo, Java, Natuna Islands), compared to values observed between the mainland and West Javan populations (see Fig. 5; Supplemental Table S6; Janečka et al. 2008).
Fluctuating sea levels during the Pliocene and Pleistocene, along with other dynamic environmental changes, produced many isolating mechanisms for speciation events to occur throughout the Southeast Asian archipelago (Harrison et al. 2006). Among these changes were the creation of many major river systems as well as expansions of dry savannah habitat. Colugos are obligately arboreal mammals and cannot survive in a savannah-like ecosystem, nor can they traverse large rivers safely (Lim 2007). These geographic barriers likely generated at least four important refugia for arboreal mammals within the present-day landscape on the Southeast Asian continent (the “Sunda Shelf”): (1) central and northern Borneo; (2) Malay Peninsula including Sumatra; (3) Mentawai Islands; and (4) Western Java (Harrison et al. 2006). A belt of dry woodland and savannah probably extended from southern and eastern Borneo south to Eastern Java, effectively isolating Western Java, which may account for the large observed genetic divergence between colugos from both West and East Java (Fig. 5). Borneo was similarly subdivided into two distinct ecological regions: the tropical refugia of the north and west, and the contrasting dry savannah of the southeast. Borneo is a very mountainous landmass, with ranges running from the center to the northeastern tip of the island, and contains many major river systems dissecting the southern portion of the island in particular. The abundance of geographic isolating mechanisms would have provided many opportunities for population subdivision and speciation and supports the unexpected phylogenetic distribution and deep divergence of the Bornean specimens examined here. Further sampling of individuals from throughout Borneo, as well as the entire Sunda Shelf, will allow for more precise delimitation of taxonomic boundaries and allow for better elucidation of biogeographic scenarios and the role different isolating mechanisms have played in the divergence and radiation of colugos.
The elusiveness of colugos, their absence in zoos, and their very broad geographical distribution make them an extremely difficult group of species to obtain detailed population-wide sampling without using museum material. This is also the case for numerous poorly known, threatened, and endangered species throughout the world, particularly those in the tropical rainforests of Southeast Asia. Our results illustrate the extreme power of harnessing untapped genetic data within archived museum specimens of unknown genetic divergence by cross-species capture hybridization, coupled with NGS, to make genetic inferences that otherwise would be difficult or logistically improbable. It is likely that further genetic sampling of colugo specimens from throughout the Southeast Asian mainland, within southeastern Sumatra, southern and western Borneo, Eastern Java, Natuna Islands, and the recently described population from Laos (Ruggeri and Etterson 1998) may provide additional evidence for deeply divergent colugo lineages that may warrant species level distinction. Broad application of this approach to other taxa will further enhance our ability to accurately estimate the true number of species on Earth, a necessary step toward preserving living biodiversity.
Methods
Museum DNA extraction
Small pieces (∼5 mg) of dried adherent soft tissue were sampled from crania, nasal cavities, or cartilage of museum specimens deposited in the Division of Mammals in the Smithsonian Institution's National Museum of Natural History (abbreviation USNM). Museum specimens were digested overnight in 200 μL of Lysis Buffer (QIAGEN) with 100 μg of proteinase K, followed by protein precipitation and isopropanol precipitation of genomic DNA following the general guidelines of the Gentra/Puregene DNA isolation protocol (QIAGEN). The final elution volume for the DNA was 40 μL. DNA extractions and all pre-selection procedures were performed in a dedicated pre-PCR laboratory space for historic specimens, removed from the PCR/molecular biology laboratory.
Blunt ending of museum-DNA extracts
DNA extracts were blunt-ended for adapter ligation using a 1 × 20 μL master mix of 9.5 μL of H2O, 8 μL of 5× reaction buffer for T4 DNA polymerase, 2.5 U of T4 DNA polymerase (Fermentas) in the presence of 0.4 mM dNTPs. Twenty microliters of the master mix was added to 20 μL of museum DNA extract and incubated for 10 min at 70°C. Following incubation, extracts were purified with CentriSpin 20 columns (Princeton Separations) returning ∼32 μL of product from a 40-μL elution, stored on ice, and immediately followed by ligation of adapters.
Adapter ligation and amplification of museum DNA
Ligation of double-stranded adapters (generated by self-ligation of two oligos ORM-28 and ORM-29) (Peterson 1998) to 32 μL of blunt-ended museum DNA extracts was performed in a 150-μL ligation volume, using 2.5 U of T4 DNA ligase (Fermentas) and 1× T4 DNA ligase buffer + ATP (Fermentas), incubating 19 h at 16°C. PCR amplification of the resulting adapter-ligated museum fragment libraries was performed using the ORM-28 primer (0.5 μM) in 5 × 50 μL reactions with 2.5 U of platinum Taq DNA polymerase (Invitrogen), 1× PCR buffer, 1.5 mM MgCl2, and 0.8 mM dNTPs. The PCR profile included a 1-min hot start at 95°C, followed by 20–30 cycles (depending on the starting concentration of DNA) of denaturing for 15 sec at 94°C, annealing for 20 sec at 58°C, and extension for 1 min at 72°C, followed by a final 5-min extension at 72°C. PCR reactions from each individual were pooled, purified in a MicroconPCR device (Millipore), and resolved on 1% agarose gels to examine fragment size distribution.
Capture probe generation
A high-quality West Java colugo tissue specimen (GVA5) served as the source of DNA for generating our mtDNA probe. To increase the probability that the mtDNA amplicons were of mitochondrial origin, rather than a nuclear mtDNA pseudogene (numt), we used a mitochondrial enrichment procedure to generate template DNA (Jones et al. 1988), modified to allow for small-scale extractions. 1.5 mg of liver was homogenized in 1 mL of prechilled homogenization buffer (30 mM Tris-HCl, 1 mM EDTA, 2.5 mM CaCl2, 0.25 M sucrose) in a 1.5-mL tube using a pestle. The homogenate was spun at 3500 rpm for 15 min in a 4°C microcentrifuge to pellet the nuclear debris. Following transfer of the supernatant to another tube, the nuclear pellet was resuspended in a second 600-μL aliquot of cold homogenization buffer and spun at 4000 rpm for 10 min. The supernatants were combined and spun at 13,400 rpm for 30 min at 4°C to pellet the mitochondria, and the nuclear pellet was preserved at −80°C. The mitochondrial pellet was resuspended in 200 μL of Cell Lysis Buffer (QIAGEN), digested overnight at 56°C with 100 μg of Proteinase K, and cooled for 7 min on ice. Sixty-seven microliters of protein precipitation solution (QIAGEN) was added, vortexed for 20 sec, and spun for 5 min at 8000 rpm. The supernatant was transferred into a fresh tube to which 200 μL of 100% isopropanol was added, inverted 50 times, and spun for 15 min at 12,000 rpm. The DNA pellet was washed with 200 μL of 70% ethanol, spun for 2 min at 12,000 rpm, and allowed to air-dry for 5–10 min. The DNA was eluted in 20 μL of Elution Buffer (QIAGEN). The enriched mtDNA extract served as a template for amplifying 19 ∼1–1.4 kb, overlapping fragments (in triplicate) that span the colugo mtDNA genome (Fig. 4; Supplemental Table 8). The PCR profile was as follows: 2 min hot start at 94°C, followed by 40 cycles of denaturing for 15 sec at 94°C, annealing for 30 sec at 60°C–50°C, and extension for 1 min at 72°C, followed by a final 2-min extension at 72°C. During the first 10 cycles the annealing temperature was decreased from 60°C to 50°C by 2°C increments every two cycles. A 2-min final extension at 72°C completed the reaction. PCR products for each amplicon were pooled and purified individually using MicronPCR devices (Millipore) and quantified. Fragments were then pooled based on concentration and fragment length to obtain equal representation of each base pair in the mitochondrial genome. The pooled mtDNA probe was labeled by biotin-nick translation (Roche) or biotin-High Prime (Roche) random priming procedures, following manufacturer protocols.
Capture hybridization and selection
Capture hybridization follows a modified version of Del Mastro and Lovett's (1997) protocol, originally described for cDNA selection with a genomic probe, with minor changes. Approximately 500 ng to 1 μg of amplified, adapter-ligated DNA was combined with 100 ng of biotin-labeled mtDNA probe and added to an equal volume of 2× Hybridization Buffer (1.5 mM NaCl, 40 mM Sodium Phosphate Buffer [0.183 M NaH2PO4, 0.778 M Na2HPO4], 10 mM EDTA, 10× Denhardt's solution, and 0.2% SDS), not exceeding 15 μL. The sample was overlaid with 50 μL of mineral oil, denatured for 5 min at 99°C, and incubated for 50 h at 65°C–60°C, reducing ∼2°C every 24 h. This “touchdown” approach was used to enhance retrieval of more divergent DNA sequences relative to the capture probe. Following hybridization, samples were added to 1 mg of Dynabeads M-280 Streptavidin beads (Dynal), which had been washed three times in 100 μL of TEN buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 M NaCl) using a magnetic tube holder, and resuspended in a final volume of 100 μL of TEN. Two room-temperature, low-stringency (1× SSC, 0.1% SDS) and three 65°C, high-stringency (0.1× SSC, 0.1% SDS) washes were performed following DelMastro and Lovett (1997). The beads were eluted in 25 μL of 0.1 N NaOH for 20 min at room temperature, with gentle vortexing every 5 min, neutralized with 25 μL of Tris-HCl (pH 7.5), and the total 50 μL was passed through a CentriSpin 20 column (Princeton Separations). The primary selected DNA was amplified in four replicate 50-μL PCR reactions using 10 μL of DNA, 0.5 μM ORM-28 primer, 2.5 U of Invitrogen platinum Taq in 1× PCR buffer, 1.5 mM MgCl2, and 0.08 mM dNTPs, and the following PCR profile: 1 min hot start at 95°C, 35 cycles of 15 sec at 94°C, 20 sec at 58°C, 1 min at 72°C, followed by a 5-min final extension at 72°C. PCR products were pooled and purified with Montage-PCR filters (Millipore) and resolved on 1% agarose gels to examine the size distribution of selected DNA fragments. A second round of DNA capture and selection was repeated using 1 μg of 1°-selected, amplified DNA as template, and 100 ng of the biotin-labeled capture probe, using the same procedure described for the primary selection and amplification. Final amplification used 5 μL of template DNA per reaction and 30 rounds of PCR amplification.
Initial evaluation of mtDNA selected libraries using Sanger sequencing
The indexed libraries for specimens #6 and #12 were cloned into the PCR-TOPO Blunt end vector (Invitrogen) and grown on LB + ampicillin plates. Ninety-six colonies from each library were picked into 15 μL of sterile water in a 96-well PCR plate. Two microliters of this template was used in a subsequent PCR reaction using vector-borne universal primers (M13 or T3/T7). PCR products were evaluated on 1% agarose gels and sequenced (ABI Big Dye3.1) on an ABI-3730 capillary DNA sequencer (Agencourt Biosciences). The resulting DNA sequences were edited for quality and vector + adapter-trimmed in Sequencher (Genecodes, Inc.). The DNA sequences were then assembled relative to a colugo reference mitochondrial DNA genome (AJ428849).
Next-generation sequencing and sequence assembly
MtDNA selection products in the 200–600-bp range were gel-excised, and higher-intensity bands outside the main fragment smear were excised, cloned, and sequenced separately with Sanger-based sequencing to reduce bias in sequencing coverage across fragments in the library. Following standard Illumina specifications, the main fragment smear was subsequently purified and indexed by PCR with Illumina paired-end primers where 12 unique index sequences were incorporated into the paired-end adapters to distinguish sequences from different individuals. These 12 individuals were multiplexed in one lane of an Illumina GAII flowcell and later sorted computationally (Table 2). Sequences for USNM 003940 (#13), and additional reads for USNM 104600 (#9) to increase representation from low-coverage regions, were generated in a second lane of a separate run. From the first reaction, 3.5 million 84-bp reads were generated, while the addition of the colugo reads in the second run increased the total to 4.3 million reads (Table 2).
Sequences were trimmed of ORM-28 adapter sequences by removing the first 22 bp of each sequence. Sequences were trimmed by quality both in CLC Genomics Workbench with default parameters, and in EULER-SR (Chaisson and Pevzner 2008). We further queried and masked all ORM-28, ORM-29, and Illumina adapter sequences within the sequence reads. Artifacts from the various ligation procedures were identified, and sequences containing them were removed. Sequences were then imported into CLC, where all sequences <30 bp were removed.
To identify and remove human sequence contamination, the high-quality reads were mapped to a human reference sequence (HM125971) where 100% of each read must match at 98% similarity to the reference sequence. The remaining unmapped reads were then mapped under global alignment parameters to two colugo reference sequences (AJ428849 and AF460846) where 100% of each read must match at 85% similarity to the respective reference sequence. Consensus sequences and depth information were derived from these alignments in CLC. Sequence depth parameters were enforced using a custom Perl script.
Open reading frame analysis
Open reading frames (ORFs) of each individual were analyzed by translating coding domain sequence (CDS) regions and checking for premature stop codons, as well as comparing translated regions to all previously published CDS regions of a colugo mtDNA genome (AJ428849). Analyses were performed in CLC using the vertebrate mitochondrial genetic code.
Phylogenetic analysis
Sequence alignments were performed in Sequencher (vers. 4.8, GeneCodes, Inc.) and adjusted by eye. Hypervariable regions of ambiguous alignment were excluded from further analysis. Maximum likelihood trees were generated with RAxML (version 7.0.3) (Stamatakis 2006), under a GTR + gamma model of sequence evolution. Bootstrap support metrics are based on 1000 replicates. Pairwise genetic distances and other sequence statistics were generated in MEGA 5.0 (Tamura et al. 2007), using maximum composite likelihood distances (gamma corrected). To investigate the effect of depth on phylogenetic robustness (i.e., potential errors in low-coverage regions might influence phylogenetic accuracy), we constructed ML trees from different alignments where inclusion of a site in the alignment required a specific read depth: 5×, 10×, 15×, 25× (Supplemental Fig. 3). ML trees were also constructed from alignments where 30%, 50%, or 70% of individuals share a base at that base site (Supplemental Fig. 4).
Data access
Raw sequence data have been submitted to the NCBI Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under Study Accession number SRP007459, and Sample Accession numbers SRS214574 and SRS214579-SRS214590. The sequence alignment has been deposited in TreeBase: http://purl.org/phylo/treebase/phylows/study/TB2:S11695.
Acknowledgments
This work was supported in part by the National Science Foundation (EF0629849 to W.J.M.) and Texas A&M University. We thank Linda Gordon for assistance in sampling the colugo museum specimens. We thank the Genome Center at Washington University for providing sequence data and allowing use of the full mtDNA genome for GVA4.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.120196.111.
References
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ 1990. Basic local alignment tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Briggs A, Good J, Green R, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajkovic D, Kucan Z 2009. Targeted retrieval and analysis of five Neandertal genomes. Science 325: 318–321 [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA 2008. Short read fragment assembly of bacterial genomes. Genome Res 325: 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Mastro R, Lovett M 1997. Isolation of coding sequences from genomic regions under direct selection. Methods Mol Biol 68: 183–199 [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Tomsho LP, Rendulic S, Packard M, Drautz DI, Sher A, Tikhonov A, Dalén L, Kuznetsova T, Kosintsev P, et al. 2007. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science 317: 1927–1930 [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Drautz DI, Lesk AM, Ho SY, Qi J, Ratan A, Hsu CH, Sher A, Dalén L, Götherström A, et al. 2008. Intraspecific phylogenetic analysis of Siberian woolly mammoths using complete mitochondrial genomes. Proc Natl Acad Sci 105: 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison T, Krigbaum J, Manser J 2006. Primate biogeography and ecology on the Sunda Shelf Islands: A paleontological and zooarchaeological perspective. In Primate biogeography (ed. Lehman SM, Fleagle JG), pp. 331–372 Springer Science & Business Media, New York [Google Scholar]
- Hawkins D, Hon G, Ren B 2010. Next-generation genomics: an integrative approach. Nat Rev Genet 11: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W 2010. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 6: e1000834 doi: 10.1371/journal.pgen.1000834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janečka JE, Helgen KM, Lim NT-L, Baba M, Izawa M, Boeadi, Murphy WJ 2008. Evidence for multiple species of Sunda colugos. Curr Biol 18: R1001–R1002 [DOI] [PubMed] [Google Scholar]
- Jones CS, Tegelström H, Latchman DS, Berry RJ 1988. An improved rapid method for mitochondrial DNA isolation suitable for use in the study of closely related populations. Biochem Genet 26: 83–88 [DOI] [PubMed] [Google Scholar]
- Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP, Pääbo S 2010. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464: 894–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim NT-L 2007. Colugo: The flying lemur of South-east Asia. Draco Publishing and Distribution, Singapore [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ 2010. Target-enrichment strategies for next-generation sequencing. Nat Methods 7: 111–118 [DOI] [PubMed] [Google Scholar]
- Millar CD, Huynen L, Subramanian S, Mohandesan E, Lambert DM 2008. New developments in ancient genomics. Trends Ecol Evol 23: 386–393 [DOI] [PubMed] [Google Scholar]
- Miller W, Drautz DI, Ratan A, Pusey B, Qi J, Lesk AM, Tomsho LP, Packard MD, Zhao F, Sher A, et al. 2008. Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456: 387–390 [DOI] [PubMed] [Google Scholar]
- Miller W, Drautz DI, Janečka JE, Lesk AM, Ratan A, Tomsho LP, Packard M, Zhang Y, McClellan L, Qi J, et al. 2009. The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome Res 19: 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AS 1998. Direct cDNA selection. In Genome analysis: A laboratory manual (ed. Birren B et al. ), pp. 159–171 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Ruggeri N, Etterson M 1998. The first records of colugo (Cynocephalus variegatus) from the Lao PDR. Mammalia 62: 450–451 [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Summerer D 2009. Enabling technologies of genomic-scale sequence enrichment for targeted high-throughput sequencing. Genomics 94: 363–368 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Triant DA, DeWoody JA 2007. The occurrence, detection, and avoidance of mitochondrial DNA translocations in mammalian systematics and phylogeography. J Mammal 88: 908–920 [Google Scholar]
- Wilson DE, Reeder DM 2005. Mammal species of the world: A taxonomic and geographic reference 3rd edJohns Hopkins University Press, Baltimore, MD [Google Scholar]