Abstract
Cytosine methylation of DNA CpG dinucleotides in gene promoters is an epigenetic modification that regulates gene transcription. While many methods exist to interrogate methylation states, few current methods offer large-scale, targeted, single CpG resolution. We report an approach combining bisulfite treatment followed by microdroplet PCR with next-generation sequencing to assay the methylation state of 50 genes in the regions 1 kb upstream of and downstream from their transcription start sites. This method yielded 96% coverage of the targeted CpGs and demonstrated high correlation between CpG island (CGI) DNA methylation and transcriptional regulation. The method was scaled to interrogate the methylation status of 77,674 CpGs in the promoter regions of 2100 genes in primary CD4 T cells. The 2100 gene library yielded 97% coverage of all targeted CpGs and 99% of the target amplicons.
DNA methylation is a known mechanism for epigenetic modulation of gene expression. Current evidence points to its importance in the dysregulation of proto-oncogenes and tumor suppressors in cancer (Cheung et al. 2009; Ehrlich 2009) as well as in the regulation of temporal gene expression during embryonic development (Geiman and Muegge 2010; Laurent et al. 2010). Additionally, aberrant DNA methylation patterns have been implicated in a number of chronic diseases, including autoimmune, inflammatory, and metabolic disorders (Grolleau-Julius et al. 2010; Villeneuve and Natarajan 2010).
While the importance of DNA methylation has been underscored by the conditions to which it has been linked, the recent advent of high-throughput sequencing has allowed for enhanced discovery and characterization of methylation sites via techniques such as bisulfite conversion, methylated DNA immunoprecipitation (MeDIP), methylated CpG island recovery assay (MIRA), reduced representation bisulfite sequencing (RRBS), and methylation sensitive restriction enzyme sequencing (MRE-seq) (Fouse et al. 2010; Huang et al. 2010). Bisulfite conversion is a chemical modification converting unmethylated cytosines to uracils (Shapiro et al. 1970), allowing the determination of which cytosines are methylated upon sequencing. Bisulfite conversion has historically been the gold standard for DNA methylation analysis, yet deep sequencing of an entire bisulfite-converted genome is cost prohibitive and bioinformatically challenging. Other methods help to overcome these issues. For example, MeDIP entails the genome-wide immunoprecipitation of methylated DNA fragments with an anti-methylcytosine antibody, permitting enrichment of methylation sites but does not yield single CpG resolution. However, MeDIP allows for methylation analysis at a fraction of the cost of bisulfite sequencing. Similarly, MIRA is an alternative that employs methyl-binding protein domains to precipitate methylated DNA. Unlike MeDIP, this method does not require DNA denaturation, and it is reportedly useful for regions with low CpG densities. Finally, methylation-sensitive restriction enzymes (MREs) have been used for several methods in combination with next-generation sequencing, including MRE-seq and RRBS. Such methods are advantageous over MeDIP and MIRA in that they allow for single CpG resolution (Fouse et al. 2010). However, with all these methods, large-scale targeted analysis is not possible (Gu et al. 2010).
Previous strategies for targeted methylation analysis have relied upon PCR, bead arrays, or microarrays to amplify or isolate regions of interest. One disadvantage with the array-based methods is the inability to interrogate multiple closely apposed CpGs individually. Bisulfite conversion coupled with PCR and sequencing overcomes this limitation, but single-plex PCR of multiple genes and gene promoters has historically been a time-consuming process (Fouse et al. 2010). To address this issue, several groups have developed methods focused on combining different selection techniques with next-generation sequencing (Deng et al. 2009; Hodges et al. 2009; Varley and Mitra 2010). Hodges et al. combined array-based capture of bisulfite-converted DNA with next-generation sequencing, allowing for the capture of 25,000 CpGs in 324 CpG islands (CGIs) spanning 300 kb of sequence (Hodges et al. 2009). However, a major drawback of this method is the large amount of bisulfite-converted DNA required (20 μg). Deng et al. used a modified padlock probe capture approach to select for 66,000 CpG in 2,020 CGI from 200 ng of bisulfite-converted DNA (Deng et al. 2009). This method yields a high degree of coverage from a small amount of starting material; however, there is significant variability in the capture efficiency for each of the probes used, and the synthesis of 30,000 probes of 150 bp each is expensive. While the authors demonstrate that the variability could be normalized with suppressor oligos, this represents an additional costly and time-consuming process. The recent study by Varley and Mitra demonstrated multiplex PCR amplification of DNA for 94 amplicons across a number of clinical samples (Varley and Mitra 2010). Although these methods all differ in their selective mechanisms, they all require multiple rounds of traditional PCR amplification (20–35 cycles) that could introduce amplification bias and take several days to complete the selection of targeted genes.
To create a workflow that would allow for targeted, large-scale bisulfite sequencing, we chose to utilize the microdroplet PCR amplification system developed by RainDance Technologies (Tewhey et al. 2009). This system allows the user to set up 1.5 × 106 parallel microdroplet amplifications in a single reaction in under an hour. The nature of microdroplet emulsion PCR significantly decreases PCR amplification bias (Hori et al. 2007; Tewhey et al. 2009). In an initial proof of principle study, we demonstrate targeted methylation analysis of nearly 400 amplicons across 50 gene promoters by combining microdroplet-based PCR amplification of bisulfite-converted DNA with next generation sequencing. This technique yielded successful mapping of 3697 CpGs and demonstrated strong correlation between CpG methylation and gene expression. Following validation of the method, we expanded our primer library to target 3500 amplicons covering 77,674 CpGs in 2127 genes in primary CD4 T cells. This library yielded high-quality sequence data for 97% of the targeted CpGs.
Results
Preparation of bisulfite-converted DNA
Our methodology for microdroplet PCR-based targeted bisulfite sequencing begins with bisulfite conversion of genomic DNA (Fig. 1A), for which there are several commercially available kits. To determine which protocol would be optimal for our needs, we tested four of these kits using genomic DNA from Jurkat cells and assessed yield and DNA degradation. All four kits were comparable in yield (60%–70% of input) (Supplemental Fig. S1A). Each of the kits demonstrated varying degrees of degradation of the single-stranded DNA (ssDNA) product (Supplemental Fig. S1B). While three of the kits were suitable for the downstream amplification of 200–600-bp amplicons, all subsequent experiments were carried out using the QIAGEN EpiTect kit.
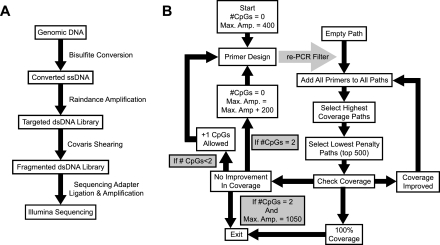
Figure 1.
Targeted MethylSeq workflow. (A) Flowchart for sample preparation. (B) Flowchart for primer design algorithm.
Development of primer design algorithm
The second step in this method is targeted amplification of regions of interest by microdroplet PCR (Fig. 1A). To determine the appropriate design parameters for PCR of bisulfite-converted DNA, we first investigated the coverage attainable in silico for bisulfite-converted genomic loci derived from the Sequenom Standard EpiPanel (Supplemental Table S1). Utilizing a simple version of the final design algorithm, we generated primers targeting 69 loci spanning 142.6 kb with various design restrictions. To ensure that performance was generalizable across the genome, this first 69-loci test set had only 10% overlap with the experimental set of 50 genes tested with Jurkat cells. Coverage was determined across a range of melting temperatures (Tm) and maximum amplicon lengths with a minimum amplicon length of 100 bp. The number of CpGs per primer was limited to ≤1, and primer GC content varied from 20%–80%. For primers covering a CpG, a degenerate base (C/T or G/A, depending on the strand) was used at that location to account for the presence of either a methylated or an unmethylated cytosine. In order to minimize amplification bias due to degeneracy, primers were designed so that CpGs were kept outside the last five bases of the 3′ end. Primer-primer and self-complementarity was restricted by thermodynamic parameters in Primer3 software (Rozen and Skaletsky 2000), and off-target amplification was restricted by electronic PCR (Fig. 1B). Maximum coverage was attained at the highest allowable amplicon size with 53°C and 58°C primers affording similar coverage (Supplemental Fig. S2). A temperature of 58°C was selected as the final target Tm, reasoning that primer stringency could be more easily controlled at higher Tm. Subsequent PCR stringency testing suggested off-target amplification could be reduced by restricting primer GC content to 35%–65% (data not shown). In order to reduce sample preparation problems for sequencing due to fragmentation and barcode ligation, minimum amplicon size was raised to 200 bp in subsequent designs.
To validate the ability of targeted microdroplet PCR to interrogate methylation status, we selected an initial set of 50 genes (Supplemental Table S2) based upon their differential expression in resting and 48-h activated primary human CD4 T cells (data not shown). We defined the regions of interest as 1 kb upstream of and 1 kb downstream from the transcription start site (TSS) and targeted each region with seven to nine primer sets (Supplemental Table S2), resulting in 393 amplicons spanning 147.1 kb. These 393 amplicons covered 100% of the targeted regions with 23% overlap.
To address the ability of the primer library to amplify methylated and unmethylated DNA, wild-type Jurkat DNA and methyltransferase-treated Jurkat DNA (hypermethylated) were bisulfite-converted and subjected to microdroplet PCR. The resulting PCR library was sheared to 200 bp and ligated to sequencing adapters. Each sample was sequenced on a single lane of an Illumina GAIIx flow cell, resulting in more than 25 million reads per lane (Fig. 2A).
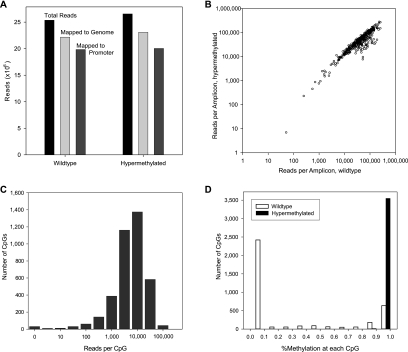
Figure 2.
Effectiveness of custom primer design and highly multiplexed PCR. (A) Targeted loci in wild-type and methyltransferase-treated (hypermethylated) Jurkat DNA were amplified, sequenced, and mapped with Novoalign. Over 85% of reads mapped to the genome, and a similar fraction of mapped reads were uniquely mapped to amplicon regions. (B) Custom primers yielded generally high read counts across 393 amplicons and minimal methylation-induced bias. (C) High read depth at each CpG allows accurate measure of methylation state and suggests that greater multiplexing can be applied. (D) Bimodal (wild-type) or unimodal (hypermethylated) distribution of methylation across all assayed CpG.
Analysis of microdroplet PCR library bisulfite sequencing
Mapping bisulfite-converted DNA to a reference genome presents several complications over standard short-read mapping. First, detecting methylation at a given CpG locus requires differentiating between a C and a T read at that locus, necessitating a strategy that allows unbiased mapping of an ambiguous base. Second, since bisulfite conversion of C to U (and subsequently to T during PCR) destroys standard Watson-Crick base-pairing, a given read must be mapped to either the modified forward or reverse strand or that strand's reverse complement. Several methods have been implemented to address these issues (Lister et al. 2009; Zemach et al. 2010); we chose to employ the Novoalign mapping program (http://www.novocraft.com) as it natively handles both ambiguous bases in the reference genome and the four-strand index required for mapping to a bisulfite-converted genome. Using this method, we successfully mapped over 86% of ∼25 million raw reads to the genome (Fig. 2A). Almost 90% of these reads mapped to targeted amplicon regions, indicating the success of primer design and mapping. Moreover, nearly all of the 393 amplicons had greater than 10,000 reads in both the wild-type and hypermethylated samples (Fig. 2B). The high read count per amplicon and the high correlation between the two samples indicates successful amplification of the intended targets with the RainDance platform and the designed primers.
We further analyzed coverage of each promoter region by examining the read count at each CpG. Figure 2C illustrates that most CpGs (>96%) in the 50 targeted regions have at least 100 reads mapped, giving high confidence estimates of methylation at each gene locus. As expected, virtually every CpG was methylated in the hypermethylated sample. In contrast, CpGs in a given promoter in the wild-type sample tended to be either fully methylated or fully unmethylated, with only a small fraction showing intermediate levels of methylation, indicative of variance across the population of cells assayed or possibly allele-specific methylation (Fig. 2D).
A fortuitous consequence of the primer design is that overlap of positive- and negative-strand amplicons allowed us to measure methylation symmetry. Where a CpG is present in these overlap regions but is not in a primer binding site, we accurately captured methylation of the CpG on both the positive and negative strands. Across 433 such CpGs in 35 genes, we confirm that methylation is highly symmetric (Fig. 3A): the percentage of C vs. T reads in the forward strand is highly correlated with the percentage of C vs. T reads in the negative strand.
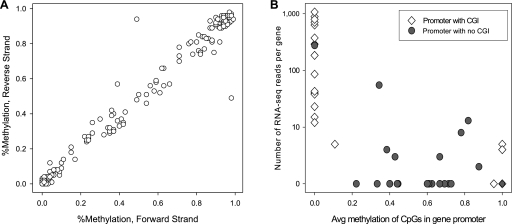
Figure 3.
Biology of methylation. (A) Methylation state of CpG probed in both forward and reverse strand amplicons (read depth ≥ 100; no mismatches; n = 433) indicates methylation at CpG is symmetric across DNA strands. (B) Methylation of CpG islands (CGI) is homogeneous and predictive of expression. In promoters containing an annotated CGI where ≥20 CpG were assayed (open diamonds; n = 24), methylation of CpG in the CGI was either near 100% or near 0%; furthermore, methylated CGI indicated low or no expression, while genes with unmethylated promoter CGI were expressed over a broad dynamic range. For genes with no annotated CGI (n = 23), methylation of all CpG across the 2-kb promoter region showed no strong correlation with expression.
Finally, we compared promoter methylation to gene expression determined by RNA-seq of Jurkat cells. First, we split our 50 genes into those with and without an annotated CpG island. For genes with a CGI and for which at least 20 CpGs were probed (n = 24), we took the average methylation across all CpGs in the CGI and compared it to the number of RNA-seq reads mapped per gene. This reveals a striking bimodal distribution (Fig. 3B, open diamonds), wherein the CGIs are either fully methylated (n = 4) or fully unmethylated (n = 20); moreover, genes with methylated CGIs show low or no expression, while genes with unmethylated CGIs show expression across a broad dynamic range. In contrast, for genes with no CGIs (n = 23, filled circles), the average methylation across all CpGs in the promoter region varies widely and is only weakly correlated with gene expression (Fig. 3B; Schubeler et al. 2000; Jones and Chen 2006). To validate the methylation patterns we identified, single CGI-containing amplicons for eight genes were selected for Sanger sequencing (three with >80% methylation, one with 50%, and three with >15%) (Supplemental Table S4). In all cases, the Sanger and high-throughput microdroplet PCR sequencing results were fully correlated.
Large-scale application of targeted microdroplet DNA methyl-seq
Following validation of this method with the 50-gene library, we increased the primer library to target ∼2100 genes. The targeted genes were chosen based upon their differential expression in resting and 48-h activated primary human naive and memory CD4 T cells (data not shown). The strong correlations seen between promoter CGI and gene expression in the original 50-gene set (Fig. 3B) suggested that the choices of targeted regions for genes with promoter CGIs could be restricted to the region containing the CGI. As such, the 2100-gene library targeted only the CGIs for genes containing a promoter CGI but continued to cover 1 kb upstream of and downstream from the TSS for genes without a promoter CGI. Application of the primer design algorithm with a minimum amplicon size of 200 bp yielded 3520 primer pairs targeting 1.35 Mb of sequence across 2127 genes. This encompassed 77,674 targeted CpGs and 1954 CGIs. To assess the efficiency of this new 2100-gene library, primary human naive CD4 T cells were isolated from a single donor. Genomic DNA was isolated, bisulfite-treated, and amplified with the 2100-gene library.
While 100% of the amplicons were mapped in the proof-of-principle experiment with the 50-gene library, a buildup of reads at the ends of the amplicons was observed (Supplemental Fig. S3). This is likely due to the nature of the shearing protocol employed prior to sequencing adapter ligation. To resolve the uneven distribution of sequencing reads across the amplicons, the microdroplet PCR-amplified product was concatenated prior to shearing (see Methods). Sequencing adapters were ligated to the sheared product, and sequencing was performed in a single lane of an Illumina GAIIx.
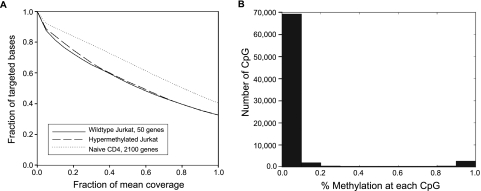
Employing Novoalign, 79% of 36.6 million reads were successfully mapped to the genome. The aligned reads accounted for 2111 (99%) of the targeted genes, with at least 100 reads for 75,180 (97%) of the targeted CpGs. Demonstrating the value of including the concatenation step, the coverage at 50% mean read depth was 67% for the primary naive CD4 DNA amplified with the 2100-gene library compared to 53% for the nonconcatenated wild-type Jurkat DNA and 54% for the hypermethylated Jurkat DNA amplified with the 50-gene library (Fig. 4A). The improvement in read depth following concatenation results from more consistent coverage of the entire amplicon, as shown in detail for two genes in Supplemental Figure S3. Similar to the bimodal distribution of wild-type Jurkat DNA (Fig. 2D), CpGs in given promoters were predominantly either fully methylated or fully unmethylated, with the majority of the CpGs being unmethylated (Fig. 4B).
Figure 4.
(A) Coverage summary for targeted BiS-seq. The fraction of bases with coverage greater than the specified fraction of mean read depth is plotted for 50-promoter assay in Jurkat wild-type (solid line; mean read depth = 17,011), Jurkat hypermethylated (dashed line; mean = 17,274), and 2,000-promoter assay in primary naive CD4 T cells (dotted line; mean = 1,320). The CD4 assay included amplicon concatenation before shearing, which increased coverage at 50% of mean read depth to 67% of the targeted amplification region. (B) Bimodal distribution of methylation across all assayed CpG.
Discussion
Regulation of gene expression is a critical cellular function, and many mechanisms exist to control it at the transcriptional and translational level. DNA methylation is one mechanism of transcriptional regulation and has been implicated in controlling gene expression in embryonic development (Geiman and Muegge 2010; Laurent et al. 2010), tumorigenesis (Cheung et al. 2009; Ehrlich 2009), autoimmune and inflammatory diseases (Grolleau-Julius et al. 2010), and metabolic disorders (Villeneuve and Natarajan 2010). While many methods for DNA methylation analysis exist (Fouse et al. 2010), there is a need to develop a targeted, high-throughput, single CpG resolution method that is cost-effective. While development of second-generation sequencing technology has allowed for whole genome bisulfite sequencing of human stem cells and fibroblasts (Laurent et al. 2010; Lister et al. 2009), it remains cost prohibitive for studying large numbers of samples. MRE-seq and RRBS utilize restriction enzymes and size selection to reduce the complexity and size of sequencing libraries and represent a more cost-effective alternative to whole genome bisulfite sequencing (Meissner et al. 2008; Fouse et al. 2010). However, these methods reduce coverage in a nontargeted fashion. Recent methods developed for targeted bisulfite sequencing have utilized solid array-based hybridization (Hodges et al. 2009), padlock probe strategies (Deng et al. 2009), and bisulfite patch PCR (Varley and Mitra 2010). While these methodologies are more cost-effective than whole genome bisulfite sequencing and have various strengths and weaknesses, they all rely on multiple rounds of traditional PCR amplification and are, thus, subject to amplification bias.
Here, we combine microdroplet PCR with bisulfite sequencing to create an effective and efficient end-to-end method for large-scale epigenetic analysis of targeted regions of the genome. One potential limitation of this approach is the ability to design primers that target bisulfite-converted DNA. To address this, a primer design algorithm was developed to optimize the selection of primers for amplification of 200–400-bp regions of bisulfite-converted DNA while minimizing the number of CpGs included in the primers. The success of this protocol is demonstrated by the fact that 99% of the targeted amplicons were detected in both the 50-gene and 2100-gene libraries, indicating that PCR-based amplification bias does not have an effect on target selection. Moreover, our primer design strategy coupled with the microdroplet PCR technology yielded over 100 reads per CpG for both the 50- and 2100-gene libraries in a single lane of an Illumina GAIIx flow cell. Thus, the number of samples per flow cell lane can be increased by multiplexing, particularly as sequencing technology continues to advance and allow for a significant reduction of costs and starting material required.
In its current form, this method requires 2 μg of bisulfite-converted DNA. While obtaining this quantity of DNA from cell lines or large tissue samples may not be problematic, it may prove more difficult for studying methylation patterns from small clinical samples. For the purposes of calibration, our starting DNA amount of 4 μg is equivalent to the yield of ∼2 mL of whole blood or 2 million cells. While this is well below the 20 μg of starting material used for array-based selection (Hodges et al. 2009), it is substantially more than the 50–250 ng of starting material used for the bisulfite patch PCR- (Varley and Mitra 2010) or padlock probe- (Deng et al. 2009) based methods. However, our method targets 53 times the amount of sequence compared to the bisulfite patch PCR method (Varley and Mitra 2010). In addition to multiplexing sequencing reactions, ongoing studies are focused upon optimizing the microdroplet PCR protocols for smaller amounts of starting material. The padlock probe method, as described, targeted a similar amount of sequence and CpGs as our method but required the production of ∼30,000 150-bp probes on programmable printed arrays (Deng et al. 2009). Our method involves the production of ∼7000 20–30-bp primers by standard oligonucleotide synthesis creating droplet libraries for over 1000 sample runs. The total cost of profiling the CpG methylation status of 2100 genes, including the RainDance chip, sequencing library materials, and multiplexing on an Illumina HiSeq2000 is comparable to the current cost of commercial microarrays. However, these cost estimates will change as rapidly as the technology evolves.
Sequencing and mapping of targeted bisulfite-treated regions in 50 genes confirmed the symmetry of methylation of CpG palindromes and, in the case of CGIs, strong correlation with gene expression. Such strong correlations to gene expression suggest that the targeted regions for genes with promoter CGIs can be restricted to the region containing the CGI. This vastly reduces the size of the targeted region for many genes and increases the number of genes that can be targeted in a single droplet library. This strategy allowed us to expand the primer library from ∼400 amplicons targeting 50 genes to ∼3500 amplicons targeting ∼2100 genes. While this is currently toward the upper limit for the number of regions than can be amplified with the RainDance microdroplet PCR platform, future multiplexing strategies may allow for a further expansion in the number of amplicons targeted in a single reaction.
In conclusion, we describe a new protocol for targeted DNA methyl-seq that combines second-generation deep sequencing and high throughput microdroplet PCR. At a scale per run of more than 2100 genes performed in a fully automated, closed circuit instrument, with the flexibility to select and design custom libraries for any selected gene set, this approach should facilitate many new investigations of the role of epigenetic changes in transcriptional regulation.
Methods
Ethics statement
All the studies in this manuscript were covered by Human Subjects Research Protocols approved by the Center's Institutional Review Board and by the IRB of The Scripps Research Institute. Informed written consent was obtained from all study subjects in the study.
Bisulfite-conversion kit testing
One μg of Jurkat genomic DNA (New England Biolabs) was bisulfite-treated using the Epitect Bisulfite kit (QIAGEN), BisulFlash kit (Epigentek), Imprint DNA Modification kit (Sigma), or the EX DNA Methylation Gold kit (Zymo Research) following the manufacturers' protocols. Converted ssDNA was quantified using the Quant-iT ssDNA Assay kit (Invitrogen) and a Qubit fluorometer (Invitrogen). Two hundred ng converted ssDNA was run on a 2% TAE agarose gel and stained with SYBR Gold (Invitrogen).
Primer design algorithm details
The algorithm begins by extracting the targeted region plus 450 bp of upstream and downstream flanking sequence from an in silico bisulfite-converted SNP-masked genome. All dbSNP130 SNPs validated by frequency are masked from primer design. In silico bisulfite conversion transforms all nonCpG cytosines to thymine and converts all CpG cytosines, including cytosines preceding a SNP where either allele is a guanine, into unknown (N) nucleotides. Primer3 (Rozen and Skaletsky 2000) is used as the engine to produce as many primer pairs as possible from both the plus and minus strand. The full array of Primer3 conditions used to design primers is presented in Supplemental Table S3. Briefly, amplicon size is initialized at 200–450 bp, the number of allowable N nucleotides is initialized at zero, Tm is allowed to range from 56°C–60°C, primer GC content is allowed to range between 35%–65%, self- complementarity, primer-primer complementarity, and hairpin formation is restricted for stable structures with melting temperatures above 4°C. Primers meeting these conditions are then scanned against an in silico bisulfite-converted genome, by NCBI reverse e-PCR (Schuler 1997). Primer pairs mapping within 50–2000 bp of one another, allowing two mismatches during scanning, are filtered out from further consideration. Remaining primers are tiled against the target region.
Amplicon tiling proceeds by initializing coverage paths first through consideration of all possible pairs of amplicons as potential paths to the highest coverage. Each base in the target region, excluding the flanking nucleotides for primer design, is considered covered if at least one amplicon, from either strand overlaps the base. All paths achieving lower coverage than the path(s) producing the highest coverage are discarded (the primers themselves are not discarded). In the case that over 500 independent paths achieve the same coverage, the top 500 paths, as determined by their lowest average penalty score calculated for each primer by Primer3, are kept, while the rest of the paths are discarded. The paths are then extended by one amplicon by generating all possible combinations of the remaining paths with all primers not already contained within each path. These new paths are then filtered through the above procedure. This process is iterated indefinitely until maximum coverage does not increase or until any path hits 100% coverage. By extending all paths in parallel, the shortest possible path attaining 100% coverage is selected. If multiple shortest paths achieving 100% coverage are produced, the path containing primers with the lowest average penalty is selected. If no path achieves 100% coverage, the algorithm proceeds through further rounds of primer design.
The additional rounds of primer design proceed by iteratively increasing the number of allowable CpGs per primer pair and the maximum amplicon size. First, primers are generated allowing one CpG per primer pair, filtering the primer pairs through the same reverse e-PCR procedure, and attempting to extend the previously generated highest coverage paths with the new amplicons. If 100% coverage is not attained, this process is repeated allowing, one CpG per primer (two CpGs total per primer pair). If 100% coverage is still not achieved at the initial maximum amplicon size, maximum amplicon size is increased by 200 bp, and the number of allowable CpGs is reset to 0. This cycle of primer design and tiling repeats until 100% coverage is attained, or until primers are designed for 1050-bp amplicons with one CpG per primer, after which point no further attempts are made to achieve 100% coverage. Most genomic regions tested hit 100% coverage well before this point.
Isolation of human naive CD4 T cells
Peripheral blood was collected from healthy donors, and peripheral blood mononuclear cells (PBMC) were collected by centrifugation through a histopaque gradient. Naive CD4 T cells were negatively selected from PBMCs using the Naive CD4 T cell isolation kit II (Miltenyi Biotec) following the manufacturer's directions. DNA was isolated from purified naive CD4 T cells using an All Prep kit (QIAGEN) following manufacturer's directions.
Preparation of bisulfite-converted DNA
Jurkat DNA, Jurkat CpG DNA (New England Biolabs), and primary naive CD4 T cell DNA was bisulfite-treated using the Epitect Bisulfite kit (QIAGEN) following the manufacturer's protocol. Two μg of DNA was treated per column, and purified DNA was eluted in 20 μl elution buffer. We determined that a total starting amount of 4 μg of genomic DNA was sufficient to yield the 2 μg bisulfite-treated DNA required for the next steps. To concentrate the bisulfite-treated DNA, eluted DNA for each sample was pooled together. Five μl 10× PCR buffer (Applied Biosystems) was added, and samples were brought to a final volume of 50 μl with distilled water. Ninety μl Agencourt AMPure XP beads (Beckman Coulter Genomics) was added, and samples were incubated at room temperature for 5 min on an orbital shaker or rocker. Samples were placed on a Dynal magnet for 2 min, the supernatant was removed, and the beads were washed with 200 μl 70% EtOH two times. DNA was eluted in 20 μl 10 mM Tris-HCl pH 8.0 (Sigma). Single-stranded DNA was quantitated on a Nanodrop 1000 (Thermo Fisher) or using the Quant-iT ssDNA Assay kit (Invitrogen) and a Qubit fluorometer (Invitrogen).
Genomic DNA template mix
To prepare the input DNA template mixture for targeted amplification, 2 μg of the bisulfite treated genomic DNA was added to 4.7 μL 10× High-Fidelity Buffer (Invitrogen), 1.8 μL of 50 mM MgSO4 (Invitrogen), 1.7 μL of 10 mM dNTP (New England Biolabs), 3.6 μL of 4 M Betaine (Sigma), 3.6 μL of RDT Droplet Stabilizer (RainDance Technologies), 1.8 μL dimethyl sulfoxide (Sigma) and 0.7 μL of 5 U/μL Platinum High-Fidelity Taq (Invitrogen). Samples were brought to a final volume of 25 μL with nuclease free water (Teknova, Fisher).
Droplet library construction
PCR droplets were generated on the RDT1000 (RainDance Technologies) using the manufacturer's recommended protocol. To process a single sample, a tube containing 25 μL of Genomic DNA Template Mix, a custom primer droplet library (RainDance Technologies), and a disposable microfluidic chip (RainDance Technologies) were placed onto the RDT1000. The custom primer droplet library consists of a collection of individual primer droplets, where each primer droplet contains matched pairs of forward and reverse primer (5.2 μM per primer) for each amplicon defined within the primer library. Each primer is present in the PCR reaction at a final concentration of 1.6 μM. For primers covering a CpG, a degenerate base (C/T or G/A, depending on the strand) was used at that location. The concentration of primers containing degenerate bases was adjusted such that primers with each degenerate base were present at 0.8 μM in a single droplet. The RDT1000 generated each PCR droplet by pairing a single gDNA template droplet with a single primer droplet. Paired droplets flow past an electrode embedded in the chip and are instantly merged together. All resulting PCR droplets were automatically dispensed as an emulsion into a single PCR tube and transferred to a standard thermal cycler for PCR amplification. Each sample generated more than 1 million singleplex PCR droplets.
PCR amplification
Samples were amplified in a Bio-Rad PTC-225 thermocycler as follows: initial denaturation at 94°C for 2 min, followed by 55 cycles of: 94°C, 15-sec denaturation; 56°C, 20-sec primer annealing; 68°C, 40- sec extension; and a final extension at 68°C for 10 min. Reactions were then held at 4°C until further processing.
Breaking emulsion
After PCR amplification, PCR droplet emulsions were broken to release each individual amplicon contained within the droplets. For each sample, an equal volume of RDT 1000 Droplet Destabilizer was added to the emulsion of PCR droplets, the sample was vortexed for 15 sec, and then spun in a microcentrifuge at 12,000 × g for 5 min. The oil from below the aqueous phase was carefully removed and disposed of, and the remaining sample was carried into the PCR product clean-up step.
PCR product clean-up
Each sample was purified over a MinElute column (QIAGEN) following the manufacturer's recommended protocol. Samples were eluted from the columns with 11 μL Elution Buffer. Purified amplicon DNA was then analyzed on an Agilent Bioanalyzer by adding 1 μl of each sample per well on a DNA chip to quantify amplicon yield.
Fragmentation of RainDance PCR product
RainDance PCR products were fragmented in 100 μl using a Covaris S2 with a duty cycle of 10%, an intensity of 5200 cycles per burst, and 20 60-sec bursts. Fragmentation was assessed on an Agilent Bioanalyzer by adding 1 μl of each sample per well on an HS DNA chip. Fragmented DNA concentration was calculated with a Quant-iT dsDNA HS Assay kit (Invitrogen) and a Qubit Fluorimeter.
Concatenation of RainDance PCR product
Four hundred ng of RainDance PCR product was end-repaired with 50 U T4 polynucleotide kinase (Enzymatics), 5 U Klenow fragment (Enzymatics), 15 U T4 DNA polymerase (Enzymatics), 400 μM dNTP mix (Enzymatics), and 1X T4 DNA ligase buffer (Enzymatics) at 30°C for 20 min. End-repaired products were purified over a DNA Clean & Concentrator-5 column (Zymo Research). Purified products were concatenated with 600 U rapid T4 DNA ligase (Enzymatics) in 1× rapid ligation buffer (Enzymatics) at 20°C for 15 min. Concatenated products were purified over a DNA Clean & Concentrator-5 column. Purified, concatenated products were fragmented using a Covaris S2 with a duty cycle of 10%, an intensity of 5, 200 cycles per burst, and three 60-sec bursts. Fragmentation was assessed on an Agilent Bioanalyzer by adding 1 μl of sample to a well on a HS DNA chip.
Preparation of sequencing libraries and deep sequencing
Sequencing libraries were constructed following manufacturer's instructions with slight modifications. Briefly, 100 ng of fragmented PCR product was end-repaired and A-tailed. Adapters were ligated, and ligation product was purified on Agencourt AMPure XP beads followed by size selection from 2% agarose. Purified product was amplified with 15 cycles of PCR, followed by a final round of size selection from 2% agarose. Libraries were analyzed on an Agilent Bioanalyzer by adding 1 μl of each sample per well on a DNA chip and quantitated using the Quant-iT dsDNA BR Assay kit and a Qubit Fluorimeter. Purified libraries were used directly for cluster generation and sequencing on an Illumina GAIIx system, according to the manufacturer's instructions. One hundred-bp single-end or paired-end reads were generated for three samples of wild-type Jurkat DNA or two samples of Jurkat CpG DNA with a single sample per lane.
Alignment and analysis
Reads were mapped to NCBI Build 36 (hg18), downloaded from the UCSC Genome Browser FTP server (ftp://hgdownload.cse.ucsc.edu/goldenPath/). Mapping was performed using Novoalign (http://www.novocraft.com) using the manufacturer's specifications.
In contrast to mapping strategies based on a separate forward- and reverse-strand in silico bisulfite-converted reference genome, Novoalign uses a four-strand index of a single converted genome with an ambiguous base (YpG vs. CpG), resulting in a single output file (in SAM format). As our amplicons are strand-specific, downstream analysis relies on determining to which strand a given read maps. This is done by considering the proportion of C and G in the read as it is recorded in the SAM file: reads with a high proportion of G relative to C are forward-strand, and vice versa. We counted the number of C and G in the first 40 bases of each read and classified each read on the basis of these counts; no reads had equal numbers of C and G. Mapped reads were split into forward-mapping and reverse-mapping SAM files and analyzed for mapping to promoter regions and individual amplicons using SAMtools (http://samtools.sourceforge.net).
To assay methylation symmetry, we identified all CpGs covered by both forward- and reverse-strand amplicons. CpGs in an amplicon primer region were excluded, as the mix of C- and T-containing primers introduced quantitation artifacts. Given the extraordinary read depth generated by our amplification and sequencing strategy, we were able to use a strict read mapping quality filter with virtually no loss of coverage. Briefly, for quantitative analysis, we accepted only those reads with a mapping quality score ≤10 (UQ field in the SAM file; UQ is the sum of the PHRED scores of the mismatches in the read). This yielded ∼5 million reads with virtually no mismatches, resulting in effectively zero FDR across the results presented here. This “perfect match” subset was used for all downstream analyses. Methylation was defined as C/(C + T) forward-strand reads at the C locus in a given CpG and G/(G + A) reverse-strand reads at the G locus.
In correlating promoter methylation with gene expression, assayed genes were split into two groups. For genes with an annotated CpG island (annotations for hg18 downloaded from the UCSC Genome Browser) in the promoter region we assayed (n = 26), average methylation across the CpGs in the CGIs was measured. For genes with no CGIs (n = 24), average methylation across all CpGs in the 2-kb amplified region was measured. Methylation at a given CpG was measured as either C/(C+T) forward-strand reads at the C locus or G/(G+A) reverse-strand reads at the G locus, whichever had a greater number of reads (most CpGs were measured by only forward- or reverse-strand reads, not both). Individual CpG methylation percentages were averaged across all CpGs assayed in the region. The gene expression data used for this analysis was generated by RNA-seq of Jurkat cells done on the Illumina GAIIx instrument. Amplified cDNA was prepared from 100 ng total RNA using the Ovation RNA-seq System (Nugen). One hundred ng cDNA was treated with S1 Nuclease (Promega), and sequencing libraries were prepared as described above.
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE30179.
Acknowledgments
This research was supported by funds from the National Institutes of Health: U19 A1063603-06 (D.R.S., G.T.H.), T32DK007022-30 (H.K.K.), UL1 RR025774-04 (S.A.L.), the Molly Baber and Verna Harrah Research Funds supporting the Salomon laboratory, and National Institutes of Health National Center for Research Resources grant UL1 RR025774 (A.T.). The informatics tool development work was supported by an unrestricted research grant to A.T., and RainDance contributed their time, instrument use, and PCR droplet library construction as part of this scientific collaboration. This is manuscript # 20950 from The Scripps Research Institute.
The authors from RainDance Technologies declare competing financial interests; RainDance Technologies is a for-profit company offering a sequence enrichment product for bisulfite-converted DNA.
Authors' contributions: This work was conceptualized originally by D.R.S., D.R.L., P.L.L., H.K.K., and S.A.L. Experimental strategies and details were developed and refined by all authors. Experiments were performed by H.K.K., S.A.L., S.K., and J.O., and the informatics and analysis work was done by A.T., G.T.H., and J.W. H.K.K., S.A.L., G.T.H., A.T., and D.R.S. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116863.110.
References
- Cheung HH, Lee TL, Rennert OM, Chan WY 2009. DNA methylation of cancer genome. Birth Defects Res C Embryo Today 87: 335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M 2009. DNA hypomethylation in cancer cells. Epigenomics 1: 239–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Nagarajan RP, Costello JF 2010. Genome-scale DNA methylation analysis. Epigenomics 2: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Muegge K 2010. DNA methylation in early development. Mol Reprod Dev 77: 105–113 [DOI] [PubMed] [Google Scholar]
- Grolleau-Julius A, Ray D, Yung RL 2010. The role of epigenetics in aging and autoimmunity. Clin Rev Allergy Immunol 39: 42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A 2010. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods 7: 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, et al. 2009. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res 19: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori M, Fukano H, Suzuki Y 2007. Uniform amplification of multiple DNAs by emulsion PCR. Biochem Biophys Res Commun 352: 323–328 [DOI] [PubMed] [Google Scholar]
- Huang HC, Zheng S, VanBuren V, Zhao Z 2010. Discovering disease-specific biomarker genes for cancer diagnosis and prognosis. Technol Cancer Res Treat 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Jones B, Chen J 2006. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J 25: 2443–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schubeler D, Lorincz MC, Cimbora DM, Telling A, Feng YQ, Bouhassira EE, Groudine M 2000. Genomic targeting of methylated DNA: Influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol Cell Biol 20: 9103–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD 1997. Sequence mapping by electronic PCR. Genome Res 7: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Servis R, Welcher M 1970. Reactions of uracil and cytosine derivatives with sodium bisulfite. J Am Chem Soc 92: 422–424 [Google Scholar]
- Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB, Larson JW, et al. 2009. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol 27: 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Mitra RD 2010. Bisulfite Patch PCR enables multiplexed sequencing of promoter methylation across cancer samples. Genome Res 20: 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve LM, Natarajan R 2010. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol 299: F14–F25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]