Abstract
The human body contains thousands of unique cell types, each with specialized functions. Cell identity is governed in large part by gene transcription programs, which are determined by regulatory elements encoded in DNA. To identify regulatory elements active in seven cell lines representative of diverse human cell types, we used DNase-seq and FAIRE-seq (Formaldehyde Assisted Isolation of Regulatory Elements) to map “open chromatin.” Over 870,000 DNaseI or FAIRE sites, which correspond tightly to nucleosome-depleted regions, were identified across the seven cell lines, covering nearly 9% of the genome. The combination of DNaseI and FAIRE is more effective than either assay alone in identifying likely regulatory elements, as judged by coincidence with transcription factor binding locations determined in the same cells. Open chromatin common to all seven cell types tended to be at or near transcription start sites and to be coincident with CTCF binding sites, while open chromatin sites found in only one cell type were typically located away from transcription start sites and contained DNA motifs recognized by regulators of cell-type identity. We show that open chromatin regions bound by CTCF are potent insulators. We identified clusters of open regulatory elements (COREs) that were physically near each other and whose appearance was coordinated among one or more cell types. Gene expression and RNA Pol II binding data support the hypothesis that COREs control gene activity required for the maintenance of cell-type identity. This publicly available atlas of regulatory elements may prove valuable in identifying noncoding DNA sequence variants that are causally linked to human disease.
A single genome gives rise to a multitude of cell types, each with its own specialized pattern of gene expression. These programs are partly governed by DNA-encoded regulatory elements. Unlike protein coding genes, DNA regulatory elements are not easy to identify in linear DNA sequence. While nearly 70% of bases in protein-coding DNA are evolutionary constrained, only half of all the regulatory elements identified in the ENCODE pilot project harbored constrained bases at all, and among these, only 10% of the bases were constrained (The ENCODE Project Consortium 2007). As part of the ENCODE effort (The ENCODE Project Consortium 2007, 2011), we have continued our development of DNase-seq (Crawford et al. 2006b; Boyle et al. 2008a; Song and Crawford 2010) and FAIRE-seq (Formaldehyde Assisted Isolation of Regulatory Elements)(Giresi et al. 2007; Giresi and Lieb 2009) to identify regulatory sites across the genome. DNase-seq employs the DNaseI enzyme to preferentially digest nucleosome-depleted sites, also known as DNaseI hypersensitive (HS) sites (Wu et al. 1979). FAIRE-seq enriches nucleosome-depleted DNA using formaldehyde fixation and phenol-chloroform extraction.
Here, we call the regions identified by DNaseI or FAIRE “open chromatin.” These open chromatin regions often correspond to nucleosome-depleted regions (NDRs) (Hogan et al. 2006; Giresi et al. 2007; Kim et al. 2007), which are often associated with regulatory factor binding. Several studies have shown that open chromatin is associated with all known classes of active DNA regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions (Gross and Garrard 1988; Cockerill 2011). We used DNase-seq and FAIRE-seq (Giresi et al. 2007; Xi et al. 2007; Boyle et al. 2008a; Gaulton et al. 2010; Stitzel et al. 2010) to generate genome-wide open chromatin maps spanning seven diverse human cell types, thereby greatly expanding the number of human regulatory elements with experimental support.
Results
A coordinated mapping pipeline for data generation, processing, and quality control
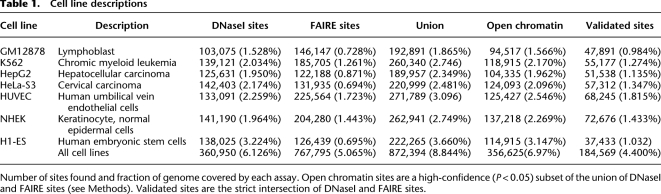
DNase-seq and FAIRE-seq were performed on seven cell lines in duplicate or triplicate (Table 1) using material from cells grown in a single batch at the same location (see Methods). DNA libraries were sequenced on an Illumina sequencer, and the resulting data were collected and processed using a standard pipeline (see Methods). ChIP-seq data were generated using the same fixed cells used for FAIRE-seq and analyzed using the same pipeline. In this way, experimental and data processing differences among the assays were minimized (Fig. 1A). Comparisons of multiple independent growths and with results from tiled microarrays on the same material support the quality of these data (Supplemental Methods; Supplemental Table S1).
Table 1.
Cell line descriptions
Figure 1.
Identification of open chromatin in seven human cell lines. (A) A schematic representation of the experiment and analysis design. (B) DNaseI (y-axis fixed at Parzen signal value 0.15) and FAIRE (y-axis fixed at 0.04) data from seven cell lines surrounding the HNF4A locus (145 kb; UCSC Genome Browser) shows both ubiquitous and cell-type selective open sites that are especially prevalent in HepG2 cells. Pol II, CTCF, and MYC ChIP-seq peaks that overlap open chromatin are highlighted.
A continuous range of signal intensities from DNase-seq and FAIRE-seq was observed across the genome (Fig. 1B), and from this signal, we identified discrete peaks (see Methods). For each cell type, the number of DNase-seq or FAIRE-seq peaks ranged from ∼100,000 to 225,000, covering between 0.65% and 2.99% of the genome (Table 1). In total, over 870,000 DNaseI and FAIRE sites were identified from the seven cell lines, covering nearly 9% of the genome. While more FAIRE peaks than DNaseI peaks were identified, FAIRE peaks tended to span fewer bases (Table 1).
DNase-seq and FAIRE-seq identify an overlapping set of open chromatin sites, but each also identifies unique chromatin features
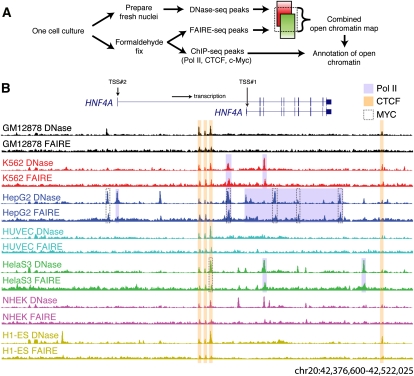
DNaseI and FAIRE assays both identify sites in the genome that tend to be nucleosome-free or nucleosome-depleted (Hogan et al. 2006; Giresi et al. 2007; Cockerill 2011). To quantitatively evaluate the overlap between these assays, we compared rank-ordered peaks from each assay, with rank based on the maximum signal intensity within each peak. We calculated the number of overlapping DNaseI and FAIRE sites for four different peak intensity cutoffs (Fig. 2A; Supplemental Fig. S1). In each cell type, ∼30%–40% of the top 100,000 (100K) DNaseI and FAIRE peaks overlapped. The amount of overlap did not change substantially when comparing the top 10,000 (10K) peaks from each assay (20%–40% overlap). About 80% of the top 10K peaks from either assay are found within the top 100K of the alternate assay (Fig. 2A; Supplemental Fig. S1).
Figure 2.
DNase-seq and FAIRE-seq identify overlapping and unique sets of open chromatin. (A) Comparisons of the top 10K, 25K, 50K, and 100K DNase-seq and FAIRE-seq peaks from a single cell line (GM12878), with overlap indicated below each Venn diagram. (B) Average percentage of DNaseI and/or FAIRE peaks, as well as permuted coordinates, in defined positional categories based on their relationship to annotated genes. Error bars represent the standard deviation over seven cell types. Several categories deviated significantly from random (Supplemental Table S2). (C) The percentage of CTCF ChIP-seq peaks that overlap DNaseI and/or FAIRE sites in all seven cell types. The x-axis values indicate different signal thresholds for calling sets of CTCF peaks, where the threshold is increasingly more stringent from left to right. (D) The same as C, except for MYC ChIP-seq data. (E) Percentage of TSSs with overlapping Pol II ChIP-seq, DNaseI, and/or FAIRE peaks in seven cell types. x-axis represents expression values for corresponding genes indicating high (7+), medium (5–7), or low/no (0–5) expression.
We consider sites detected by both DNase-seq and FAIRE-seq to be “cross-validated” and to represent high-confidence open chromatin sites. Across the seven cell lines, there are more than 180,000 high-confidence sites covering nearly 4.5% of the genome (Table 1). Though we found a significant enrichment of these high-confidence sites within 2 kb of an annotated transcription start site (TSS) (P-value < 2 × 10−5), most (80%) were far from gene starts (see Methods) (Fig. 2B).
In a given cell type, many sites were detected by only one of the two assays. Multiple lines of evidence suggest that these DNase-only and FAIRE-only sites are biologically relevant and that the assays differentially detect real chromatin features. First, ∼50% of DNase-only and 40% of FAIRE-only sites were detected in multiple cell types (Supplemental Fig. S2) and across multiple independent growths (Supplemental Table S1), which would be unlikely if the peaks were spurious. Second, the differences between DNase-seq and FAIRE-seq arise from distinct genomic regions, which may be interrogated differentially by each method. DNaseI-only sites were enriched within 2 kb of a TSS (P-value < 4 × 10−6) and within 5′ exons and introns (P-value < 3 × 10−6) compared to FAIRE-only sites, while FAIRE-only sites were found preferentially in internal introns and exons and nonpromoter intergenic regions (P-value < 3 × 10−6 and 2 × 10−5, respectively) relative to DNase-only sites (Fig. 2B; Supplemental Table S2). Third, we found that both DNase-only and FAIRE-only sites were enriched for H3K4 mono-methylation (H3K4me1), a mark associated with enhancers (Heintzman et al. 2009; Ernst et al. 2011), along with H3K4 tri-methylation (H3K4me3) and H3K9 acetylation (H3K9ac); both marks of a TSS (Supplemental Fig. S3; Heintzman et al. 2009; Ernst et al. 2011). DNase-only sites were more strongly associated with H3K4me3 and H3K9ac, and FAIRE-only sites with H3K4me1. Lastly, DNase-only and FAIRE-only sites corresponded differentially to specific transcription factor binding sites, as explained in the next section.
Together, DNase-seq and FAIRE-seq identify most of the sites bound by regulatory factors
DNase-seq and FAIRE-seq data were compared to ChIP-seq data generated from the same cell lines using antibodies to CTCF, MYC, and Pol II (see Methods) (Supplemental Fig. S4). Over 96% of the strongest CTCF and MYC ChIP sites were identified by one or both assays (Fig. 2C,D). About 30% of CTCF and 15% of MYC sites were captured by DNase-only or FAIRE-only sites. At any given ChIP-seq peak cut-off, ChIP-seq signal intensity was the strongest for peaks detected by both DNaseI and FAIRE, was weaker in sites detected by only one assay, and the weakest for sites that overlapped neither assay (Supplemental Fig. S5).
We examined the correspondence of published ChIP-seq data in matching cell types (Fujiwara et al. 2009; Motallebipour et al. 2009; Frietze et al. 2010; Kouwenhoven et al. 2010; Raha et al. 2010) with our open chromatin data. DNase-seq and FAIRE-seq captured >80% of sites (>90% of the strongest sites) for TP63 in NHEK, FOXA1, and FOXA3 in HepG2, and GATA1 in K562 (Supplemental Fig. S6), and ∼70% of the ZNF263 sites in K562. We note that FOXA1, FOXA3, and GATA1 were better identified by FAIRE-seq, while ZNF263 was found more often by DNase-seq.
We next evaluated our Pol II ChIP-seq data in conjunction with RNA expression data generated from the same cells. For each gene with RNA data in each cell line, we determined whether there was a significant signal for Pol II binding and/or open chromatin in the region 1000 bases upstream of and 500 bases downstream from an annotated transcription start site. We found that 81% of all TSSs harbored accessible chromatin, consistent with previous estimates that 70%–80% of all genes are either active or poised (Guenther et al. 2007). We divided genes into highly expressed (46% of genes, log2 RNA > 7; Methods), moderately expressed (29%, log2 RNA between 5 and 7), and lowly or not expressed (25%, log2 RNA < 5). We found that nearly all highly expressed genes had Pol II binding and open chromatin at their TSS (Fig. 2E). About 60% of the moderately expressed genes showed Pol II and open chromatin signals, while an additional 30% showed just open chromatin signal. About half of the lowly or nonexpressed genes showed evidence of Pol II or open chromatin, while the remaining half had no evidence of either signal. In all, open chromatin identifies the TSS of nearly all of expressed genes and indicates that a large fraction of the remaining genes may be poised for transcription.
A combined open chromatin atlas reveals chromatin similarities between functionally related cell types
To take advantage of the strengths of each assay, we created a combined annotation for each of the seven cell lines by integrating data from DNaseI and FAIRE (see Methods). Our open chromatin atlas contains sites strongly identified by both assays, high confidence peaks present in only one assay, and lower confidence peaks supported by both assays (Table 1). The number of combined significant open chromatin sites ranged from 100,000 to 125,000 (P < 0.05; Methods) for each cell line. Between any two cell types, ∼30%–40% of open chromatin sites are shared (Supplemental Table S3).
Using open chromatin sites, we performed hierarchical clustering of the cell lines (see Methods) (Supplemental Fig. S7A). The clustering appears to reflect functional and lineage similarities in cell types and almost perfectly matches cell-line clustering based on gene expression data (Supplemental Fig. S7B). For example, we find that the two cell types of hematopoietic lineage, GM12878 (lymphoblastoid cell line) and K562 (chronic myeloid leukemia), clustered together using either expression or chromatin data. Embryonic stem cells do not have a considerably different number of open chromatin sites and do not contain a superset of open chromatin sites found in other more differentiated cell types. However, embryonic stem cell open chromatin sites tended to be larger and covered a greater fraction of the genome than other cell types (Table 1).
The discovery of human regulatory elements by open chromatin mapping is far from saturation
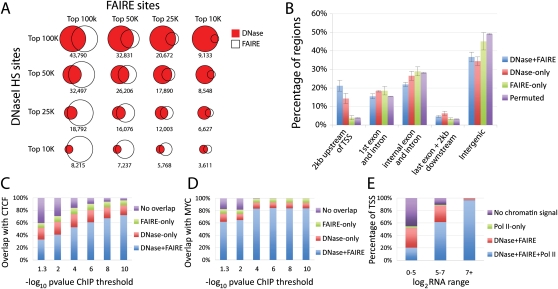
We created union sets for every possible combination of 2, 3, 4, 5, 6, and 7 cell types and plotted the rate at which new sites appeared (Fig. 3A). Regardless of the threshold used to call the sites, the number of new sites identified does not abate as the number of cell lines analyzed increases. In contrast, performing the same analysis on DNase-seq data from lymphoblastoid lines derived from seven different individuals (McDaniell et al. 2010) shows clear signs of saturation after the third cell line (Supplemental Fig. S8). This indicates that testing additional cell types is necessary and will continue to uncover new open chromatin sites.
Figure 3.
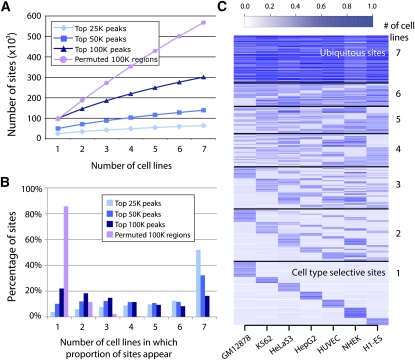
Distribution of open chromatin regions across cell types. (A) Saturation of total open chromatin sites discovered as a function of the number of cell types tested (x-axis). The rate of new top 25K sites per cell type was lower than for top 50K and 100K sites, likely reflecting more ubiquitous sites in this top fraction. (B) Percentage of the top 25K, 50K, and 100K combined open chromatin sites (y-axis) detected in one to seven of the cell types tested (x-axis). Over 50% of the top 25K open chromatin sites were ubiquitous, while more top 50K and 100K peaks were cell type selective. (C) Top 100K combined open chromatin sites partitioned by number of cell types in which they appear (y-axis). Color intensity indicates strength of open chromatin signal in that cell type.
Open chromatin sites found in all seven cell types are characterized by high signal
The number of open chromatin sites held in common among these seven cell lines, referred to as ubiquitous sites, was much higher than would be expected by chance (Fig. 3B). These sites tended to have the strongest signals in some or all cell types. For example, of the union set of the 25K strongest combined open chromatin peaks in each cell type (64,400 sites total), nearly 52% (33,466) were present in each of the seven cell lines (Fig. 3B; Supplemental Table S4). Of the union set of the top 50K (139,133 total) and 100K (301,235 total) sites, 32% (44,750) and nearly 16% (49,009) were ubiquitous, respectively (Fig. 3B; Supplemental Table S4). In contrast, when we randomly permuted the genomic coordinates of the top 100K sites for each cell line across the genome, over 85% appeared in only a single cell type, while <0.02% were ubiquitous (Fig. 3B; Supplemental Table S4). Overall, open chromatin sites with the strongest signals tend to be detected across cell types, while cell-type selective sites tend to produce weaker signals (Fig. 3C). Indeed, the distributions of –log10(P-values) for open chromatin sites categorized by the number of cell lines in which the sites appeared were found to be correlated (Supplemental Fig. S9), with ubiquitous sites having significantly higher amplitude than all other categories (P < 10−16; pairwise T-tests). No single cell type dominates the cell-type selective signal (Fig. 3C).
Ubiquitous open chromatin sites tend to be near TSSs and are often bound by CTCF, while cell-type selective sites are distal with little CTCF binding
We examined the location of ubiquitous and cell-type selective open chromatin relative to genes, considering for this analysis the union of the top 100K combined open chromatin sites across all cell types. We found that ∼30% of ubiquitous sites was located near transcription start sites, ∼35% was within intergenic regions, and ∼35% was within transcribed regions (see Methods for definitions of categories) (Fig. 4A). Ubiquitous open chromatin sites were much more likely to occur near TSSs, while cell-type selective sites were rarely found near TSSs (Fig. 4A). This suggests that cell-type selective gene regulation is controlled through distal regulatory elements.
Figure 4.
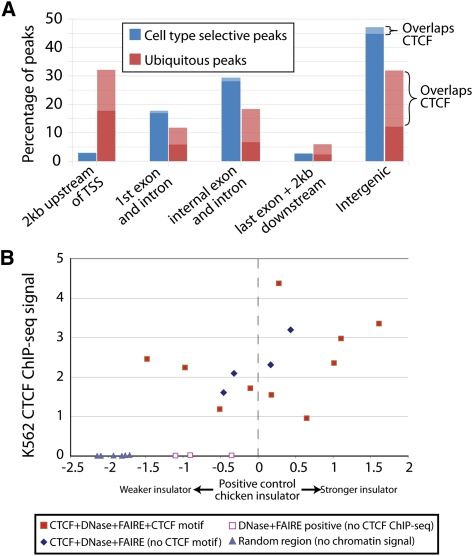
Ubiquitous and cell-type selective sites differ related to transcription start sites and presence of CTCF. (A) Percentage of ubiquitous and cell-type selective open chromatin sites in positional categories relative to annotated genes. Light bars represent open sites overlapping CTCF. (B) Insulator assays performed on sites with (1) DNase-seq, FAIRE-seq, and CTCF ChIP-seq signal and a CTCF motif (filled red squares); (2) signal in all three assays but without a CTCF motif (blue diamonds); (3) DNase-seq and FAIRE-seq signal, but not CTCF ChIP-seq (open red squares); and (4) no signal in any assay (gray triangles). y-axis indicates the signal from CTCF ChIP-seq in K562 cells. Enhancer blocking values (x-axis) were calculated as described (Supplemental Methods), with a value of zero equaling the measured activity of a known insulator.
CTCF has been shown to perform diverse regulatory functions, but it is primarily identified as an insulator that blocks interaction between promoters and enhancers. Overall, CTCF binding occurred in 28% of this union set of open chromatin sites but was bound to >55% of ubiquitous open chromatin sites (Fig. 4A). In contrast, CTCF bound to <5% of cell-type selective open chromatin sites. We hypothesized that open chromatin sites with evidence of CTCF binding should be more likely to function as insulators than open chromatin sites without CTCF. We tested 15 open chromatin sites with strong CTCF ChIP-seq signals for insulator activity (see Methods) (Supplemental Table S5). Seven of the sites were previously described (Xi et al. 2007). Five were chosen specifically based on the absence of the 20-bp CTCF binding motif to test for insulator function even in the absence of this motif. We also tested three open chromatin sites that lacked evidence of CTCF binding and six sites with no strong evidence of CTCF binding or open chromatin in K562 cells (Supplemental Table S5).
The majority of open chromatin sites also bound by CTCF displayed insulator activity (Fig. 4B). The absence of a canonical CTCF binding motif did not appear to impact insulator function. To support this finding, we deleted the CTCF motif(s) from three potent insulators to determine the effect of removing the known binding site on insulator activity. We found that for one construct, the deletion of the CTCF site completely ablated insulator activity. However, deleting the CTCF binding sequence from the other two sites had no significant effect on insulator activity (Supplemental Fig. S10). At these sites, CTCF may bind to DNA through an unknown motif or protein intermediate. It is also possible that another unknown protein or chromatin configuration provides insulator function at these sites.
Cell-type selective open chromatin harbors DNA sequence motifs corresponding to master regulators of cell identity
Regulatory elements far from the TSS have been shown, typically through single-gene experiments, to control tissue-specific and cell-type selective gene expression (Deal et al. 2006). Using the top 100K combined open chromatin sites, we used two approaches to characterize and discover DNA sequence motifs in cell-type-selective DNase-seq and FAIRE-seq sites located away from the TSS.
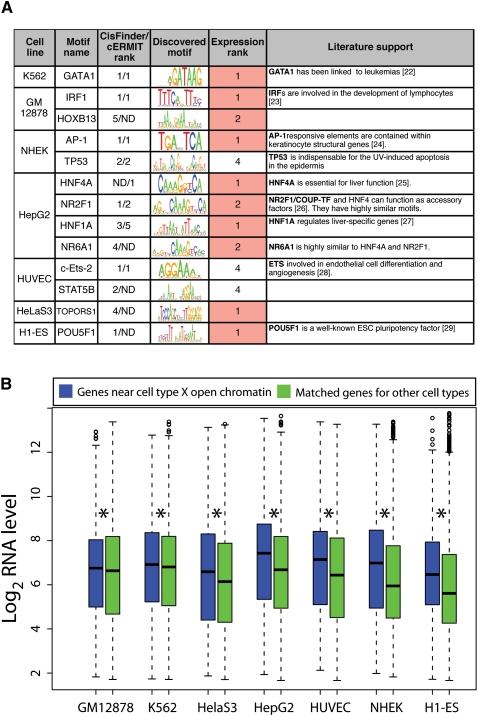
Known transcription factor binding motifs were identified genome-wide using publicly available position weight matrices (PWMs) from the Transfac database (Matys et al. 2006). For each cell type, we determined the top 10 PWMs that were significantly enriched in distal open chromatin sites specific to that single cell type (see Methods) (Supplemental Table S6). Among these are PWMs that match the binding specificities of factors known to function in processes that occur in the corresponding cell type. For example, the binding sites for IRF1 and IRF7 and the interferon-stimulated responsive element ISRE are enriched in lymphoblasts, which are interferon-responsive. Other examples include GATA family members, which regulate hematopoiesis, in K562; BACH1, necessary for DNA repair and linked to several cancers, in HelaS3; HNF1A and HNF4A, transcription factors critical for liver development, in HepG2; ELK1, involved in endothelial cell differentiation, in HUVEC; and PLXNA2 family members critical for pluripotency and SP1 in embryonic stem cells.
We also performed de novo motif finding using the cERMIT (Georgiev et al. 2010) and CisFinder (Sharov and Ko 2009). We analyzed the top five motifs returned from each cell line by each algorithm. Using the software STAMP (Mahony and Benos 2007), we searched for motifs in the Transfac (Matys et al. 2006), JASPAR (Bryne et al. 2008), UniPROBE (Newburger and Bulyk 2009), and hPDI (Xie et al. 2010) databases that corresponded to each discovered motif (P-value < 10−9) (Fig. 5A). Consistent with our analysis above, both cERMIT and CisFinder detected the GATA1 motif in K562, HNF1/4A motifs in HepG2, FOS and TP53 in NHEK, and ETS/ELK motifs in HUVEC. CisFinder identified a highly enriched POU5F1 motif in the embryonic stem cell line.
Figure 5.
Distal cell-type selective open chromatin contains functionally relevant motifs and is linked to cell-type specific expression. (A) Top motifs enriched (P-value < 1 × 10−9) in cell-type selective open chromatin. Expression rank reflects the transcription factor's expression level in that cell type relative to all other cell types. (B) Distribution of expression values for genes closest to distal cell-type selective open chromatin sites (>2 kb from a TSS) from each cell type (x-axis) for that cell type (blue box plots). Similar distributions were calculated for these genes in the six other cell types lacking the distal open chromatin sites (green box plots). Asterisk indicates significant difference (pairwise T-tests).
For each of the transcription factors corresponding to the discovered motifs, we determined their RNA expression levels in each of the seven cell types (Supplemental Table S7). In 10 of 13 instances, the cell type in which the motif was identified expressed that gene at the highest or second highest level among all seven cell types (Fig. 5A). In nearly all cases, the associated transcription factor has been functionally linked to the corresponding cell type (Fig. 5A; Hall et al. 1995; Nichols et al. 1998; Jessen et al. 2000; Taniguchi et al. 2001; Dejana et al. 2007; Shimizu et al. 2008; Bolotin et al. 2010). Both cERMIT and CisFinder also identified motifs that did not match any motifs in the databases we used—for example, a CCCCT motif in H1-ES (a stress-responsive element in yeast) and a CCAGCCTGG motif in HelaS3 cells, a core sequence in Alu repeats.
We repeated the above motif analyses in distal FAIRE-only or DNase-only sites and found biologically relevant motifs. For example, DNase-only sites are most enriched for the same motifs as the combined open chromatin set in K562, GM12878, NHEK, and HepG2 (GATA1, STAT1, AP-1, and HNF4A, respectively). Likewise, FAIRE-only sites are enriched for many of the same motifs detected in the combined open chromatin set in HepG2, HelaS3, and H1-ES (HNFs, PITX2, and POU5F1, respectively). In ES cells, the POU5F1 motif was found in FAIRE-only sites but not in DNase-only sites. Furthermore, not every motif found in the FAIRE-only or DNase-only sets matched those from the union set. For example, the K562 FAIRE-only sites are enriched for a well-defined NFE2L2 motif (Chen et al. 2010) that does not get picked up by DNase-only or the union set. In summary, the most highly enriched motifs are held in common between the assays, but each assay provides independent, biologically relevant information.
Open chromatin specific to a cell type occurs near genes that are expressed specifically in that cell type
If a substantial fraction of open chromatin sites function as positive regulators of transcription, one would expect increased expression of genes near open chromatin sites. To test this, we first identified distal cell-type selective sites present in only one of the tested cell types that were further than 2 kb from a TSS of a nearby gene. For each cell type, we calculated the average and median expression values of all genes mapped to each distal site. In all cases, expression levels of genes linked to distal cell-type selective sites for that cell type were significantly higher than in the remaining cell types (pairwise T-tests) (Fig. 5B). This was similarly true for six of seven cell types when considering proximal cell-type selective open chromatin sites within 2 kb of a TSS (Supplemental Fig. S11).
Cell-type selective open chromatin often occurs near genes that govern cellular identity and function
A number of cell-type selective open chromatin sites reside near genes encoding transcription factors that play critical roles in cell-type specific gene expression and function. In many cases, the motifs recognized by these proteins were themselves enriched in cell-type selective open chromatin (Fig. 5A). For example, in H1 embryonic stem cells, we identified a number of ES-cell specific open chromatin sites around the POU5F1 and NANOG genes, which are known to control pluripotency (Supplemental Fig. S12). The upstream NANOG open chromatin site was recently identified as a poised enhancer, which is consistent with our assays (Rada-Iglesias et al. 2010). Other notable examples include a K562 cell-type selective open chromatin site upstream of the GATA1 transcription factor gene (Supplemental Fig. S13) and a number of HepG2 cell-type selective open chromatin sites within and around the HNF4A transcription factor gene (Fig. 1B). In this latter case, we detect open chromatin at the two annotated TSSs (Fig. 1B), both of which are utilized in FT0-2B, another hepatocellular carcinoma cell line (Thomas et al. 2001). These are singular examples, but our chromatin atlas identifies tens of thousands of such putative regulatory elements in each of the cell types we have studied.
Clusters of open regulatory elements coordinate cell-type selective gene expression
Our analyses above focused on individual cell-type selective open chromatin sites and their relation to genes with a known function. However, we noticed regions in which multiple open chromatin sites in a given genomic region were coordinately present or absent across one or more cell types (Fig. 1B; Supplemental Figs. 12, 13). This was consistent with previous reports of clustered FAIRE sites called clusters of open regulatory elements (COREs) (Gaulton et al. 2010). To detect COREs systematically in our data set, we calculated pairwise correlations of open chromatin signals using data from all seven cell types (see Methods) (Fig. 6A,B). Using these correlations, we designed a hidden-Markov model to define 181 high-confidence COREs (see Methods) (Supplemental Table S9; Supplemental COREs figures; Supplemental COREs table). COREs varied in size from 32 kb to 6.6 Mb.
Figure 6.
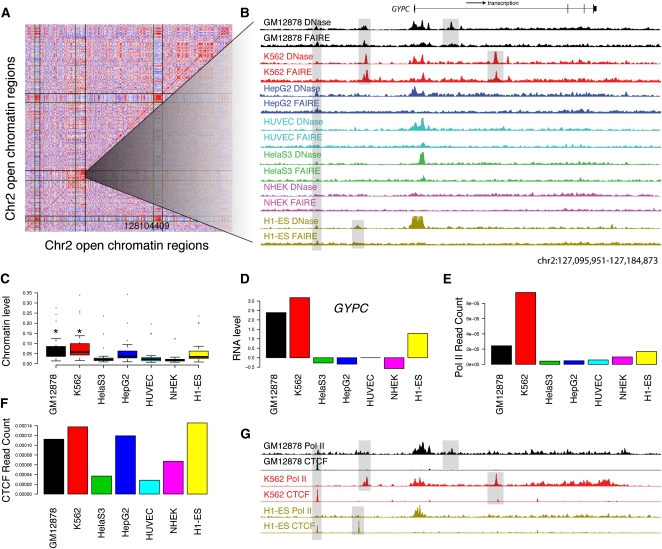
Open chromatin patterns form clusters of open regulatory elements (COREs). (A) Pairwise correlations between 500 open chromatin sites from chromosome 2 show three blocks of correlated sites (see Methods). Each row and column represents an open chromatin region found by both DNase-seq and FAIRE-seq in at least one of the seven cell types. Red indicates high correlation, white indicates no correlation, and blue indicates negative correlation. Vertical and horizontal lines show CORE boundaries. (B) DNase-seq (y-axis fixed at 0.1) and FAIRE-seq (y-axis fixed at 0.04) signals for a 90-kb subsection of CORE 98 containing the GYPC gene. GYPC is the only gene in this CORE. Highlighted are open chromatin sites found in all cell types, only GM12878 and K562 together, and GM12878 and K562 individually. (C) Boxplots show the distributions of open chromatin levels within open chromatin sites with CORE 98. GM12878 and K562 both have significantly higher levels of open chromatin (*; Mann-Whitney Wilcoxon rank sum test). (D) Relative expression levels (y-axis) of GYPC show increased expression in GM12878 and K562 cell lines. (E) Open chromatin sites within CORE 98 also show higher normalized Pol II ChIP-seq read counts in GM12878 and K562 cell types. (F) Normalized CTCF ChIP-seq read counts do not show significant differences between GM12878 and K562 and other cell types CORE98. (G) Pol II and CTCF signals in this 90-kb region (shown in B) provide preliminary annotations of similar and differential open chromatin sites.
We hypothesized that COREs represent coordinated nucleosome depletion events caused by multiple regulatory elements participating in the regulation of a nearby gene or genes. We determined in which cell line(s) each CORE was active using the Mann-Whitney Wilcoxon rank sum test (see Methods) (Fig. 6C). Ninety-five COREs (52%) had increased open chromatin within primarily one cell-type, while 78 COREs (43%) were characterized by increased open chromatin within at least two cell types (Supplemental Table S9). In the remaining eight COREs (5%), no cell type had significantly more open chromatin signal relative to the others, as defined by our threshold. We examined genes inside or within 10 kb of each CORE and found that, for 75 of the 114 COREs with at least one gene with expression data, the cell type with the highest expression also contained significantly more open chromatin (Supplemental Table S9). Among the 67 COREs not associated with any genes for which we had expression data, the aggregate Pol II signal was greatest in a cell type with significantly enriched open chromatin (Supplemental Table S9). Considering all COREs, the highest cumulative CTCF signal was in a cell type with enriched open chromatin 77% (140/181) of the time. These relations with expression and Pol II and CTCF binding also hold when COREs are analyzed in aggregate (Supplemental Fig. S14). In 18 of 175 COREs, open chromatin levels did not correspond to gene expression, CTCF binding, or Pol II binding. For these cases, we may not have correctly identified a distant target gene associated with the CORE, or the open chromatin regions may not be acting as enhancers.
To demonstrate the utility of identifying COREs, we show one typical CORE in detail (Fig. 6A). This CORE extends over 1.2 Mb, but GYPC is the only annotated gene found in this region, so we focused on the 90 kb surrounding the gene (Fig. 6B). GYPC encodes both glycophorin-C and glycophorin-D, which function in membrane stability of human erythrocytes and lymphocytes (Walker and Reid 2010). Open chromatin is detected at the GYPC TSS in nearly all cell types. However, nucleosome-depletion events unique to GM12878 and K562 occur ∼10 kb upstream of the TSS and within the transcribed region of GYPC (Fig. 6B). These are accompanied by increased FAIRE and DNaseI signal (Fig. 6C), expression of GYPC (Fig. 6D), and higher Pol II signal (Fig. 6E–G). Thus, this CORE is typical, showing coordinated nucleosome depletion, gene expression, and transcription factor binding, and identifies several key regions that may be responsible for the cell-type specific regulation of this gene.
Many other COREs reveal putative regulatory elements surrounding genes with cell-type specific functions. For example, CORE 70 defines a 600-kb region on chromosome 1 that contains several genes with specific functions in keratinocytes, including “late cornified envelope” (LCE) genes and small proline rich proteins (SPRR). Within CORE 70, NHEK cells exhibit significantly more nucleosome depletion, higher expression of these genes, greatly increased amounts of Pol II signal, and greater CTCF signal (Supplemental COREs figures). In other cases, unannotated genes within COREs can be associated with a specific cell type. For example, CORE 60 is a 325-kb region encompassing the sparsely annotated RNF152 (ring finger protein 152) gene. Increased nucleosome depletion, expression of this gene, and Pol II signal are observed specifically in HUVEC and NHEK cells (Supplemental COREs figures.).
Discussion
We produced maps of open chromatin in seven diverse human cell types using DNase-seq and FAIRE-seq. DNase-seq and FAIRE-seq are independent methods that provide strong cross-validation. Performing both assays on cells collected from the same culture in each replicate helps to ensure that the differences we observe are due to the assay specificities, rather than experimental variation. We present evidence that sites detected by a single assay are biologically relevant and functional. In each cell type, we identify 100,000–200,000 open chromatin regions covering 1%–2% of the genome.
Differences in DNase-seq and FAIRE-seq may be due to the specific regulatory complexes bound at each open chromatin site, which could affect the ability of DNaseI to cut or formaldehyde to crosslink. DNase-only sites tended to occur at transcription start sites while FAIRE-only sites were more often found in distal regions. It is possible that FAIRE cannot detect some nucleosome-depleted regions that are bound very tightly by nonhistone proteins if those complexes support a level of crosslinking similar to that of a nucleosome. However, FAIRE appears to capture chromatin structures away from promoters that the DNaseI enzyme cannot easily cut.
Most binding sites of regulatory proteins we examined were within open chromatin sites, suggesting that, in general, open chromatin sites are indicators of regulatory proteins operating in each cell type. Between any two cell types, 23%–48% of open chromatin sites are shared. Among the seven cell types, nearly 9% of the genome was identified as an open chromatin site by DNase-seq and/or FAIRE-seq, and the identification of open chromatin sites is not complete. Continued experimentation on new cell types is a cost-effective strategy to find new regulatory elements and determine the fraction of the genome associated with a regulatory function.
DNase-seq and FAIRE-seq cannot directly reveal the function of the identified nucleosome-depleted regions, or the regulatory proteins that are bound to them. ChIP-seq, such as we performed for Pol II, MYC, and CTCF, provides a degree of functional annotation, as does association with certain histone modifications. Motif enrichment can provide guidance for selecting specific transcription factors to confirm by ChIP. We and others have shown that DNase-seq identifies DNaseI footprints, which reveal the location and identity of a bound motif with great accuracy (Hesselberth et al. 2009; Boyle et al. 2010; Pique-Regi et al. 2010). DNaseI footprinting will be critical in inferring binding at individual open chromatin sites, but it cannot identify proteins that associate indirectly with DNA. As more proteins are mapped by ChIP-seq in relevant cell types, we can more fully annotate the open chromatin sites we report here.
Another challenge is mapping regulatory elements to the genes they regulate. Assuming that the nearest gene is the most likely target is clearly naïve, as demonstrated by counter-examples in the literature (i.e., Spilianakis et al. 2005). We showed that clusters of open regulatory elements defined by multiple sites spanning tens of thousands of bases show good correspondence between open chromatin and levels of gene expression, Pol II signal, and/or CTCF signal. Analysis suggests that COREs encompass noncoding DNA elements that act coordinately to regulate genes important for cell type identity and function. In many cases, the identification of COREs can guide candidate target gene selection, and methods like 3C, 4C, 5C, Hi-C, and ChIA-PET (van Steensel and Dekker 2010) will continue to be important in solving this difficult problem. Finally, low-throughput functional assays (similar to our insulator assays) will continue to be critical for understanding the biological activity of open chromatin sites, but assays that can test thousands of DNA segments in parallel will be required to make any significant progress in characterizing how these regulatory sites work together in a given biological context.
There is a nearly inexhaustible number of combinations of human cell types, genotypes, disease states, and environmental conditions. Genome-wide association studies (GWAS) have linked variation in numerous noncoding regions with different diseases (Gaulton et al. 2010; Hindorff et al. 2009). It is likely that many of these associations are due to polymorphisms that affect gene regulation. Our atlas can be used immediately to guide further characterization of regulatory elements that may be causally linked to disease risk (Gaulton et al. 2010; Stitzel et al. 2010). Furthermore, we have shown previously that identification of open chromatin sites in the same cell type derived from different individuals can identify individual-specific gene regulatory elements (McDaniell et al. 2010). Larger studies with hundreds or thousands of individuals will allow identification of connections between DNA sequence variation, chromatin organization, transcriptional regulation, and disease risk on a population level.
Methods
Cell culture
Vendor information and standard cell growth protocols can be found at the UCSC ENCODE site (http://genome.ucsc.edu/ENCODE/cellTypes.html) (see Supplemental Methods).
Experimental protocols
DNase-seq (Song and Crawford 2010), DNase-ChIP (Crawford et al 2006a; Shibata and Crawford 2009), FAIRE (Giresi et al. 2007; Giresi and Lieb 2009), ChIP (Bhinge et al. 2007; The ENCODE Project Consortium 2007), and the insulator/enhancer blocking (Bell et al. 1999) assays were performed as previously described with slight modifications (see Supplemental Methods). Exon arrays were processed following a standard protocol for the ENCODE Consortium (see Supplemental Methods).
Data processing
For sequence data from all experiments, (1) sequences were aligned to the human reference genome (NCBI Build 36) using MAQ (Li et al. 2008), and (2) filtered to remove artifacts, (3) replicates were compared for reproducibility, then combined, and (4) base-pair signal was generated using F-seq (Boyle et al. 2008b) and discrete peaks called (see Supplemental Methods). Gene-relative categories were defined as follows: (1) promoter: overlaps 2 kb upstream of any TSS; (2) 5′: overlaps first exon or first intron; (3) intragenic region: overlaps internal exon or intron; (4) 3′: overlaps last exon or 2 kb downstream from end of transcription; and (5) intergenic: not within any previous category. Sites were assigned to the first category whose criterion was satisfied. Cell type selective and ubiquitous open chromatin sites were calculated using the top 100K sites from each cell type. Combined union sets of DNase-seq and FAIRE-seq sites were created with significance calculated using Fisher's combined probability test (Fisher 1925). Affymetrix Exon 1.0 ST array data (available at GEO, GSE15805) was processed using the Affymetrix Expression Console (see Supplemental Methods).
Motif finding
Cell-type selective distal peaks were defined as present in a single cell type, at least 2 kb upstream of any TSS, and downstream from 5′ exons and introns. Enriched motifs were determined in two ways: (1) Motif scanning was performed using public transcription factor position weight matrices from Transfac (version 7.0; (Matys et al. 2006) with enrichment defined as the ratio of predicted binding site frequency per kb in peaks from one cell type vs. cell-type selective peaks in the other six cells, and significance determined using a χ2 test (P-value < 6.6 × 10−3); and (2) de novo motif finding was performed using cERMIT (Georgiev et al. 2010) and CisFinder (Sharov and Ko 2009). For cERMIT, the combined open chromatin –log(P-value) was used as experimental evidence. The union set of cell-type selective peaks from the other six cell lines served as the background. The online version of CisFinder (http://lgsun.grc.nia.nih.gov/CisFinder/) was used with default parameter settings, except “clustered” motifs rather than the “elementary” ones. FASTA files of cell-type selective distal peaks were the foreground set and the union set of distal peaks from the other six cell lines was the background set. Top motifs from cERMIT and CisFinder were annotated using the STAMP web server (http://www.benoslab.pitt.edu/stamp/ [Mahony and Benos 2007]), with the “selected eukaryotic” option, and included Transfac (Matys et al. 2006), JASPAR 2010 (Portales-Casamar et al. 2009), UniPROBE (http://the_brain.bwh.harvard.edu/uniprobe), and a few organism-specific databases. Additional comparisons were done to the hPDI database, (Xie et al. 2010) using a custom PWM file, and to the “predicted human” database in STAMP (Mahony and Benos 2007).
Clusters of open regulatory elements
Open chromatin sites found by both DNase-seq and FAIRE-seq in at least one cell type were considered. Each site was represented by a vector of seven combined open chromatin values, one for each cell type and quantile-normalized across cell types. Pairwise correlations were calculated between each site and 500 surrounding sites. One hundred and eighty-one high-confidence COREs were defined by a two-state HMM, using the average correlation across five adjacent open regions as the observable. Transition and emission probabilities were set manually. In each CORE, cell types with significantly more open chromatin signal were determined using pairwise Mann-Whitney tests (P-value < 0.05 vs. at least four cell types). Genes (UCSC knownGene annotation) overlapping and within 10 kb were assigned to each CORE for expression comparisons. CTCF and Pol II ChIP-seq signal were calculated as normalized sequence read counts mapped inside COREs. To eliminate background signals, only reads located within the top 20K Pol II or top 10K CTCF peaks were considered.
Data access
All data from this atlas are publicly available on the UCSC Genome Browser (http://genome.ucsc.edu; [Kent et al. 2002]), the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, GSE30227 [Edgar et al. 2002]), and the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/, SRP007348, SRP007349, SRP007350, SRP002002, SRP004453 [Wheeler et al. 2008]).
Acknowledgments
We would like to thank Lisa Bukovnik, Tonya Severson, and Fangfei Ye from the Duke Sequencing Core facility; Sridar Chittur, Marcy Kuentzel, and Scott Tenenbaum for RNA expression data; and Joe Lucas for help with exon array expression analysis.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121541.111. Freely available online through the Genome Research Open Access option.
References
- Bell AC, West AG, Felsenfeld G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bhinge AA, Kim J, Euskirchen GM, Snyder M, Iyer VR 2007. Mapping the chromosomal targets of STAT1 by Sequence Tag Analysis of Genomic Enrichment (STAGE). Genome Res 17: 910–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM 2010. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology 51: 642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE 2008a. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS 2008b. F-Seq: A feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS 2010. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res 21: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Lin WC, Kuo CN, Lu FJ 2010. Role of redox signaling regulation in propyl gallate-induced apoptosis of human leukemia cells. Food Chem Toxicol 49: 494–501 [DOI] [PubMed] [Google Scholar]
- Cockerill PN 2011. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J 19: 2182–2210 [DOI] [PubMed] [Google Scholar]
- Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS 2006a. DNase-chip: A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods 3: 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. 2006b. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 16: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM 2006. Distant regulatory elements in a Sox10-βGEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev Dyn 235: 1413–1432 [DOI] [PubMed] [Google Scholar]
- Dejana E, Taddei A, Randi AM 2007Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim Biophys Acta 1775: 298–312 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2011. A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol 9: e1001046 doi: 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA 1925. Statistical methods for research workers. Oliver and Boyd, Edinburgh, Scotland, UK [Google Scholar]
- Frietze S, Lan X, Jin VX, Farnham PJ 2010. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J Biol Chem 285: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 36: 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. 2010. A map of open chromatin in human pancreatic islets. Nat Genet 42: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev S, Boyle AP, Jayasurya K, Ding X, Mukherjee S, Ohler U 2010. Evidence-ranked motif identification. Genome Biol 11: R19 doi: 10.1186/gb-2010-11-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD 2009. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods 48: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT 1988. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57: 159–197 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RK, Sladek FM, Granner DK 1995. The orphan receptors COUP-TF and HNF-4 serve as accessory factors required for induction of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Proc Natl Acad Sci 92: 412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD 2006. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet 2: e158 doi: 10.1371/journal.pgen.0020158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen BA, Qin Q, Rice RH 2000. Functional AP1 and CRE response elements in the human keratinocyte transglutaminase promoter mediating Whn suppression. Gene 254: 77–85 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Song SH, Brand M, Dean A 2007. Nucleosome and transcription activator antagonism at human beta-globin locus control region DNase I hypersensitive sites. Nucleic Acids Res 35: 5831–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, et al. 2010. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet 6: e1001065 doi: 10.1371/journal.pgen.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Benos PV 2007. STAMP: A web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35 (Web Server issue): W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, et al. 2010. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motallebipour M, Ameur A, Reddy Bysani MS, Patra K, Wallerman O, Mangion J, Barker MA, McKernan KJ, Komorowski J, Wadelius C 2009. Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq. Genome Biol 10: R129 doi: 10.1186/gb-2009-10-11-r129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML 2009. UniPROBE: An online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res 37: D77–D82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK 2010. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A 2009. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Ko MS 2009. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res 16: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Crawford GE 2009. Mapping regulatory elements by DNaseI hypersensitivity chip (DNase-Chip). Methods Mol Biol 556: 177–190 [DOI] [PubMed] [Google Scholar]
- Shimizu R, Engel JD, Yamamoto M 2008. GATA1-related leukaemias. Nat Rev Cancer 8: 279–287 [DOI] [PubMed] [Google Scholar]
- Song L, Crawford GE 2010. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SC, Boyle AP, Scott LJ, et al. 2010. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab 12: 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19: 623–655 [DOI] [PubMed] [Google Scholar]
- Thomas H, Jaschkowitz K, Bulman M, Frayling TM, Mitchell SM, Roosen S, Lingott-Frieg A, Tack CJ, Ellard S, Ryffel GU, et al. 2001. A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum Mol Genet 10: 2089–2097 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Dekker J 2010. Genomics tools for unraveling chromosome architecture. Nat Biotechnol 28: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PS, Reid ME 2010. The Gerbich blood group system: A review. Immunohematol 26: 60–65 [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. 2008. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36: D13–D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC 1979. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell 16: 797–806 [DOI] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. 2007. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136 doi: 10.1371/journal.pgen.0030136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Hu S, Blackshaw S, Zhu H, Qian J 2010. hPDI: A database of experimental human protein-DNA interactions. Bioinformatics 26: 287–289 [DOI] [PMC free article] [PubMed] [Google Scholar]