Abstract
Objectives
Regulation of gene expression is important for the development and function of the nervous system. However, the transcriptional programs altered in psychiatric diseases are not completely characterized. Human gene association studies and analysis of mutant mice suggest that the transcription factor specificity protein 4 (SP4) may be implicated in the pathophysiology of psychiatric diseases. We hypothesized that SP4 levels may be altered in the brain of bipolar disorder (BD) subjects and regulated by neuronal activity and drug treatment.
Methods
We analyzed messenger RNA (mRNA) and protein levels of SP4 and SP1 in the postmortem prefrontal cortex and cerebellum of BD subjects (n = 10) and controls (n = 10). We also examined regulation of SP4 mRNA and protein levels by neuronal activity and lithium in rat cerebellar granule neurons.
Results
We report a reduction of SP4 and SP1 proteins, but not mRNA levels, in the cerebellum of BD subjects. SP4 protein and mRNA levels were also reduced in the prefrontal cortex. Moreover, we found in rat cerebellar granule neurons that under non-depolarizing conditions SP4, but not SP1, was polyubiquitinated and degraded by the proteasome while lithium stabilized SP4 protein.
Conclusions
Our study provides the first evidence of altered SP4 protein in the cerebellum and prefrontal cortex in BD subjects supporting a possible role of transcription factor SP4 in the pathogenesis of the disease. In addition, our finding that SP4 stability is regulated by depolarization and lithium provides a pathway through which neuronal activity and lithium could control gene expression suggesting that normalization of SP4 levels could contribute to treatment of affective disorders.
Keywords: bipolar disorder, cerebellar granule neurons, lithium, postmortem cerebellum, postmortem prefrontal cortex, SP4/SP1 transcription factor
Despite the personal and social burden of bipolar disorder (BD), the etiology and the pathogenesis underlying this condition are not completely understood. Structural abnormalities in brain areas of BD subjects have been reported in magnetic resonance imaging studies, including alterations in the cerebellum and prefrontal cortex (1–4). An increasing body of evidence indicates that, in addition to its classical motor coordination function, the cerebellum is also involved in psychological alterations and in the modulation of affective, cognitive, and perceptual functions (5–10). Morphologic abnormalities in cerebellum and medial temporal limbic structures, as well as alterations that disrupt the anterior limbic network and the prefrontal-striatal-thalamic pathways have been observed in BD (11, 12). Furthermore, a diffusion tensor imaging study in BD has reported white matter abnormalities along the tracts communicating such areas, a finding consistent with a model of BD that implicates dysregulation of the cortico-subcortical and cerebellar regions (13). Taken together, this evidence suggests that alterations in brain structures including the cerebellum and prefrontal cortex may contribute to the cognitive and psychotic symptoms of the disease. Although several molecular alterations have been identified in the prefrontal cortex of BD patients, little is known about the molecular changes that occur in the cerebellum (14–18).
Treatment with lithium has been successfully used as a mood stabilizer in BD for decades (19). Currently, lithium is the most efficient drug for long-term preventive treatment and it also has an anti-suicidal effect (20). Notably, the therapeutic effects of lithium appear after weeks of treatment (19). This latency of response may be due to changes in gene expression that could contribute to long-term cellular remodeling. Lithium is known to inhibit glycogen synthase kinase 3 (GSK-3) and inositol monophosphatase (IMPase), leading to the activation of β-catenin dependent-transcription and blockage of the phosphatydilinositol cycle due to the depletion of inositol, respectively (21–23). In addition, other signaling pathways have been found to be regulated by lithium dependent or independent of GSK-3 and/or IMPase, e.g., cyclin-dependent kinase-5, protein phosphatase 1, protein phosphatase 2A, and MEK/ERK (24–27). Furthermore, changes in gene expression in rodent brain upon lithium treatment revealed its regulation of many cellular processes (28, 29). Nonetheless, the transcriptional programs and neuronal processes regulated by lithium in psychiatric diseases are not fully understood.
Transcription factor specificity protein 4 (SP4) and the closely related factor SP1, have recently been reported to be involved in psychiatric diseases. Polymorphisms in the genomic locus of human SP4 have been associated with major depression (30, 31), schizophrenia and BD (32); and rare copy number variants and deletions in the SP4 gene have been associated with schizophrenia (33, 34). Altered SP1 messenger RNA (mRNA) levels have also been reported in lymphocytes and some brain areas of patients with schizophrenia (35). Sp4 is broadly expressed in neurons of the central nervous system with higher levels in the cerebellum and hippocampus (36, 37), while SP1 is expressed ubiquitously in the organism. SP4 and SP1 regulate expression of genes implicated in a variety of biological processes including neuronal development and function (38, 39). We have previously reported that the transcription factor SP4 is required for activity-dependent dendritic pruning and limiting of branching during neuronal morphogenesis (40, 41). Further, mice with reduced expression of SP4 exhibit behavioral abnormalities associated with psychiatric diseases, such as defects in prepulse inhibition, which are reversed upon restoration of SP4 levels (42).
Although a role for specificity protein transcription factors in psychiatric disorders is becoming appreciated, little is known about how SP4 and SP1 protein levels are regulated in brains of subjects with psychiatric disorders. Here we report that protein, but not mRNA, levels of SP4 are reduced in the postmortem cerebellum of BD patients. In addition, SP4 protein and mRNA levels are reduced in the prefrontal cortex. We show that in rat cerebellar granule neurons the SP4 protein is destabilized in the absence of membrane depolarization and we also show that, under these conditions, SP4 is partially stabilized by lithium. The finding that SP4 protein is downregulated in the brains of BD subjects raises the possibility that alterations in the neuronal transcription factor SP4 may contribute to the pathogenesis of BD. Our study also provides a mechanism by which depolarization controls SP4 protein levels. In addition, we suggest that the stabilization of SP4 protein by lithium treatment could lead to changes in gene expression and, hence the normalization of SP4 levels could be a relevant pharmacological strategy in the treatment of mood disorders.
Methods and materials
Brain tissue samples
Postmortem human brain tissue from the cerebellum and prefrontal cortex of subjects with BD and healthy controls was obtained from the Brain Tissue Collection of the University of Basque Country, Bizkaia, Spain (see Table 1 for detailed demographic data). The study was developed in compliance with policies of research and ethical review boards for postmortem brain studies at the Basque Institute of Legal Medicine, Bilbao, Spain, and samples were obtained at autopsy by forensic pathologists. All deaths were subjected to retrospective analysis for previous medical diagnosis and treatment.
Table 1.
Demographic characteristics of subjects with bipolar disorder (BD) (n = 10) and controls (n = 10)
| Paired subjects | Gender/Age (yrs) | Postmortem delay (hrs) | Cause of death | Treatmenta | ||||
|---|---|---|---|---|---|---|---|---|
| BD | Control | BD | Control | BD | Control | BD | Control | |
| 1 | M/72 | M/72 | 22 | 43 | Suicide | Accident | Drug free | Drug free |
| 2 | F/44 | F/49 | 19 | 40 | Suicide | Accident | APS, BZD, mirtazapine | Drug free |
| 3 | M/58 | M/54 | 10 | 23 | Suicide | Accident | APS, lamotrigine | BZD |
| 4 | M/40 | M/43 | 17 | 11 | Suicide | Accident | BZD | Drug free |
| 5 | M/27 | M/30 | 10 | 11 | Suicide | Accident | Drug free | Cannabis |
| 6 | M/63 | M/60 | 8 | 4 | Suicide | Natural | BZD | Drug free |
| 7 | M/64 | M/61 | 23 | 23 | Suicide | Accident | BZD, venlafaxine | Drug free |
| 8 | M/57 | M/55 | 22 | 22 | Natural | Natural | Drug free | Drug free |
| 9 | M/63 | M/67 | 31 | 19 | Natural | Natural | Drug free | NSAID |
| 10 | F/73 | F/74 | 3 | 19 | Suicide | Accident | Drug free | NSAID |
M = male; F = female; APS = antipsychotics; BZD = benzodiazepines; NSAID = nonsteroidal anti-inflammatory drug. BD group (mean ± SD): age: 56 ± 15 yrs; postmortem delay (PMD): 16 ± 9 hrs; pH in the cerebellum (CB): 6.74 ± 0.24; pH in the prefrontal cortex (PFC): 6.59 ± 0.14; RNA integrity number (RIN) in the CB: 7.52 ± 0.46; RIN in the PFC: 6.40 ± 1.03. Control group (mean ± SD): age: 56 ± 14 yrs; PMD: 21 ± 12 hrs; pH in the CB: 6.77 ± 0.26; pH in the PFC: 6.92 ± 0.32; RIN in the CB: 7.90 ± 0.53; RIN in the PFC: 6.85 ± 0.40.
Treatment indicates results of toxicology analysis performed by the National Institute of Toxicology, Madrid, Spain.
A total of 20 brains from subjects who died by suicide (n = 8), accident (n = 7), or natural causes (n = 5) were selected. Subjects with antemortem criteria for BD according to the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) (8 suicide victims and 2 non-suicide subjects) were matched to control subjects who died by accidental or natural causes in a paired design, based on gender, age, and post mortem delay (PMD). All cases were subjected to a retrospective search for previous medical diagnoses and treatments by using medical examiner’s information and records. Toxicological screening for antipsychotics, antidepressants, and other drugs such as lithium was performed at the National Institute of Toxicology, Madrid, Spain.
Specimens of the cerebellum and dorsolateral prefrontal cortex were dissected extending from the pial surface to white matter, taking special care to include only gray matter. No differences were observed between BD and control subjects for tissue integrity parameters [pH and RNA integrity number (RIN)]. Samples were immediately stored at −70°C until homogenization.
Cerebellar granule neuron culture
Cerebellar granule neurons were obtained from postnatal day 6 rat pups and cultured as previously described (40). Animal experimentation protocol was approved by the Institutional Animal Care and Use Committees of the University of Barcelona, Barcelona, Spain, and Harvard Medical School and Tufts University School of Medicine, Boston, MA, USA. For the biochemical experiments, cerebellar granule neurons were switched to basal essential medium with 5 mM KCl or 25 mM KCl with or without 10% fetal calf serum at the indicated days in vitro (DIV). In some experiments, cells were treated with 10 μM or 100 μ MG132 (Calbiochem), 80 μM z-VAD-fmk (Calbiochem), LiCl, or DMSO for one hour before and after switching the medium. Granule neurons were treated with Cycloheximide (Calbiochem) 50 μg/ml where indicated. Granule neurons were pretreated with GSK-3 inhibitors or 5 mM LiCl for five hours before and one hour after switching to basal essential medium plus 10% fetal calf serum with 25 mM KCl or 5 mM KCl.
Protein extraction
Human brain samples were dissected from frozen brain slices. A sample of 100 mg of tissue was homogenized on ice in a glass douncer with lysis buffer [50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 2 mM EDTA, 2 mM EGTA, 10 mM Na-β-glycerophosphate, 5 mM sodium pyrophosphate, 1 mM Na3VO4, 1% β-mercaptoethanol, 1 mM phenylmethylsulphonylfluoride, 50 mM NaF, 25 mM N-ethylmaleimide, and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA)]. Protein lysates from rat cerebellar granule neurons were extracted in the lysis buffer described above. Protein extracts were incubated on ice for 30 min, sonicated and centrifuged at 18,200 g for 15 min at 4oC. Protein concentration was determined using the Bradford assay (BioRad).
Immunoblotting
Equal amount of total protein extracts (50 μg) were resolved by SDS/PAGE electrophoresis and immunoblotted with antibody against SP4 (Santa Cruz Technology), SP1 (Upstate), glyceraldehyde-3 phosphate dehydrogenase (GAPDH) (Chemicon International), PHF-1 that recognizes phosphorylated tau (provided by Dr. P. Davies, Albert Einstein College of Medicine, New York, NY, USA) or ubiquitinylated proteins (BioMol International). Densitometric quantification of Sp proteins was performed using Image NIH software and Quantity One software (BioRad) in duplicate samples. Values were normalized to GAPDH and referred to a standard sample (healthy subject) to normalize between different sets of samples and between repetitions.
Real-time quantitative RT-PCR (RT-qPCR)
Total RNA was extracted using Trizol reagent (Sigma-Aldrich or Qiagen). First strand cDNA was synthesized using SuperScript III (Invitrogen) in human samples or iScript™ cDNA Synthesis kit (BioRad) for granule neurons from the initial 2 μg of RNA. For human brain samples, Applied Biosystem Taqman master mix formulation for gene expression and probe and primers were used for quantitative real-time PCR (Applied Biosystem). Assay identification primers for target genes were Hs00162095_m1 for SP4 and Hs00916521_m1 for SP1. Assay identification primers for reference genes were Hs00183533_m1 for importin 8 (IP08), Hs00265254_m1 for fibroblast growth factor 1 (FGF1), and Hs00427620_m1 for TATA-binding protein (TBP). Beta-glucuronidase (GUSB) and beta-2-microglobuline (B2M) were also used as reference genes (Applied Biosystem). Amplification was performed on an Applied Biosystem model 7500 real-time PCR system. For cerebellar granule neurons, the following forward and reverse primers were used: SP4 (forward: AGCGATCAGAAGAAGGAGGAG; reverse: TCCTGGTGAAAATCAAGCAAC), Gapdh (forward: ATGACCACAGTCCATGCCATC; reverse: CCAGTGGATGCAGGGATGATGTTC). RT-qPCR products were monitored by quantification of SYBR Green I fluorescence using an iQ5 Real-Time PCR detection system (BioRad).
The relative quantification of SP4 and SP1 expression in each experimental condition was performed using the comparative cycle threshold (ΔΔCT) method. Briefly, for cerebellar granule neurons, the CT of SP4 was calibrated against that of the housekeeping gene GAPDH (CTSP4 − CTGAPDH = ΔCT). Then, ΔCT from each condition was subtracted from ΔCT from KCl 25 mM, fold change = 2−ΔΔCT. In human brain samples, the fold change was determined from the following calculations: ΔCT = CTGENE (sample) − CTGENE (healthy subject). Then, the fold change = 2−ΔCT(GENE)/2−ΔCT(NF), where NF is the normalization factor. Partial fold change (2−ΔCT) was calculated for a reference gene panel (TBP, GUSB, B2M, FGF1, IP08). After performing a stability analysis with GeNorm and NormFinder softwares (43, 44) (Supplementary Fig. 1), a normalization factor of a geometric mean of three genes was selected for the cerebellum: GUSB, TBP, and FGF1; and for the prefrontal cortex: GUSB, TBP, and B2M.
Statistical analysis
Nonparametric statistical techniques were chosen due to the lack of normality of the analyzed variables for the analysis of human samples. Association between SP4 or SP1 levels and BD were computed with the Wilcoxon signed rank test for paired samples. Association analysis of other factors such as age, gender, PMD, and the presence of psychotropic drugs was controlled by testing bivariate associations between SP4, SP1, and other variables of interest. These associations were carried out according to the type of the data, using the test of Kruskal-Wallis for more than two categories group, Mann-Whitney test for two groups, and the r of Spearman correlation when both variables were quantitative. Statistical differences in rat cerebellar granule neuron experiments were determined by Student t-test or ANOVA. For every condition, quantitation was performed in at least three independent experiments with similar results. Statistical analysis was performed with GraphPad Prism version 5.00, with the significance level set to 0.05.
Results
SP1 and SP4 proteins are downregulated in postmortem cerebellum of patients with BD
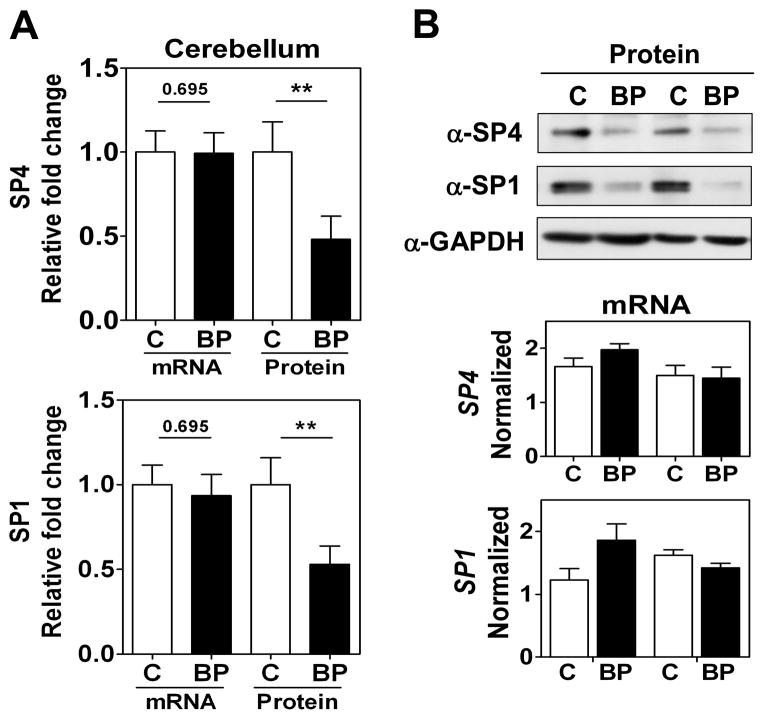
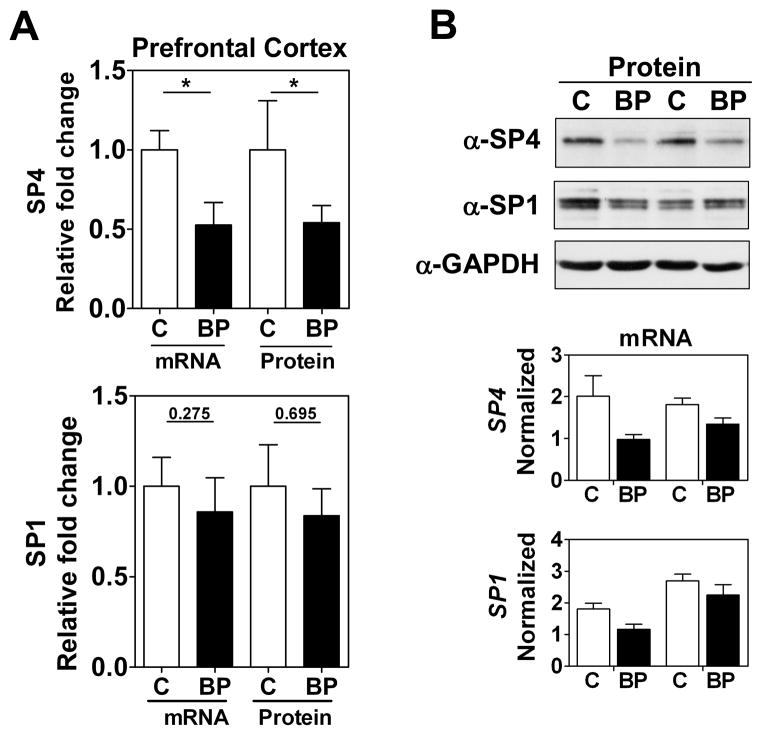
We focused our study on SP4 and SP1 proteins from the postmortem brain tissue of BD subjects. We analyzed SP4 protein levels in the postmortem cerebellum and prefrontal cortex of 10 BD subjects and 10 matched healthy controls (Table 1). We observed that SP4 protein levels were significantly decreased in both the cerebellum (Figs. 1A and 1B, top) and prefrontal cortex (Figs. 2A and 2B, top) of BD subjects. In contrast, SP4 mRNA levels were not altered in the cerebellum of BD subjects compared to controls; while in the prefrontal cortex, SP4 mRNA levels were reduced in BD subjects (Figs. 1A top, 1B bottom and 2A top, 2B bottom). In addition, we also analyzed the closely related transcription factor SP1 in the cerebellum and prefrontal cortex. SP1 protein, but not mRNA, levels were also significantly reduced in the cerebellum, while in the prefrontal cortex SP1 was not significantly altered (Figs. 1 and 2). In these human samples, we observed a range in Sp protein levels that overlapped in both areas between the BD and control groups (Supplementary Fig. 2). Association analysis of other variables in the study revealed that the PMD interval and RIN values correlated with reduced SP4 protein levels and mRNA levels in the prefrontal cortex, respectively (Table 2). However, our comparison analysis of paired healthy and BD samples with similar PMDs and the absence of significant difference in RIN values between groups excludes the influence of these confounding variables on SP4 levels. These results demonstrate that SP4 transcription factor is downregulated in the cerebellum and prefrontal cortex of BD subjects, and indicate that the primary mechanism leading to reduced SP4 levels in the cerebellum is post-transcriptional.
Fig. 1.
SP4 and SP1 protein levels are reduced in postmortem cerebellum in bipolar disorder (BD) subjects. (A) Protein and mRNA levels for SP4 (top) and SP1 (bottom) in the cerebellum. Protein extracts from cerebellar postmortem tissue of healthy individuals (C, n = 10), and individuals with BD (n = 10) were immunoblotted for SP4, SP1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The resultant bands were quantified by densitometry. SP4 and SP1 were normalized to GAPDH values and referred to a standard sample (healthy subject). Each value represents the mean of two independent analyses. mRNA levels for SP4 and SP1 from the same subject samples were determined RT-qPCR and normalized to a reference healthy control sample and the geometric mean of three reference genes: beta glucuronidase (GUSB), TATA-binding protein (TBP), and fibroblast growth factor 1 (FGF1). Each value represents the mean of at least three independent analyses performed in duplicate. Statistical analysis was performed using Wilcoxon signed rank test for paired values (**p < 0.01). (B) Representative Western blots images for SP4, SP1, and GAPDH from four individuals (top), and representative RT-qPCR data for the same four individuals (bottom).
Fig. 2.
SP4, but not SP1, protein levels are reduced in postmortem prefrontal cortex from bipolar disorder (BD) subjects. (A) Protein and mRNA levels for SP4 (top) and SP1 (bottom) in the prefontal cortex. Protein extracts from prefrontal cortex postmortem tissue of healthy individuals (C, n = 10), and individuals with BD (n = 10) were immunoblotted for SP4, SP1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The resultant bands were quantified by densitometry and SP4 and SP1 were normalized to GAPDH values and referred to a standard sample (healthy subject). Each value represents the mean of two independent analyses. mRNA levels for SP4 and SP1 from the same subject samples were determined by RT-qPCR and normalized to a reference healthy control sample and the geometric mean of three reference genes: beta glucuronidase (GUSB), TATA-binding protein (TBP) and beta-2-microglobuline (B2M). Each value represents the mean of at least three independent analyses performed in duplicate. Statistical analysis was performed using Wilcoxon signed rank test for paired values (**p < 0.01). (B) Representative Western blot images for SP4, SP1, and GAPDH from four individuals (top), and representative RT-qPCR data for the same four individuals (bottom).
Table 2.
Association analysis of other variables in the study (n = 22)
| Cerebellum | Prefrontal cortex | |||
|---|---|---|---|---|
| SP4 (protein) | SP1 (protein) | SP4 (protein) | SP4 (mRNA) | |
| Age (years) | ||||
| Spearman’s r | 0.2536 | −0.144 | −0.420 | −0.252 |
| p-value | 0.2807 | 0.546 | 0.065 | 0.284 |
| Gender (male/female) | ||||
| Mann-Whitney’s p-value | 0.6707 | 0.3211 | 0.6707 | 0.2376 |
| PMD (hours) | ||||
| Spearman’s r | −0.281 | −0.432 | 0.618 | 0.073 |
| p-value | 0.230 | 0.057 | 0.004 | 0.761 |
| Toxicology | ||||
| Kruskal-Wallis p-value | 0.9583 | 0.8581 | 0.2947 | 0.8562 |
SP4 and SP1 protein levels in the cerebellum and prefrontal cortex were compared to subjects’ age, gender, postmortem delay (PMD), and toxicology in order to detect if any of these variables was influencing the data. RNA integrity number was also analyzed as a possible variable influencing SP4 mRNA in the prefrontal cortex (r = 0.597, p = 0.005). Correlation among quantitative variables was analyzed by r of Spearman. Qualitative variables were analyzed by Mann-Whitney’s test for two groups and Kruskal-Wallis for more than three groups.
SP4 protein stability is regulated by neuronal activity in cerebellar granule neurons
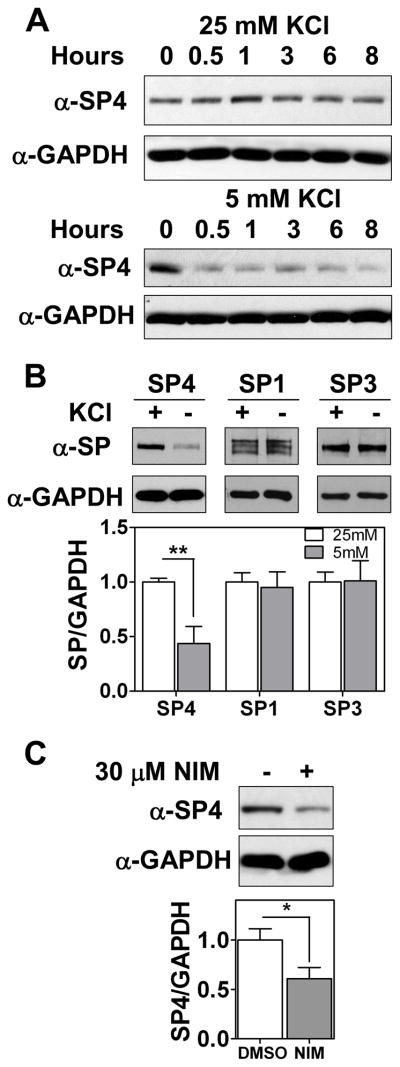
To further investigate the mechanisms by which SP4 protein levels are regulated in the cerebellum, we used primary cultures of rat cerebellar granule neurons. To investigate whether SP4 is directly regulated by neuronal depolarization, SP4 protein levels were monitored in a time course upon potassium depletion. SP4 levels were reduced to 50% within as little as 30 min, in both the presence and absence of serum (Fig. 3A and Supplementary Fig. 3A), at a time when viability of the neurons was unaffected (Supplementary Fig. 3B). Furthermore, levels of SP4, but not the related SP1 or SP3 proteins, decreased within one hour of switching cerebellar granule neurons from depolarizing (25 mM) to non-depolarizing (5 mM) concentrations of potassium chloride (Fig. 3B). L-type voltage sensitive calcium channels (L-VSCC) are major effectors of calcium influx upon membrane depolarization in cerebellar granule neurons (45). Pharmacological inhibition of L-VSCC with nimodipine led to a delayed reduction of SP4 levels in cultured cerebellar granular neurons at 12 hours and in cerebellar slices from P9 pups at 24 hours (Fig. 3C and Supplementary Fig. 4). Shorter incubations with nimodipine in dissociated cerebellar granule neuron culture did not show a decrease in SP4 levels (data not shown). These studies reveal that SP4 protein levels are regulated in response to membrane depolarization.
Fig. 3.
SP4 protein stability is regulated by depolarization. Cerebellar granule neurons were obtained from P6 rat pups and maintained in culture for 7 days in vitro (DIV-7). (A) Neurons were incubated in medium with 25 mM or 5 mM KCl in the presence of serum for the indicated times. Protein extracts were probed with antisera against SP4 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (B) Granule neurons were incubated in medium with 25 mM or 5 mM KCl in the presence of serum for one hour. Protein extracts were resolved by SDS/electrophoresis and probed with a polyclonal antibody against Sp proteins or GAPDH as indicated. Sp protein levels were analyzed by densitometry and normalized to GAPDH levels. Values represent mean ± standard deviation. (C) At DIV-7, cerebellar granule neurons were incubated with 30 μM nimodipine or the vehicle (DMSO) for 12 hours. SP4 protein levels were analyzed by densitometry and normalized to GAPDH levels. Values represent mean ± standard deviation of three independent experiments. Statistical analysis was performed using two-tailed t-test (*p < 0.05; **p < 0.01).
SP4 protein is polyubiquitinated and degraded by the proteasome in non-depolarizing conditions
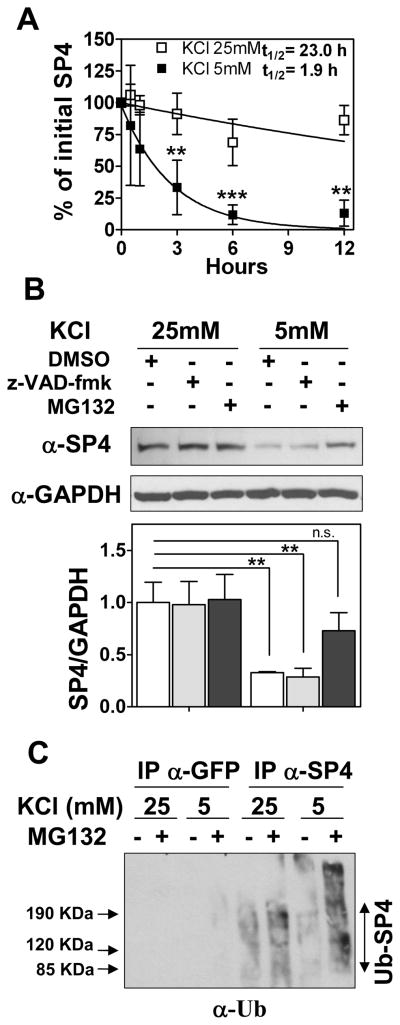
In order to determine the basis for the rapid change in SP4 protein levels, we determined the half-life of SP4 under depolarizing and non-depolarizing conditions in cerebellar granular neurons. We observed that the half-life of SP4 under depolarizing conditions was approximately 23 hours, while SP4 half-life was reduced to 1.8 hours in non-depolarizing conditions (Fig. 4A). We used pharmacological inhibitors to investigate whether proteolytic cleavage of SP4 contributes to the reduced stability of SP4 under non-depolarizing conditions. Addition of the caspase inhibitor z-VAD-fmk did not prevent SP4 degradation, but high levels of SP4 protein were maintained in non-depolarizing conditions in the presence of the proteasome inhibitor MG132 (Fig. 4B). Similar results were obtained in the presence of serum and with lower concentrations of MG132 (10 μM) (Supplementary Fig. 5). Consistent with the regulation of SP4 by the ubiquitin/proteasome pathway, when endogenous SP4 was immunoprecipitated from cerebellar granule neurons treated with MG132, we observed an increase in high molecular weight ubiquitinated proteins in the SP4 immunoprecipitate in non-depolarizing conditions that were also positive for SP4 immunodetection (Fig. 4C and data not shown). Together, these results demonstrate that in non-depolarizing conditions SP4 is rapidly degraded by the ubiquitin proteasome pathway.
Fig. 4.
SP4 is polyubiquitinated and degraded via the proteasome in non-depolarizing conditions. (A) Cerebellar granule neurons were incubated in media containing serum and 50 μg/ml cycloheximide and 25 mM (open symbols) or 5 mM (closed symbols) KCl. Cell lysates were prepared at the indicated times and immunoblotted with anti-SP4 antisera. Quantitation was performed by densitometry and half-life (log10 50%) of SP4 was estimated by adjusting the percentage of initial SP4 percentage values to an exponential decay curve. Values represent the mean ± standard deviation of four independent experiments. Statistical analysis was performed using ANOVA and Tukey post-hoc test (***p < 0.001; **p < 0.01). (B) Granule neurons were exposed to control vehicle (DMSO), 100 μM MG-132 (proteasome inhibitor), or 80 μM z-VAD-fmk (broad range caspase inhibitor) one hour before and after changing into a culture medium with 25 mM or 5 mM KCl in the absence of serum. After one hour of incubation, cells were lysed and protein extracts were immunoblotted with antibody against SP4 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Values represent mean ± standard deviation of three independent experiments. Statistical analysis was performed using two-tailed t-test (**p < 0.01; n.s = not significant). (C) Granule neurons were incubated and treated with and without 10 μM MG-132 as in (B). Cell lysates were subjected to immunoprecipitation with antisera against SP4 or GFP. Immunoprecipitates were probed with an antibody against ubiquitin (α-Ub). An increase in high molecular weight species in the presence of MG132, particularly in low KCl, was also detected in anti-SP4 immunoblots (data not shown).
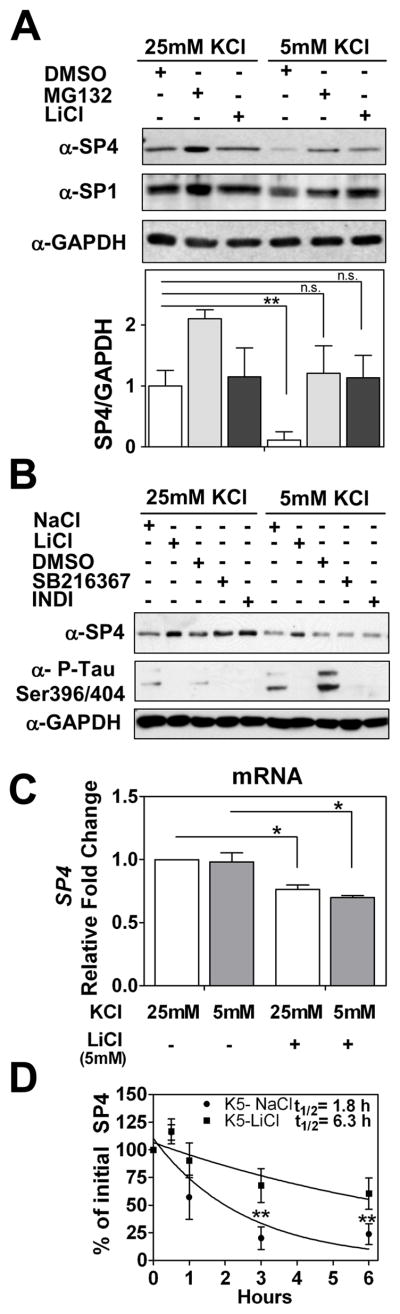
Lithium partially stabilizes SP4 protein in non-depolarizing conditions
To further investigate the signaling pathway that regulates SP4 protein stability, we treated neurons in non-depolarizing conditions with the mood stabilizer drug LiCl. We observed that addition of LiCl (5 mM) blocked the reduction in SP4 levels in non-depolarizing conditions (Fig. 5A). Interestingly, the increase in SP4 levels upon lithium treatment was only observed in the presence of serum (data not shown). Levels of the closely related transcription factor SP1 were not substantially altered under non-depolarizing conditions, while lithium treatment increased SP1 levels in depolarizing media (Fig. 5A and Supplementary Fig. 4).
Fig. 5.
Lithium stabilized SP4 protein levels in non-depolarizing conditions through a GSK-3 independent mechanism. (A) Cerebellar granule neurons at 7 days in vitro (DIV-7) were treated with 100 μM MG132, 5 mM LiCl, or DMSO before and after switching cell medium to 25 mM or 5 mM KCl with serum. Cell lysates were analysed with antisera against SP4, SP1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). SP4 levels were analyzed by densitometry and normalized to GAPDH levels. Values represent mean ± standard deviation of three independent experiments. Statistical analysis was performed using two-tailed t-test (**p < 0.01; n.s. = not significant). (B) Granule neurons at DIV-7 were incubated with 6 μM SB216367, 5 μM Indirubin-3′-oxime (INDI), DMSO vehicle, 5 mM NaCl or 5 mM LiCl for five hours prior to and one hour after switching cells into media containing serum and 25 mM or 5 mM KCl. Extracts were immunoblotted with antibodies against SP4, GAPDH, or the GSK-3 substrate Tau phospho Ser396/Ser404 (PHF-1). (C) Granule neurons at DIV-7 were incubated with 5 mM LiCl or 5 mM NaCl for five hours before and one hour after switching into medium containing serum and 25 or 5 mM KCl for one hour. mRNA was obtained and quantitative RT-PCR reactions for SP4 and GAPDH were performed. Data represents mean ± standard deviation of two independent reactions performed in four biological repeats. Statistical analysis was performed using one-tailed t-test (*p < 0.05; n.s. = not significant). (D) Granule neurons were incubated with 5 mM LiCl or 5 mM NaCl for five hours and then switched into medium containing serum, 50 μg/ml cycloheximide, and 25 or 5 mM KCl for the indicated times. Immunoblot analysis were performed as described for Panel A. Half-life (log10 50%) of SP4 was estimated by adjusting the logarithm of initial SP4 percentage values to a linear regression. Values represent the mean ± standard deviation of four independent experiments. Statistical analysis was performed using ANOVA and Tukey post-hoc test (**p < 0.01)
To investigate whether lithium mediates its effect on SP4 stability through GSK-3, we treated neurons with either LiCl or the specific inhibitors of GSK-3, SB216367 and Indirubin-3′-oxime (46, 47). All three drugs effectively inhibited GSK-3 activity as judged by reduced phosphorylation of Tau protein in the PHF-1 domain (Ser396/404). However, only LiCl maintained SP4 levels in non-depolarizing conditions while the GSK-3-specific inhibitors did not affect SP4 levels (Fig. 5B). In addition, overexpression of a constitutively active form of GSK-3 (mutated in serine 9) in a neuroblastoma cell line did not alter SP4 protein levels (data not shown). Together, these results indicate that lithium maintains SP4 levels in non-depolarizing conditions independently of effects on GSK-3 activity.
In order to further characterize the pathway by which lithium antagonizes the reduction of SP4 protein levels normally seen in non-depolarizing media, we analyzed lithium effects on Sp4 mRNA levels and SP4 protein stability. We performed RT-qPCR analysis in neurons maintained in depolarizing and non-depolarizing conditions in the absence or presence of LiCl. No significant change in Sp4 mRNA levels was observed upon the switch from 25 mM to 5 mM KCl, supporting the view that membrane depolarization predominantly acts post-transcriptionally to regulate SP4 levels. Treatment with LiCl did reduce Sp4 mRNA levels (Fig. 5C), however this reduction cannot account for the increase in SP4 protein observed in lithium treated neurons in non-depolarizing conditions (Fig. 5A). We performed a time course analysis of SP4 protein levels in neurons in the presence of cycloheximide in non-depolarizing culture medium treated with sodium chloride or LiCl. We observed that lithium treatment led to a 3.5-fold of increase in the half-life of SP4 in non-depolarizing conditions from 1.8 to 6.3 hours (Fig. 5D). Thus, under conditions where SP4 is rapidly degraded by the ubiquitin proteasome system, the mood stabilizing drug lithium increased SP4 stability, leading to greater steady state levels of this transcription factor.
Discussion
Our study shows a significant reduction in levels of transcription factor SP4 in the cerebellum and prefrontal cortex and SP1 in the cerebellum of human subjects with BD. A number of observations suggest that changes in gene regulation contribute to the development and treatment of psychiatric disorders (48, 49). However, transcriptional mechanisms altered in BD are largely unknown. Recent findings point to an association of SP4 gene to psychiatric diseases including BD (30–33), while altered SP1 expression has been reported in patients with schizophrenia (35). Analysis of mice expressing a hypomorphic allele of Sp4 revealed that reduced expression of Sp4 led to memory defects characteristic of psychiatric disorders (42). SP4 and SP1 transcription factors share high homology in the DNA binding domain (39). SP4 can function as either an activator or a repressor of transcription, thus, SP4 and SP1 may function independently, cooperate, or even compete to regulate genes through a common DNA element. Binding sites for SP1/SP4 have been shown to be important for expression of several genes implicated in psychiatric disease (16, 50, 51) including, for example, the finding that polymorphisms in the SP1/SP4 binding site of the GRK3 promoter are associated with BD (52). These studies together with our finding that SP4 and SP1 are misregulated in postmortem brains of BD subjects suggest that both Sp protein factors may be relevant transcriptional regulators whose alterations contribute to the pathogenesis of BD and other psychiatric diseases.
Our data strongly support altered post-transcriptional regulation of SP4 in BD in the cerebellum, however, transcription of SP4 may be affected in the prefontal cortex. We observed significant changes in BD in the cerebellum for SP4 and SP1 proteins, but not mRNA. Since the cerebellar cortex is a homogeneous region composed mainly of granule neurons, this probably corresponds to changes in regulation of SP4 and SP1 at the post-transcriptional level in these cells. Furthermore, our finding that SP4 protein is regulated at the post-transcriptional level in both rat cerebellar granule neurons in response to neuronal activity and in the cerebellum of BD subjects, suggests a common underlying mechanism.
Our studies have revealed that SP4 is a neuronal activity regulated transcription factor. The expression of many neuronal genes important for development and function of the nervous system has been shown to be regulated by neuronal activity (53, 54). In addition to the important roles of activity-regulated transcriptional programs in the promotion of dendritic growth, synapse development, and neuronal plasticity, alterations in several activity-regulated target genes have been found to confer susceptibility to psychiatric, neurological, and neurodegenerative disorders (55). Our studies indicate that at least one mechanism that controls SP4 activity in response to extracellular signals is the regulation of protein stability by ubiquitin-mediated proteasomal degradation. We found that levels of SP4, but not SP1, were regulated by membrane depolarization, indicating that regulation of SP4 levels by depolarization-dependent pathways is one mechanism that contributes to functional specificity in the Sp protein transcription factor family. Interestingly, the levels of both SP4 and the related SP1 proteins have been reported to be reduced under conditions of excitotoxic stress imposed by high glutamate levels in cortical neurons (56). How these different neuronal signals regulate specific members of the specificity protein transcription factor family is thus of significant interest. Of the diverse mechanisms that regulate transcription factors in response to calcium signaling pathways (57), to our knowledge regulated degradation by the proteasome has not been previously described.
SP4 is a novel transcriptional regulator controlled by lithium as we report that lithium partially stabilized SP4 protein, leading to a significantly delayed degradation under non-depolarizing conditions. Delayed therapeutic response to lithium has been proposed to be a result of changes in gene expression and cellular reprogramming (19). In fact, microarray studies performed with rodent brain areas after chronic treatment with lithium have shown that lithium treatment significantly alters the expression of genes associated with a wide range of cellular processes, including synaptic transmission, apoptosis, transport, and protein degradation (28, 29). These microarray studies raise the possibility that lithium controls SP4 through altered expression of components in the ubiquitin/proteasome pathway such as proteasome subunit beta type 5 (PSMB5). Surprisingly, we found that Sp4 mRNA levels were reduced upon lithium treatment (Fig. 5C), although this reduction cannot account for the increase in SP4 protein levels observed in lithium treated neurons in non-depolarizing conditions (Fig. 5A). One possible mechanism to account for the opposite regulation of SP4 gene expression and protein stability by lithium proposes that a feedback regulation of SP4 gene expression occurs upon an excess of SP4 protein. Since SP4 gene promoter contains GC-boxes and SP4 could act as a repressor, it is possible that high levels of SP4 protein could repress its own promoter (41, 58). The observation that lithium also increased SP1 protein levels in cerebellar granule neurons under conditions of membrane depolarization (Supplementary Fig. 6) suggests that lithium treatment could counteract the decrease observed for both factors in cerebellum of BD subjects and thus both these Sp protein factors may be susceptible to be targeted by new therapeutic approaches to BD. In our cell culture studies, we have used a higher dose of lithium than that which is recommended for therapy in humans. Further studies will be needed to investigate the effects of lithium on SP4 and SP1 levels in vivo at therapeutic doses.
Functional magnetic resonance studies have revealed connectivity defects in patients with schizophrenia and BD that could contribute to the cognitive and psychotic symptoms of the disease (2, 59–61). Aberrant dendritic arbors appear in neurons of patients with psychiatric disorders (62, 63). The formation of dendritic arbors is one of the later stages of neural development which is regulated by intrinsic transcriptional programs and extrinsic signals such as neuronal activity (40, 64–66). We have previously identified an essential role for SP4 in regulation of dendritic morphology during development (40, 41). Thus, reduced levels of SP4 may contribute to altered dendritic patterning in some BD subjects. Here, we report that lithium restores SP4 levels, suggesting that normalization of SP4 levels could contribute to the effects of lithium in therapeutic mechanisms on affective disorders. Interestingly, treatment with lithium or the antidepressant fluoxetine was reported to modulate dendritic branching (67, 68).
The fact that many of the brain samples analyzed in this study came from subjects that also committed suicide raises the possibility that alterations in SP1 and SP4 levels could be associated with suicide per se and not to the fact that the subjects had BD. Importantly, we have not found differences in SP4 and SP1 protein and mRNA between control individuals versus suicide subjects without BD diagnosis in the cerebellum and prefrontal cortex (n = 8) (Supplementary Fig. 7). Nevertheless, further investigations of SP4 and SP1 will be needed to determine whether SP4 and SP1 are altered in subjects with suicide. In addition, the extension of this study to a larger population will help to confirm whether reduced levels of SP4 and SP1 transcription factors in brain are a consistent molecular alteration among BD subjects.
Supplementary Material
Acknowledgments
We acknowledge support from The Spanish Ministry of Education/Fulbright (Secretary of State and Education and Universities, and The European Social Fund), CIBERSAM, Marie Curie IRG [EU Framework Seventh-RTD-REG/T.2(2007)D/530573], and Plan Nacional de Investigación (MCINN-Spain) BFU2008-01103 to BR; SAF2009-08460 to JJM; The Canadian Institutes for Health Research to JL; Predoctoral Fellowship from ISCIII and Fondo de Investigación Sanitaria (PFIS) to RP; and the National Institutes of Health (HD043364) to GG. We thank Raquel Iniesta for assistance with the statistical analysis, Parizad M. Bilimoria for assistance with cerebellar slice culture, and Dr. Lavarino and Dr. de Torres for their advice on real-time PCR assays. We also thank Dr. Meijer, CNRS, France for the gift of indirubin-3′-oxime; and Dr. Davies, Albert Einstein College of Medicine, New York, NY, USA for the gift of anti-phospho tau PHF-1.
Footnotes
The authors of this paper do not have any commercial associations that might pose a conflict of interest in connection with this manuscript.
References
- 1.Haldane M, Frangou S. New insights help define the pathophysiology of bipolar affective disorder: neuroimaging and neuropathology findings. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:943–960. doi: 10.1016/j.pnpbp.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 2.Savitz J, Drevets WC. Bipolar and major depressive disorder: neuroimaging the developmental-degenerative divide. Neurosci Biobehav Rev. 2009;33:699–771. doi: 10.1016/j.neubiorev.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajek T, Carrey N, Alda M. Neuroanatomical abnormalities as risk factors for bipolar disorder. Bipolar Disord. 2005;7:393–403. doi: 10.1111/j.1399-5618.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- 4.Beyer JL, Krishnan KR. Volumetric brain imaging findings in mood disorders. Bipolar Disord. 2002;4:89–104. doi: 10.1034/j.1399-5618.2002.01157.x. [DOI] [PubMed] [Google Scholar]
- 5.Bolbecker AR, Mehta C, Johannesen JK, et al. Eyeblink conditioning anomalies in bipolar disorder suggest cerebellar dysfunction. Bipolar Disord. 2009;11:19–32. doi: 10.1111/j.1399-5618.2008.00642.x. [DOI] [PubMed] [Google Scholar]
- 6.Schmahmann JD. An emerging concept. The cerebellar contribution to higher function. Arch Neurol. 1991;48:1178–1187. doi: 10.1001/archneur.1991.00530230086029. [DOI] [PubMed] [Google Scholar]
- 7.Schmahmann JD, Weilburg JB, Sherman JC. The neuropsychiatry of the cerebellum–insights from the clinic. Cerebellum. 2007;6:254–267. doi: 10.1080/14734220701490995. [DOI] [PubMed] [Google Scholar]
- 8.Schutter DJ, van Honk J. The cerebellum on the rise in human emotion. Cerebellum. 2005;4:290–294. doi: 10.1080/14734220500348584. [DOI] [PubMed] [Google Scholar]
- 9.Katz DB, Steinmetz JE. Psychological functions of the cerebellum. Behav Cogn Neurosci Rev. 2002;1:229–241. doi: 10.1177/1534582302001003004. [DOI] [PubMed] [Google Scholar]
- 10.Baillieux H, De Smet HJ, Paquier PF, De Deyn PP, Mariën P. Cerebellar neurocognition: insights into the bottom of the brain. Clin Neurol Neurosurg. 2008;110:763–773. doi: 10.1016/j.clineuro.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Strakowski SM, Delbello MP, Adler CM. The functional neuroanatomy of bipolar disorder: a review of neuroimaging findings. Mol Psychiatry. 2005;10:105–116. doi: 10.1038/sj.mp.4001585. [DOI] [PubMed] [Google Scholar]
- 12.Adler CM, DelBello MP, Jarvis K, Levine A, Adams J, Strakowski SM. Voxel-based study of structural changes in first-episode patients with bipolar disorder. Biol Psychiatry. 2007;61:776–781. doi: 10.1016/j.biopsych.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 13.Mahon K, Wu J, Malhotra AK, et al. A voxel-based diffusion tensor imaging study of white matter in bipolar disorder. Neuropsychopharmacology. 2009;34:1590–1600. doi: 10.1038/npp.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McQueen MB, Devlin B, Faraone SV, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakatani N, Hattori E, Ohnishi T, et al. Genome-wide expression analysis detects eight genes with robust alterations specific to bipolar I disorder: relevance to neuronal network perturbation. Hum Mol Genet. 2006;15:1949–1962. doi: 10.1093/hmg/ddl118. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Shachar D, Karry R. Neuroanatomical pattern of mitochondrial complex I pathology varies between schizophrenia, bipolar disorder and major depression. PLoS One. 2008;3:e3676. doi: 10.1371/journal.pone.0003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatemi SH, Laurence JA, Araghi-Niknam M, et al. Glial fibrillary acidic protein is reduced in cerebellum of subjects with major depression, but not schizophrenia. Schizophr Res. 2004;69:317–323. doi: 10.1016/j.schres.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Choi KH, Elashoff M, Higgs BW, et al. Putative psychosis genes in the prefrontal cortex: combined analysis of gene expression microarrays. BMC Psychiatry. 2008;8:87. doi: 10.1186/1471-244X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenox RH, Hahn CG. Overview of the mechanism of action of lithium in the brain: fifty-year update. J Clin Psychiatry. 2000;61 (Suppl 9):5–15. [PubMed] [Google Scholar]
- 20.Shastry BS. Bipolar disorder: an update. Neurochem Int. 2005;46:273–279. doi: 10.1016/j.neuint.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 22.Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 24.Mora A, Sabio G, Risco AM, et al. Lithium blocks the PKB and GSK3 dephosphorylation induced by ceramide through protein phosphatase-2A. Cell Signal. 2002;14:557–562. doi: 10.1016/s0898-6568(01)00282-0. [DOI] [PubMed] [Google Scholar]
- 25.Jorda EG, Verdaguer E, Canudas AM, et al. Implication of cyclin-dependent kinase 5 in the neuroprotective properties of lithium. Neuroscience. 2005;134:1001–1011. doi: 10.1016/j.neuroscience.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 26.Pardo R, Andreolotti AG, Ramos B, Picatoste F, Claro E. Opposed effects of lithium on the MEK-ERK pathway in neural cells: inhibition in astrocytes and stimulation in neurons by GSK3 independent mechanisms. J Neurochem. 2003;87:417–426. doi: 10.1046/j.1471-4159.2003.02015.x. [DOI] [PubMed] [Google Scholar]
- 27.Kopnisky KL, Chalecka-Franaszek E, Gonzalez-Zulueta M, Chuang DM. Chronic lithium treatment antagonizes glutamate-induced decrease of phosphorylated CREB in neurons via reducing protein phosphatase 1 and increasing MEK activities. Neuroscience. 2003;116:425–435. doi: 10.1016/s0306-4522(02)00573-0. [DOI] [PubMed] [Google Scholar]
- 28.Fatemi SH, Reutiman TJ, Folsom TD. The role of lithium in modulation of brain genes: relevance for aetiology and treatment of bipolar disorder. Biochem Soc Trans. 2009;37:1090–1095. doi: 10.1042/BST0371090. [DOI] [PubMed] [Google Scholar]
- 29.Chetcuti A, Adams LJ, Mitchell PB, Schofield PR. Microarray gene expression profiling of mouse brain mRNA in a model of lithium treatment. Psychiatr Genet. 2008;18:64–72. doi: 10.1097/YPG.0b013e3282fb0051. [DOI] [PubMed] [Google Scholar]
- 30.Shi J, Potash JB, Knowles JA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shyn SI, Shi J, Kraft JB, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry. 2011;16:202–215. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Tang W, Greenwood TA, et al. Transcription factor SP4 is a susceptibility gene for bipolar disorder. PLoS One. 2009;4:e5196. doi: 10.1371/journal.pone.0005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tam GW, van de Lagemaat LN, Redon R, et al. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 2010;38:445–451. doi: 10.1042/BST0380445. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Nie Z, Roberts A, et al. Reduced NMDAR1 expression in the Sp4 hypomorphic mouse may contribute to endophenotypes of human psychiatric disorders. Hum Mol Genet. 2010;19:3797–3805. doi: 10.1093/hmg/ddq298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Shachar D, Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS One. 2007;2:e817. doi: 10.1371/journal.pone.0000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 37.Zhu H, Nguyen VT, Brown AB, et al. A novel, tissue-restricted zinc finger protein (HF-1b) binds to the cardiac regulatory element (HF-1b/MEF-2) in the rat myosin light-chain 2 gene. Mol Cell Biol. 1993;13:4432–4444. doi: 10.1128/mcb.13.7.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 39.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 40.Ramos B, Gaudilliere B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proc Natl Acad Sci USA. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos B, Valin A, Sun X, Gill G. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Mol Cell Neurosci. 2009;42:152–159. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Long JM, Geyer MA, et al. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- 43.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 45.Marchetti C, Usai C. High affinity block by nimodipine of the internal calcium elevation in chronically depolarized rat cerebellar granule neurons. Neurosci Lett. 1996;207:77–80. doi: 10.1016/0304-3940(96)12492-7. [DOI] [PubMed] [Google Scholar]
- 46.Knockaert M, Greengard P, Meijer L. Pharmacological inhibitors of cyclin-dependent kinases. Trends Pharmacol Sci. 2002;23:417–425. doi: 10.1016/s0165-6147(02)02071-0. [DOI] [PubMed] [Google Scholar]
- 47.Guengerich FP, Sorrells JL, Schmitt S, Krauser JA, Aryal P, Meijer L. Generation of new protein kinase inhibitors utilizing cytochrome p450 mutant enzymes for indigoid synthesis. J Med Chem. 2004;47:3236–3241. doi: 10.1021/jm030561b. [DOI] [PubMed] [Google Scholar]
- 48.Konradi C, Heckers S. Antipsychotic drugs and neuroplasticity: insights into the treatment and neurobiology of schizophrenia. Biol Psychiatry. 2001;50:729–742. doi: 10.1016/s0006-3223(01)01267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karege F, Vaudan G, Schwald M, Perroud N, La Harpe R. Neurotrophin levels in postmortem brains of suicide victims and the effects of antemortem diagnosis and psychotropic drugs. Brain Res Mol Brain Res. 2005;136:29–37. doi: 10.1016/j.molbrainres.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 50.Krainc D, Bai G, Okamoto S, et al. Synergistic activation of the N-methyl-D-aspartate receptor subunit 1 promoter by myocyte enhancer factor 2C and Sp1. J Biol Chem. 1998;273:26218–26224. doi: 10.1074/jbc.273.40.26218. [DOI] [PubMed] [Google Scholar]
- 51.Miyatake R, Furukawa A, Suwaki H. Identification of a novel variant of the human NR2B gene promoter region and its possible association with schizophrenia. Mol Psychiatry. 2002;7:1101–1106. doi: 10.1038/sj.mp.4001152. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X, Barrett TB, Kelsoe JR. Promoter variant in the GRK3 gene associated with bipolar disorder alters gene expression. Biol Psychiatry. 2008;64:104–110. doi: 10.1016/j.biopsych.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- 54.Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao X, Moerman AM, Barger SW. Neuronal kappa B-binding factors consist of Sp1-related proteins. Functional implications for autoregulation of N-methyl-D-aspartate receptor-1 expression. J Biol Chem. 2002;277:44911–44919. doi: 10.1074/jbc.M204292200. [DOI] [PubMed] [Google Scholar]
- 57.Hong EJ, West AE, Greenberg ME. Transcriptional control of cognitive development. Curr Opin Neurobiol. 2005;15:21–28. doi: 10.1016/j.conb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Song J, Mangold M, Suske G, et al. Characterization and promoter analysis of the mouse gene for transcription factor Sp4. Gene. 2001;264:19–27. doi: 10.1016/s0378-1119(01)00328-6. [DOI] [PubMed] [Google Scholar]
- 59.Andreasen NC, Nopoulos P, O’Leary DS, Miller DD, Wassink T, Flaum M. Defining the phenotype of schizophrenia: cognitive dysmetria and its neural mechanisms. Biol Psychiatry. 1999;46:908–920. doi: 10.1016/s0006-3223(99)00152-3. [DOI] [PubMed] [Google Scholar]
- 60.Friston KJ, Frith CD. Schizophrenia: a disconnection syndrome? Clin Neurosci. 1995;3:89–97. [PubMed] [Google Scholar]
- 61.McGuire PK, Frith CD. Disordered functional connectivity in schizophrenia. Psychol Med. 1996;26:663–667. doi: 10.1017/s0033291700037673. [DOI] [PubMed] [Google Scholar]
- 62.Miguel-Hidalgo JJ, Rajkowska G. Morphological brain changes in depression: can antidepressants reverse them? CNS Drugs. 2002;16:361–372. doi: 10.2165/00023210-200216060-00001. [DOI] [PubMed] [Google Scholar]
- 63.Rosoklija G, Toomayan G, Ellis SP, et al. Structural abnormalities of subicular dendrites in subjects with schizophrenia and mood disorders: preliminary findings. Arch Gen Psychiatry. 2000;57:349–356. doi: 10.1001/archpsyc.57.4.349. [DOI] [PubMed] [Google Scholar]
- 64.Aizawa H, Hu SC, Bobb K, et al. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- 65.Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- 66.Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 67.Watase K, Gatchel JR, Sun Y, et al. Lithium therapy improves neurological function and hippocampal dendritic arborization in a spinocerebellar ataxia type 1 mouse model. PLoS Med. 2007;4:e182. doi: 10.1371/journal.pmed.0040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou L, Huang KX, Kecojevic A, Welsh AM, Koliatsos VE. Evidence that serotonin reuptake modulators increase the density of serotonin innervation in the forebrain. J Neurochem. 2006;96:396–406. doi: 10.1111/j.1471-4159.2005.03562.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.