Abstract
In air pollution epidemiology, there is a growing interest in estimating the health effects of coarse particulate matter (PM) with aerodynamic diameter between 2.5 and 10 μm. Coarse PM concentrations can exhibit considerable spatial heterogeneity because the particles travel shorter distances and do not remain suspended in the atmosphere for an extended period of time. In this paper, we develop a modeling approach for estimating the short-term effects of air pollution in time series analysis when the ambient concentrations vary spatially within the study region. Specifically, our approach quantifies the error in the exposure variable by characterizing, on any given day, the disagreement in ambient concentrations measured across monitoring stations. This is accomplished by viewing monitor-level measurements as error-prone repeated measurements of the unobserved population average exposure. Inference is carried out in a Bayesian framework to fully account for uncertainty in the estimation of model parameters. Finally, by using different exposure indicators, we investigate the sensitivity of the association between coarse PM and daily hospital admissions based on a recent national multisite time series analysis. Among Medicare enrollees from 59 US counties between the period 1999 and 2005, we find a consistent positive association between coarse PM and same-day admission for cardiovascular diseases.
Keywords: Air pollution, Coarse particulate matter, Exposure measurement error, Multisite time series analysis
1. INTRODUCTION
Ambient particulate matter (PM) is a mixture of solid and liquid particles regulated by the Environmental Protection Agency (EPA) as one of the 6 criteria air pollutants. Under the Clean Air Act, EPA is responsible for establishing national standards for these pollutants to protect public health and the environment (Bachmann, 2007). PM can be characterized into the fine and coarse size fractions that represent distinct pollutant mixtures of different sources and properties (Wilson and Suh, 1997). Particle size is an important attribute because it governs the particle's behaviors in the atmosphere and deposition in the respiratory system. Ambient coarse PM is most often released directly as a primary pollutant through mechanical processes such as dust suspension or physical attrition involving grinding and crushing. Mineral residue resulting from combustion burn out (fly ash) also contributes to the coarse fraction. Biogenic compounds including bacterial endotoxin, pollen, and other animal/plant debris may also be present.
Protecting public health from coarse PM has endured considerable controversy in the regulatory context. EPA's current National Ambient Air Quality Standards (NAAQS) use ambient PM10 concentration (PM with aerodynamic diameter < 10 μm) as the pollutant measure to protect public health from coarse PM. Most studies also routinely use PM10 to quantify health risks and have consistently found that increased concentrations of ambient (outdoor) PM10 are associated with increased risks of various adverse health outcomes (Pope and Dockery, 2006). However, there exists persistent criticism in interpreting the health effects of coarse PM since PM10 contains both the coarse and fine fraction.
Recent studies of coarse PM have increasingly focused on exposure to inhalable coarse particles (PM10 − 2.5) of size between 2.5 and 10 μm aerodynamic diameter. While toxicological evidence supports the potential health effects of coarse PM, current epidemiological evidence is limited and mixed (Brunekreef and Forsberg, 2005). Most time series analysis of ambient PM10 − 2.5 concentrations and short-term mortality showed nonstatistically significant associations except in arid regions such as Mexico City (Castillejos and others, 2000) and Phoenix US (Mar and others, 2004). Results from both the Harvard Six Cities Study (Dockery and others, 1993) and the American Cancer Society cohort (Pope and others, 2002) also found no association between long-term exposure to coarse particles and mortality. However, studies have reported statistically significant short-term effects of ambient PM10 − 2.5 on hospital admissions (Peng and others, 2008) and mortality (Zanobetti and Schwartz, 2009). Particularly, a multisite time series analysis conducted by Peng and others (2008) found that on average across the 108 US counties, ambient PM10 − 2.5 concentration was associated with emergency admissions for cardiovascular diseases, but this association lost statistical significance when adjusted by PM2.5.
In a time series design, the health outcome is only available as daily total number of adverse health events in a community, such as a county, a city or a large metropolitan area. Unbiased risk estimates require the exposure measure to coincide with the true average exposure experienced by all at-risk individuals in the community (Zeger and others, 2003, , Sheppard2005). When the ambient pollutant concentration is spatially smooth, current practice of averaging measurements from outdoor monitors provides a reasonable surrogate measure for the population exposure due to outdoor sources. However, coarse PM concentrations often exhibit higher spatial heterogeneity compared to PM2.5 and PM10. Therefore, averaging PM10 − 2.5 values from the fixed-location monitors placed in the same community may not capture the true population exposure. Moreover because there is no national monitoring network for PM10 − 2.5, community-level daily PM10 − 2.5 concentrations are calculated based on the limited network of collocated monitor pairs where both PM10 and PM2.5 are measured at the same location.
One of the main objectives of this paper is to develop a statistical modeling approach and computationally efficient estimation procedures for estimating the health effects of air pollution accounting for exposure measurement error (ME) in multisite time series analyses. We are concerned with the error that results from assigning an incorrect exposure measure to the study population living in an area when (1) pollution concentrations are available from a small number of monitors placed within the community; and (2) the pollution concentrations are highly variable within the community. To incorporate exposure ME in risk estimates, we view monitor-level PM values as error-prone repeated measurements of the true community-level average exposure. Our approach estimates exposure ME by quantifying, on any given day, the disagreement in PM values measured across the monitoring stations located within the same community. Specifically, we develop ME models for a bivariate vector of exposure variables in order to estimate the effect of PM10 − 2.5 adjusted by PM2.5. Joint modeling also addresses the bias where the effect of one pollutant measured with more error is transferred to another pollutant measured with less error (Zidek and others, 1996). Finally, we calculate different measures of county-level ambient daily exposure to PM10 − 2.5 and investigate the sensitivity of the national average effect of PM10 − 2.5 on hospital admissions estimated in Peng and others (2008).
The exposure ME encountered in the analysis of PM10 − 2.5 is related to the statistical problem known as spatial misalignment (Gotway and Young, 1999). Spatial variation in ambient concentrations and exposure ME caused by spatial misalignment have been addressed in several studies on the long-term health effects of air pollution (Zhu and others, 2003, , Gryparis2008:Biostat). However, few have examined its effects specifically in time series analysis. Through simulation studies, Sheppard and others (2005) find minor effect attenuation when the ambient concentration varies spatially. Peng and Bell (2010) calculate county-level exposure by first interpolating PM2.5 chemical constituents concentration via spatial modeling. The authors show that the resulting county-specific short-term health effects are greater in magnitude and have larger standard errors compared to estimates that do not consider spatial variation in pollution concentration. To our knowledge, no study has investigated the effects of exposure ME in the analysis of PM10 − 2.5 and health.
Our ME approach estimates daily county-level PM10 − 2.5 exposure accounting for spatial heterogeneity without explicitly characterizing the spatial gradient. While this approach provides computational advantages, a space–time model for PM10 and PM2.5 offers an alternative approach for obtaining PM10 − 2.5 exposure measure. However, this increases model complexity significantly when considering daily PM10 − 2.5 concentrations nationally over a long study period. The ability to characterize PM10 − 2.5 spatial variation is also limited by the sparse network of collocated PM10 and PM2.5 monitors.
The remainder of this paper is organized as follows. Section 2 describes the data sets, the modeling framework and the estimation procedures. In Section 3, we calculate different measures of daily county-level ambient exposure to PM10 − 2.5 and investigate the sensitivity of the national average effect of PM10 − 2.5 on hospital admissions. Section 4 describes a simulation study that examines the impact of PM10 − 2.5 exposure ME and the performance of our proposed method. Finally, discussion and future work appear in Section 5.
2. METHODS
2.1. Data
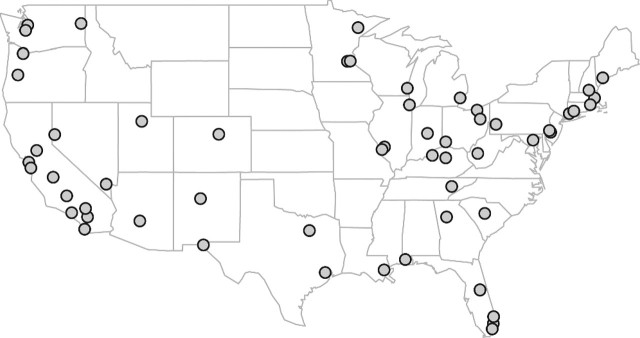
Daily average concentrations of PM10 − 2.5 and PM2.5 data for the period 1999–2005 were obtained from the EPA's National Air Pollution Monitoring Network in the air quality system (AQS). Without a national monitoring network for PM10 − 2.5, the EPA calculates PM10 − 2.5 values indirectly by subtracting PM2.5 from PM10 measurements at monitors that are physically located at the same place (collocated monitor pairs). We considered PM2.5 measurements from all AQS monitors and PM10 − 2.5 measurements from collocated PM2.5 and PM10 monitor pairs. We restricted our analysis to the 59 US counties with (1) a population greater than 200 000 based on the 2000 census; (2) at least 2 pairs of collocated PM10 and PM2.5 monitors; and (3) at least 210 daily PM10 − 2.5 measurements over the study period. Locations of the counties in our study are shown in Figure 1.
Fig. 1.
59 US counties with population greater than 200 000, at least 2 pairs of collocated PM10 and PM2.5 monitors, and at least 210 daily PM10 − 2.5 measurements over the period 1999–2005 (Honolulu, HI and Anchorage, AL not shown). 1
Time series of daily emergency hospital admissions for cardiovascular and respiratory diseases were assembled for Medicare enrollees aged 65 years or above within each county (Peng and others, 2008). Records were extracted from the National Claim History Files for the period between 1999 and 2005. Based on the International Classification of Diseases, Ninth Revision disease classification, we considered primary diagnosis of admissions due to 2 aggregated causes: (1) cardiovascular disease admissions and (2) respiratory disease admissions.
2.2. Trimmed mean exposure indicator
For county c on day t, let W1tjc and W2tjc denote the PM10 − 2.5 and PM2.5 levels at monitor j, where j = 1,…,Jc and Jt is the total number of PM2.5 monitors in county c. For some PM2.5 monitors, a collocated PM10 monitor is present and both W1tjc and W2tjc are observed. Otherwise, only W2tjc is observed, and we treat W1tjc as missing. In our study, there were 220 collocated PM2.5 and PM10 monitor pairs and an additional 173 PM2.5 monitors that did not have collocated PM10 monitors.
Under the assumption that PM10 − 2.5 and PM2.5 concentrations are spatially homogeneous across each county, the standard daily county-level PM10 − 2.5 exposure (X1tc) and PM2.5 exposure (X2tc) measures are obtained by averaging all available measurements across monitors on a particular day (Samet and others, 2000). Specifically, the 10% trimmed mean (TM) is used to exclude extreme monitor-level values that may be invalid. In this algorithm, no PM measurements are excluded in computing the mean on days with less than 3 monitor-level PM measurements and only the maximum and minimum measurements are excluded on days between 3 and 9 PM measurements.
2.3. ME model
To illustrate the impact of a spatially heterogeneous pollutant in time series studies of air pollution and health and the importance of defining appropriate population exposure indicators, first consider the following example adopted from Zeger and others (2003) and Sheppard (2005). Denote by Xit the exposure to a pollutant due to outdoor sources for an individual i on day t. For each individual in the study, we assume that the binary health outcome follows a Bernoulli distribution with probability λ0exp(βXit), where λ0 represents a baseline risk common across individuals and exp(β) represents the multiplicative change in risk associated with a unit increase in Xit.
Let Nt denote the number of at-risk individuals in a community on day t, and let Yt denote the number of hospital admissions on day t in the population that resides in the community. If the occurrence of outcome is independent across individuals, the community-level outcome count has mean equal to
 |
(2.1) |
Since the short-term relative risk of air pollution is typically very small, following Zeger and others (2003), a linear approximation to the exponential term in (2.1) gives
 |
where Xt is the average exposure of all at-risk individuals. Under these assumptions, the relative risk obtained by regressing aggregated outcome Yt on aggregated exposure Xt via log-linear regression is equivalent to the personal risk β. Therefore, Xt represents the “desired but unobserved” exposure indicator in time series analysis.
Let Xt* be a surrogate exposure indicator calculated, for example, by averaging measurements from some fixed-location monitoring sites within the community. We can rewrite (2.1) as
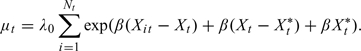 |
(2.2) |
Applying a similar linear approximation gives
| (2.3) |
Hence, we wish to have the exposure indicator calculated from monitoring data to coincide with the true average exposure (Xt = Xt*).
For coarse PM, ME can occur when the sparse PM10 − 2.5 monitoring network does not fully capture the spatial variation in PM10 − 2.5 concentrations. Here, it is also important to account for the spatial distribution of the at-risk population since individuals can be exposed to different levels of ambient PM10 − 2.5. The standard approach of defining community-level exposure by averaging monitor measurements can over- or underestimate the true average exposure. This bias can also vary temporally because the set of PM10 − 2.5 measurements used to calculate average exposure often varies between days due to missing data or different monitoring schedules.
In the ME modeling approach, we view PM measurements from different monitors on the same day within the same county as error-prone repeated measurements of the unobserved population exposure. Let Xtc = (X1tc,X2t c) denote the unobserved county-level exposure to ambient PM10 − 2.5 and PM2.5 experienced by the at-risk population. Under the classical additive ME model (Fuller, 1987), we assume for j = 1,…,J c,
 |
(2.4) |
where the 2 ME variances (σ12,c and σ22,c) capture same-day between-monitor variability for PM10 − 2.5 and PM2.5. Parameter ρc captures the correlation between MEs, which is assumed to be constant across days and monitors within the same county. A positive ρc indicates that daily monitor-level PM10 − 2.5 and PM2.5 measurement pairs tend to deviate from the true exposures in the same direction. Note that at some monitoring locations and on some days, only W2tjc is observed and W1tjc is treated as missing. Through Bayesian inference described in Section 2.5, we simultaneously address the ME problem and the imputation of missing monitor-level PM10 − 2.5 measurements.
Additionally, we assume
| (2.5) |
where Ztc denotes a covariate vector that includes indicators for month, indicators for day of the week, and calendar dates. Parameters η1c and η2c denote the corresponding vectors of regression coefficients. The logarithmic transformation accounts for the strictly positive and right-skewed PM concentration measurements. The diagonal elements of Σxc represent the residual variance for log-PM exposure and its off-diagonal element captures the correlation between daily county-level PM10 − 2.5 and PM2.5 exposure for county c. Note that we choose to model Wtjc in (2.4) without the log-transformation to allow for negative values of observed PM10 − 2.5 concentrations. Also, Xtc is then interpreted as an average of the observed monitoring data. Alternatively, Wtjc can be modeled on the log-scale where a multiplicative ME is assumed.
However, the error specification in (2.4) assumes that errors between the observed monitor-level PM10 − 2.5 concentrations and the true county-level exposure are distributed identically with the same error variance across monitors. For a spatially heterogeneous PM, true county-level exposure represents a population-weighted average exposure. Hence, measurements taken at monitors located in less populated areas, relative to the total population of the county, may tend to deviate more from the true average exposure experienced by everyone in the county.
We also consider a ME model with heteroskedastic errors between monitors where the ME variance is weighted inversely to the population living around the monitor. Let Pkc denote the population of city k in county c. Following notations from (2.4), we now assume
| (2.6) |
where  if monitor j is in city k*. For counties where PM is measured in multiple cities, αjc approximates the proportion of at-risk population in county c that are exposed to the PM concentration measured at monitor j. City population data were retrieved from the Site Descriptive Data database of EPA's AQS. Under this ME model, monitors in less populated areas contribute less to the estimation of county-level exposure. We refer to the above model as the population-weighted WME approach.
if monitor j is in city k*. For counties where PM is measured in multiple cities, αjc approximates the proportion of at-risk population in county c that are exposed to the PM concentration measured at monitor j. City population data were retrieved from the Site Descriptive Data database of EPA's AQS. Under this ME model, monitors in less populated areas contribute less to the estimation of county-level exposure. We refer to the above model as the population-weighted WME approach.
Finally, we assign the following priors for the model parameters: (1) η1c and η2c each follows a multivariate Normal distribution with dispersed variances; (2) Σwc follows  , where
, where 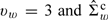 is the estimated covariance of
is the estimated covariance of  ; and (3) Σxc follows
; and (3) Σxc follows 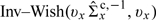 , where
, where  is the estimated residual covariance from regressing logXtc on Ztc. Following Gelman (2006), we also consider assigning a Uniform[0, 100] on the standard deviations (SDs) and a Uniform[ − 1,1] on the correlation for either Σwc or Σxc, or both. The differences in posterior inference for selected counties with small numbers of days or monitors were negligible.
is the estimated residual covariance from regressing logXtc on Ztc. Following Gelman (2006), we also consider assigning a Uniform[0, 100] on the standard deviations (SDs) and a Uniform[ − 1,1] on the correlation for either Σwc or Σxc, or both. The differences in posterior inference for selected counties with small numbers of days or monitors were negligible.
2.4. Health model
We model the expected number of admissions E(Ytc) using Poisson regression:
| (2.7) |
where Ntc is the size of the population at risk. Following Dominici and others (2006), confounders (Ct c) include seasonal trends, weather effects, and age-group effects that are modeled via natural cubic splines with degrees of freedom d. Specifically, we include: (1) calendar time (d = 8 per year), (2) current-day temperature (d = 6) and average temperature for the previous 3 days (d = 6); (3) current-day dew-point temperature (d = 3) and average dew-point temperature for the previous 3 days (d = 3); (4) age-group intercept (64–74 vs. 75 and above) and its interaction with a smooth function of calendar time (d = 1 per year); and (5) day of the week.
2.5. Risk estimation
For the standard time series analysis without ME modeling, county-level exposures (Xtc) are obtained using the 10% TM algorithm described in Section 2.2. Relative risks estimates are obtained by fitting the health model via maximum quasi-likelihood separately for each county.
Due to the complex health model and the large data set, we describe a 2-stage approach that involves 2 separate Markov chain Monte Carlo (MCMC) implementations to estimate relative risks with ME modeling. In the first stage, posterior samples of PM10 − 2.5 and PM2.5 time series (Xc) given the observed monitor-level data (Wc) are obtained by sampling from the following posterior predictive distribution:
| (2.8) |
where [Wc|X c,θ1 c] represents the “measurement model” given by (2.4) with parameters θ1c = (Σw c) and [Xc|Z c,θ2 c] represents the “exposure model” given by (2.5) with parameters θ2 = (η1c,η2 c,Σx c). Here, the posterior distribution of Xc does not depend on the health model. Stage 1 computation is carried out by using JAGS version 1.0.3 (Plummer, 2003).
At the second stage, we obtain posterior samples of [Xc,βc|Wc,Zc,Yc] by using [Xc|Wc,Zc] from Stage 1 as the prior distribution of Xc. Given health data Yc for county c, we assume
| (2.9) |
where [Yc|Xc,βc,ψc] corresponds to the “health model” from (2.7). To decrease computational burden, we treat ψc as nuisance parameters and carry out a profile sampler approach described in Lee and others (2005). Specifically, we carry out block Metroplis–Hastings updates between βc and Xc, where the acceptance probabilities are calculated using the profile likelihood. Since both Xc and βc are updated by the health data Yc, we refer to this estimation approach as the “Bayesian” approach. This approach also provides samples of [Xc|Wc,Zc,Yc], the posterior distribution of the average PM exposure incorporating the health information. Details of the estimation procedure and an example of validating the 2-stage Bayesian approach are provided in Section 1 of the supplementary material available at Biostatistics online.
We also estimate the county-specific relative risks βc, where we replace Xc in the health model by the marginal posterior mean E[Xc|Wc,Z c] from stage 1. This plug-in method resembles a regression calibration where the unobservable exposure is replaced by its best linear prediction conditional on the covariates measured without error (Ztc) and the observed error-prone measurements (Carroll and others, 2006). While this method is computationally simple, it does not fully reflect the uncertainty in the exposure measure when estimating βc. Also, unlike the Bayesian approach, there is no feedback between the health observations and the exposure estimates. However, since the acute health effects of PM are typically small (relative risk of less than 2% per 10 μg/m3 increase in PM), the information in the health model is possibly negligible in determining the posterior distribution of the true county-level PM exposure.
We pool county-specific relative risks βc = (β1c,β2c) defined in (2.7) by assuming βc∼N(μ,Σβ). Here, the parameter of interest, μ, is interpreted as the pooled (national) PM10 − 2.5 and PM2.5 effects and Σβ captures the heterogeneity in relative risks between counties. Denote the estimated county-specific relative risks by  and the corresponding covariance matrix by
and the corresponding covariance matrix by 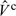 estimated either with or without exposure ME modeling as described in Section 2.5. For the Bayesian approach with ME modeling, we define
estimated either with or without exposure ME modeling as described in Section 2.5. For the Bayesian approach with ME modeling, we define  and
and 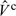 as the posterior mean and posterior covariance from the second-stage MCMC. Assuming
as the posterior mean and posterior covariance from the second-stage MCMC. Assuming 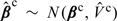 , μ and Σβ are estimated using the 2-level Normal independent sampling estimation algorithm of Everson and Morris (2000).
, μ and Σβ are estimated using the 2-level Normal independent sampling estimation algorithm of Everson and Morris (2000).
3. RESULTS
Across counties, the median number of PM2.5 monitors in a county was 5; the first quartile (Q1, 25th percentile) was 4; and the third quantile (Q3, 75th percentile) was 8. Similarly, the median number of PM2.5 and PM10 collocated monitor pairs was 3 (Q1 = 2 and Q3 = 5). Therefore, by restricting our analysis to collocated monitor pairs, the standard TM exposure for PM10 − 2.5 was calculated based on a considerably smaller number of monitors compared to PM2.5. In Figure 2, correlation of daily PM measurements at any pair of monitors in the same county is plotted versus the distance between the monitors. There was considerable larger spatial variability in PM10 − 2.5 measurements compared to PM2.5.
Fig. 2.
Correlations of monitor-level daily PM time series calculated between pairs of PM2.5 or PM10 − 2.5 monitoring locations in the same county and plotted versus the distance between monitor pair. For PM2.5, we used all available monitors without restricting to those with a collocated PM10 monitor.
Our study included approximately 5 million Medicare enrollees between the period 1999 and 2005. There were about 2.6 million admissions for cardiovascular diseases and 1.0 million admissions for respiratory diseases. Across counties, the median daily admission for cardiovascular diseases was 18.7 per 100 000 people (Q1 = 15.8 and Q3 = 21.4) and the median for respiratory diseases was 7.4 per 100 000 people (Q1 = 6.3 and Q3 = 8.8).
We considered 5 exposure measures of daily county-average PM10 − 2.5 level. For example, Figure 3 shows the marginal posterior distributions of PM10 − 2.5 exposure on July 17, 2000 in Harris County, TX. The 4 posterior distributions were obtained under different ME modeling and estimation approaches: (1) constant ME variances across monitors without using the health data (ME, [X|W,Z]); (2) constant ME variances across monitors using the health data (ME, [X|W,Z,Y]); (3) population-WME variances across monitors without using the health data (WME, [X|W,Z]); and (4) population-WME variances across monitors using the health data (WME, [X|W,Z,Y]). For (2) and (4), the relative risks associated with cardiovascular admissions were simultaneously estimated with PM10 − 2.5 exposure. Also, a vertical line is placed at the 10% TM estimate. The differences in exposure estimates reflect which monitor-level observations were used. On this particular day, there were 4 observations of PM10 − 2.5 concentration: 3 observations 18, 61, and 20 were from Houston and one observation 12 was from Deer Park. The TM PM10 − 2.5 measure excluded 12 and 61 in computing the average; the ME measure considered all values equally; and the WME measure down weighted the measurement from Deer Park which has a considerably smaller population than Houston.
Fig. 3.
Posterior distributions of the average exposure to outdoor PM10 − 2.5 concentration on July 17, 2000 in Harris County, TX. The vertical line is placed at the 10% TM estimate. The solid and dotted lines represent 4 different PM10 − 2.5 posterior distributions obtained from ME modeling.
We calculated the SD of county-level PM2.5 and PM10 − 2.5 levels across days and Table 1 gives the median, Q1, and Q3 across 59 counties for different exposure measures. First, daily variation of county-average PM10 − 2.5 levels derived from ME and WME were lower compared to TM and this decrease was less significant for PM2.5. PM daily variation decreases when ME is considered because the model assumes that the observed PM concentrations are more noisy than the true exposure. Specifically, a large decrease in time series SD reflects greater disagreement between same-day monitor-level measurements. Moreover, for both PM10 − 2.5 and PM2.5, the decrease in daily variation was more significant for the ME measures compared to the WME measures. If PM levels vary across cities of different population sizes, county-average exposure is determined mainly by measurements in cities with large populations. Since the true exposure represents a population-weighted average exposure, the WME approach can result in smaller ME because disagreement between PM measurements in cities with small populations and the true exposure is down-weighted.
Table 1.
Quantiles of county-specific SD of PM2.5 and PM10–2.5 time series using either TM, ME modeling with constant error variance (ME), or monitor-specific weighted error variance (WME). The median (25th quantile and 75th quantile) of the SD across 59 counties are given
| TM | ME | WME | |
| PM2.5 (μ g/ m3) | 7.6 (6.1, 9.0) | 7.1 (5.8, 8.7) | 7.3 (5.9, 8.6) |
| PM10–2.5 (μ g/ m3) | 7.9 (6.2, 12.2) | 6.0 (4.4, 10.3) | 7.0 (4.8, 11.7) |
Table 2 gives some pairwise correlations between PM10 − 2.5 and PM2.5 exposure measures obtained using either TM or WME across counties. Comparing rows 1 and 2, higher correlations are observed between different PM2.5 measures compared to PM10 − 2.5. This is expected since PM2.5 level is less heterogeneous spatially and the ME approach results in less calibration when the between-monitor agreement is strong. Comparing rows 3 and 4, we find that deriving PM10 − 2.5 and PM2.5 exposures via ME modeling increases the correlation between the 2 pollutants slightly. We also found very high correlation between the average exposure measures derived from the 2 ME models (ME vs. WME) for PM10 − 2.5 and PM2.5, having minimum correlation of 0.82 and 0.87, respectively, in the 59 counties (not shown in table).
Table 2.
Quantiles of correlations between different measures of county-level daily PM10–2.5 and PM2.5 exposures across 59 counties. Exposure measures for PM2.5 and PM10–2.5 are derived using either TM or WME modeling
| Correlation between | Minimum | 25% | 50% | 75% | Maximum | |
| (1) | PM10–2.5, TM , PM10–2.5, WME | 0.50 | 0.89 | 0.92 | 0.95 | 1.00 |
| (2) | PM2.5, TM, PM2.5, WME | 0.72 | 0.94 | 0.97 | 0.99 | 1.00 |
| (3) | PM2.5, TM, PM10–2.5, TM | – 0.20 | 0.05 | 0.12 | 0.23 | 0.59 |
| (4) | PM2.5, WME, PM10–2.5, WME | – 0.15 | 0.05 | 0.18 | 0.32 | 0.59 |
The ME variances, σ12,c and σ2c in (2.4) quantify the variability across monitors of the PM values. Figure 4 plots the posterior mean and 95% intervals for the ME SD versus log-transformed county land area (square kilometer). We found greater between-monitor variation for PM10 − 2.5 (black) measurements compared to PM2.5 (gray) measurements, even though the 2 pollutants had similar average concentration over the study period. The median ME SD across counties for PM10 − 2.5 is 5.6 (Q1 = 4.4 and Q3 = 8.8) and for PM2.5 is 2.3 (Q1 = 1.7 and Q3 = 3.2). In Figure 4, it also appears that larger counties were associated with greater between-monitor variation in PM measurements. We also found evidence of a weak positive association between PM2.5 and PM10 − 2.5 measurement errors at collocated monitors for some counties. The posterior means of ρc across 59 counties have a median of 0.10 (min = − 0.23, Q1 = − 0.1, Q3 = 0.4, max = 0.6) .
Fig. 4.
County-specific ME SD (σ12,c and σ22,c in (2.4) for PM10 − 2.5 (black) and PM2.5 (gray) plotted versus log county land area (cubic kilometer) for 59 counties. Each bullet denotes the posterior mean and the vertical line indicates the 95% posterior interval.
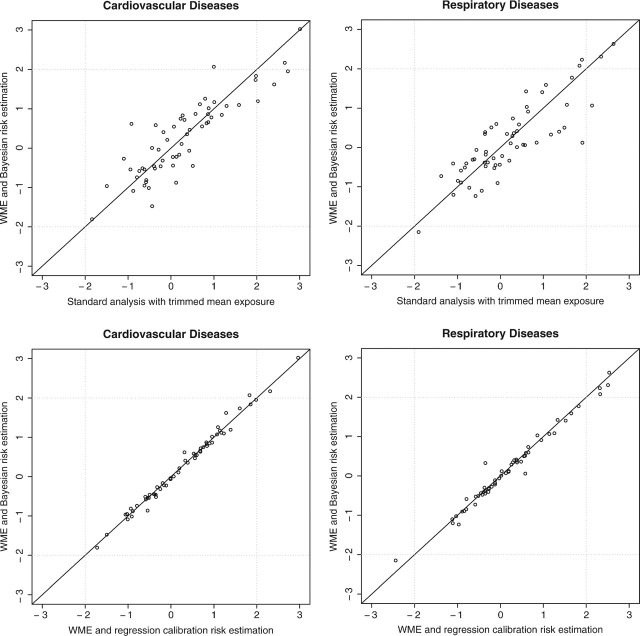
The 2 upper panels in Figure 5 plot the county-specific standardized coefficients,  to examine the strength and direction of the health effect of PM10 − 2.5 on cardiovascular and respiratory admissions estimated using different exposure measures. Comparing estimates derived from standard TM exposure and WME with the Bayesian risk estimation, we did not observe large changes in the health effects' direction. However, there is attenuation for large
to examine the strength and direction of the health effect of PM10 − 2.5 on cardiovascular and respiratory admissions estimated using different exposure measures. Comparing estimates derived from standard TM exposure and WME with the Bayesian risk estimation, we did not observe large changes in the health effects' direction. However, there is attenuation for large  possibly due to increased uncertainty in risk estimates when MEs are accounted for. From the 2 bottom panels in Figure 5, we show that in our application, standard error (SE),
possibly due to increased uncertainty in risk estimates when MEs are accounted for. From the 2 bottom panels in Figure 5, we show that in our application, standard error (SE),  for cardiovascular and respiratory admissions are very similar between those derived from regression calibration and those estimated through the Bayesian approach. For the Poisson health model, regression calibration will result in some bias in the relative risk estimates; however in the analysis of PM10 − 2.5, the uncertainty in exposure appears to dominate.
for cardiovascular and respiratory admissions are very similar between those derived from regression calibration and those estimated through the Bayesian approach. For the Poisson health model, regression calibration will result in some bias in the relative risk estimates; however in the analysis of PM10 − 2.5, the uncertainty in exposure appears to dominate.
Fig. 5.
Upper panels: scatter plot of county-specific standardized health effect estimates for PM10 − 2.5,  , comparing 2 approaches: (1) including ME modeling with monitor-specific weighted error variance (WME) versus (2) using TM as PM10 − 2.5 exposure. Lower panels: scatter plot of county-specific standardized health-effect estimates for PM10 − 2.5,
, comparing 2 approaches: (1) including ME modeling with monitor-specific weighted error variance (WME) versus (2) using TM as PM10 − 2.5 exposure. Lower panels: scatter plot of county-specific standardized health-effect estimates for PM10 − 2.5,  , using exposure derived from WME comparing Bayesian risk estimation versus regression calibration.
, using exposure derived from WME comparing Bayesian risk estimation versus regression calibration.
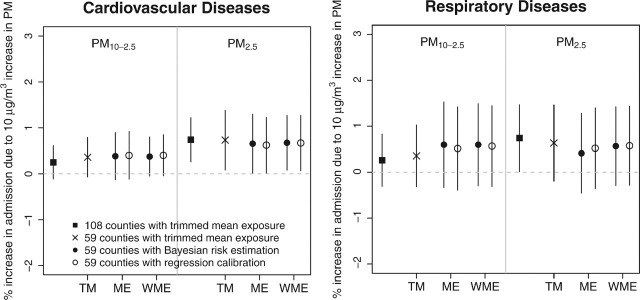
Figure 6 gives the pooled estimates of percent increase in cardiovascular and respiratory disease admissions per 10 μg/m3 increase in same-day particulate matter concentration. Exposure measures for PM2.5 and PM10 − 2.5 were derived using either TM, ME, or WME, and we considered both regression calibration and Bayesian risk estimations. The original Peng and others (2008) estimates based on 108 counties using TM exposure are also shown. We found consistent positive effects for PM10 − 2.5 and PM2.5 with different exposure measures and estimation procedures. For cardiovascular admissions, effects of PM2.5 remain statistically significant under different scenarios. The posterior intervals are wider under ME modeling compared to using the standard TM exposure. Also, when ME modeling are used, the confidence intervals are wider for Bayesian risk estimations compared to regression calibration and the bias associated with regression calibration appears negligible.
Fig. 6.
Percent increase in emergency hospital admissions rates for cardiovascular and respiratory diseases per 10 μg/m3 increase in same-day particulate matter concentration. Exposure measures for PM2.5 and PM10 − 2.5 are derived using either TM, ME modeling with constant error variance across monitors (ME) or monitor-specific weighted error variance (WME).
4. SIMULATION STUDY
This section describes a simulation study that examines the impact of PM10 − 2.5 exposure ME and the performance of our proposed method using data from Clark County, NV. Clark County contains 8 PM10 − 2.5 monitoring locations from 5 cities with population ranging from about 200 to half a million. On each day t, we do not observe the complete vector of monitor-level measurements Xt = (xt1,xt2,…,xt8)′. Across the 1337 days with at least one PM10 − 2.5 measurement, the average number of measurements per day was 4.3.
We generated 100 replicate data sets of the complete monitor-level PM10 − 2.5 values as follows. We assumed 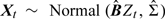 , where Zt is the p×1 vector of covariates from (2.5). Parameter
, where Zt is the p×1 vector of covariates from (2.5). Parameter 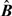 is an 8×p matrix of monitor-specific regression coefficients and parameter
is an 8×p matrix of monitor-specific regression coefficients and parameter 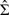 is the 8×8 residual covariance matrix. These parameters were estimated from the observed data and the between-monitor correlation ranges from 0.3 to 0.8. Given the observed PM10 − 2.5 concentrations, missing concentrations were imputed using the corresponding conditional mean and covariance. We used the complete PM data set to generate hospital admissions. However, in constructing the TM exposure measures and in carrying out ME modeling, we followed the observed missing data structure and ignored the imputed PM10 − 2.5 concentrations.
is the 8×8 residual covariance matrix. These parameters were estimated from the observed data and the between-monitor correlation ranges from 0.3 to 0.8. Given the observed PM10 − 2.5 concentrations, missing concentrations were imputed using the corresponding conditional mean and covariance. We used the complete PM data set to generate hospital admissions. However, in constructing the TM exposure measures and in carrying out ME modeling, we followed the observed missing data structure and ignored the imputed PM10 − 2.5 concentrations.
We set the total number of at-risk individuals to be 43 410. We considered 2 exposure scenarios by allocating the population to each monitor either (1) equally or (2) proportional to the city population that the monitor is in. Finally, for each simulated PM10 − 2.5 data set, we generated daily total admission with a baseline risk of 1.8×10 − 4 and a relative risk of 0.5% per 10 unit increase PM10 − 2.5 concentration following the Poisson model in (2.7) without additional confounders. Relative risk for PM10 − 2.5 was then estimated using the TM, ME, or WME exposure measures. Here, we only considered the estimation approach without using the health data ([X|W,Z]) due to computational limitation.
Table 3 gives the median bias and average 95% confidence interval length for the relative risk estimates. With the TM exposure measures, we found that attenuation occurs in both scenarios and our ME approaches (ME and WME) reduce this bias. ME modeling increases the confidence interval width when the same number of individuals are exposed to different concentrations (scenario 1). In this case, our method effectively propagates the uncertainty in population exposure when between-monitor disagreement is present. However, when the number of at-risk individuals varies across PM10 − 2.5 concentrations (scenario 2), the WME exposures does not result in wider confidence intervals. This is likely due to recovering some exposure variability that is oversmoothed by simply averaging monitor-level concentrations.
Table 3.
Simulation study results: median bias, average 95% confidence interval (C.I.) width, and root mean squared error for PM10–2.5 relative risk in Clark County, Nevada. At-risk population is allocated to each monitor either equally (Scenario 1) or proportional to the city population that the monitor is in (Scenario 2). Exposure measures considered include: TM or ME modeling without (ME) or with (WME) heteroskedastic variances
| Scenario 1 |
Scenario 2 |
|||||
| True | TM | ME | True | TM | WME | |
| Bias (× 104) | – 0.02 | – 0.39 | – 0.18 | – 0.01 | – 0.33 | – 0.16 |
| C.I. width (× 104) | 1.62 | 3.11 | 3.93 | 1.94 | 3.04 | 3.03 |
5. DISCUSSION
EPA does not regulate PM10 − 2.5 directly but continues to use PM10 as a surrogate to protect public health. In the most recent 2006 NAAQS revision for particulate matter, a 24-h PM10 − 2.5 standard was proposed but ultimately not accepted due to insufficient evidence linking short-term PM10 − 2.5 exposure and adverse health outcomes (Environmental Protection Agency, 2006). Time series analysis plays an important role in providing epidemiological evidence for the acute health effects of PM and in establishing regulatory standards (Greenbaum and others, 2001). Its popularity is due to the ability to utilize public databases to estimate the relatively small acute effects with large study populations. However, recent interest in quantifying the health effects of PM10 − 2.5 raises statistical questions regarding the time series design when the pollutant concentration varies spatially.
In this paper, we address the challenge of exposure ME due to spatial misalignment through ME modeling. The goal is to obtain risk estimates that reflect the uncertainty in PM10 − 2.5 exposure in a time series study. This differs from the past work that has focused predominantly on errors due to either (1) the discrepancy between ambient levels measured outdoors versus total personal exposure (Dominici and others, 2000) or (2) the ecological bias that results from using aggregated outcome and exposure to infer individual-level risk (Sheppard, 2005, Sheppard and others, 2005).
Computing average community-level exposure with monitor-specific weights is a common practice in time series analysis. The TM approach represents a simple way to remove extreme values observed on a particularly day; however, this may oversmooth a spatially varying exposure when the number of measurements per day is small. A similar approach taken by Zanobetti and Schwartz (2009) first removes monitors that are not well correlated with others in the same region to avoid measurements that are due to local pollution sources not reflective of the overall population exposure. However, this approach does not address the scenario when all monitors are poorly correlated with each other. In contrast, our ME method is a parametric approach that provides average exposure estimates and accounts for the same-day between-monitor variability using all available data. Moreover, by allowing error variances to be inversely proportional to the population living around each monitor, we automatically specify monitor-specific weights in computing average exposure.
While this paper is motivated by the analysis of PM10 − 2.5 and health, we note that the analysis of PM2.5 chemical constituents shares similar challenges in exposure ME. For example, the metal constituents in PM2.5 can exhibit high spatial heterogeneity and the minor components are often measured with high instrumental ME. One limitation of the proposed approach is that we need to restrict our analysis to counties with at least 2 pairs of collocated monitors. Future work will borrow information across counties by building regression models for the ME variances to predict the extent of exposure ME for counties that only have a single PM10 − 2.5 measurement each day. Our model also assumes the MEs to be independent between days and does not model the temporal correlation between pollutant concentrations. This is because PM10 − 2.5 measurements are typically only available every sixth day. But at some locations where daily measurements of PM10 − 2.5 are available, additional modeling of these temporal trends should be explored.
SUPPLEMENTARY MATERIAL
Supplementary material is available at http://biostatistics.oxfordjournals.org.
FUNDING
United States Environmental Protection Agency (R83622, RD-83241701); National Institute for Environmental Health Sciences (ES012054-03); National Institute for Environmental Health Sciences Center in Urban Environmental Health (P30 ES 03819). It has not been subjected to EPA's required peer and policy review and therefore does not reflect the views of the Agency and no official endorsement should be inferred.
Acknowledgments
We are grateful to the editor, associate editor, and one anonymous referee for their useful suggestions for improvements. Conflict of Interest: None declared.
References
- Bachmann J. Will the circle by unbroken: a history of the US National Ambient Air Quality Standards. Journal of the Air & Waste Management Association. 2007;57:652–697. doi: 10.3155/1047-3289.57.6.652. [DOI] [PubMed] [Google Scholar]
- Brunekreef B, Forsberg B. Epidemiological evidence of effects of coarse airborne particles on health. European Respiratory Journal. 2005;26:309–318. doi: 10.1183/09031936.05.00001805. [DOI] [PubMed] [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement Error in Nonlinear Models: a Modern Perspective. New York: Chapman & Hall; 2006. [Google Scholar]
- Castillejos M, Borja-Aburto VH, Dockery DW, Gold DR, Loomis D. Airborne coarse particles and mortality. Inhalation Toxicology. 2000;12:61–72. [Google Scholar]
- Dockery DW, Pope CA, Xu X, Spengler JD, Ware JH, Fay ME, Ferris BG, Speizer FE. An association between air pollution and mortality in six U.S. cities. The New England Journal of Medicine. 1993;329:1753–1759. doi: 10.1056/NEJM199312093292401. [DOI] [PubMed] [Google Scholar]
- Dominici F, Peng R, Bell M, Pham L, McDermott A, Zeger S. Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. Journal of the American Medical Association. 2006;295:1127–1134. doi: 10.1001/jama.295.10.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici F, Zeger SL, Samet JM. A measurement error model for time-series studies of air pollution and mortality. Biostatistics. 2000;1:157–175. doi: 10.1093/biostatistics/1.2.157. [DOI] [PubMed] [Google Scholar]
- Environmental Protection Agency. National ambient air quality standards for particulate matter: proposed rule. Federal Register. 2006;71(2620):2708. [Google Scholar]
- Everson PJ, Morris CN. Inference for multivariate normal hierarchical models. Journal of the Royal Statistical Society, Series B. 2000;62:399–412. [Google Scholar]
- Fuller WA. Measurement Error Models. New York: John Wiley & Sons; 1987. [Google Scholar]
- Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Analysis. 2006;1:515–534. [Google Scholar]
- Gotway CA, Yong LJ. Combining incompatible spatial data. Journal of the American Statistical Association. 2002;97:632–648. [Google Scholar]
- Greenbaum DS, Bachmann JD, Krewski D, Samet JM, White R, Wyzga RE. Particulate air pollution standards and morbidity and mortality: case study. American Journal of Epidemiology. 2001;154:S78–S90. doi: 10.1093/aje/154.12.s78. [DOI] [PubMed] [Google Scholar]
- Gryparis A, Paciorek CJ, Zeka A, Schwartz J, Coull BA. Measurement error caused by spatial misalignment in environmental epidemiology. Biostatistics. 2009;10:258–274. doi: 10.1093/biostatistics/kxn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BL, Kosorok MR, Fine JP. The profile sampler. Journal of the American Statistical Association. 2005;100:960–969. [Google Scholar]
- Mar TF, Larson TV, Stier RA, Claiborn C, Koenig JQ. An analysis of the association between respiratory symptoms in subjects with asthma and daily air pollution in Spokane, Washington. Inhalation Toxicology. 2004;16:809–815. doi: 10.1080/08958370490506646. [DOI] [PubMed] [Google Scholar]
- Peng RD, Bell ML. Spatial misalignment in time series studies of air pollution and health data. Biostatistics. 2010;11:720–740. doi: 10.1093/biostatistics/kxq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng RD, Chang HH, Bell ML, McDermott A, Zeger SL, Samet JM, Dominici F. Coarse particulate matter air pollution and hospital admissions for cardiovascular and respiratory diseases among Medicare patients. Journal of the American Medical Association. 2008;299:2172–2179. doi: 10.1001/jama.299.18.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M. Vienna: Austria; 2003. JAGS: A Program for Analysis of Bayesian Graphical Models Using Gibbs Sampling, Proceedings of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003) [Google Scholar]
- Pope CA, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. Journal of the American Medical Association. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, Dockery DW. Health effects of fine particulate air pollution: lines that connect. Journal of the Air & Waste Management Association. 2006;56:709–742. doi: 10.1080/10473289.2006.10464485. [DOI] [PubMed] [Google Scholar]
- Samet JM, Dominici F, Zeger SL, Schwartz J, Dockery DW. The National Morbidity, Mortality, and Air Pollution Study, Part I: Methods and Methodological Issues. Cambridge, MA: The Health Effects Institute; 2000. [PubMed] [Google Scholar]
- Sheppard L. Acute air pollution effects: consequences of exposure distribution and measurements. Journal of Toxicology and Environmental Health Part A. 2005;68:1127–1135. doi: 10.1080/15287390590935987. [DOI] [PubMed] [Google Scholar]
- Sheppard L, Slaughter JC, Schildcrout J, Liu L-JS, Lumley T. Exposure and measurement contributions to estimates of acute air pollution effects. Journal of Exposure Analysis and Environmental Epidemiology. 2005;15:366–376. doi: 10.1038/sj.jea.7500413. [DOI] [PubMed] [Google Scholar]
- Wilson WE, Suh HH. Fine particles and coarse particles: concentration relationships relevant to epidemiologic studies. Journal of the Air & Waste Management Association. 1997;47:1238–1249. doi: 10.1080/10473289.1997.10464074. [DOI] [PubMed] [Google Scholar]
- Zanobetti A, Schwartz J. The effect of fine and coarse particulate air pollution on mortality: a national analysis. Environmental Health Perspective. 2009;117:898–903. doi: 10.1289/ehp.0800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeger SL, Thomas D, Dominici F, Samet JM, Schwartz J, Dockery D, Cohen A. Exposure measurement error in time-series studies of air pollution: concepts and consequences. Environmental Health Perspective. 2000;108:419–426. doi: 10.1289/ehp.00108419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Carlin BP, Gelfand AE. Hierarchical regression with misaligned spatial data: relating ambient ozone and pediatric asthma ER visits in Atlanta. Environmetrics. 2003;14:537–557. [Google Scholar]
- Zidek JV, Wong H, Le ND, Burnett R. Causality, measurement error and multicollinearity in epidemiology. Environmentrics. 1996;7:441–451. [Google Scholar]