Abstract
Objective
To use the semantic and structural properties in the Unified Medical Language System (UMLS) Metathesaurus to characterize and discover potential relationships.
Design
The UMLS integrates knowledge from several biomedical terminologies. This knowledge can be used to discover implicit semantic relationships between concepts. In this paper, the authors propose a problem-independent approach for discovering potential terminological relationships that employs semantic abstraction of indirect relationship paths to perform classification and analysis of network theoretical measures such as topological overlap, preferential attachment, graph partitioning, and number of indirect paths. Using different versions of the UMLS, the authors evaluate the proposed approach's ability to predict newly added relationships.
Measurements
Classification accuracy, precision-recall.
Results
Strong discriminative characteristics were observed with a semantic abstraction based classifier (classification accuracy of 91%), the average number of indirect paths, preferential attachment, and graph partitioning to identify potential relationships. The proposed relationship prediction algorithm resulted in 56% recall in top 10 results for new relationships added to subsequent versions of the UMLS between 2005 and 2007.
Conclusions
The UMLS has sufficient knowledge to enable discovery of potential terminological relationships.
Introduction
The Unified Medical Language System (UMLS) 1 is a knowledge-rich resource composed of over a hundred biomedical terminologies. One of the important sources of knowledge is the set of semantic relationships asserted between the concepts derived from different source terminologies. Such integration of multiple sources of terminological knowledge can potentially enable discovery of implicit meaningful relationships between the concepts (unrelated at a given time). In this research we explore whether the existing knowledge in the UMLS in the form of semantics (types or categories) and structure (network topology) can be used to discover potential relationships.
The UMLS has been used in a variety of domains requiring discovery of novel terminological relationships. Zeng et al 2 used the UMLS metathesaurus to build a knowledge base for information retrieval. Butte et al 3 used the relationships in the UMLS metathesaurus to discover links between genome, phenome, and environmental concepts. Cantor et al 4 used the relationships in the UMLS metathesaurus to find gene-to-disease connections. Hristovski et al 5 used the UMLS relationships with MEDLINE to discover novel biomedical knowledge.
One of the key commonalties across all these studies is the traversal of the terminology graph from the metathesaurus to find implicit knowledge. Consider for example, Cell differentiation,† a term related to or possibly synonymous with Adipogenesis which in turn is related to or synonymous with Fat Cell which has a co-occurence relationship with diabetes mellitus. Based on this observation, we sought to investigate whether there are specific patterns or features among such indirect relationship paths that can be used to predict meaningful terminological relationships.
Our goal is to develop a generic relationship discovery framework that uses the existing knowledge in the UMLS metathesaurus to predict new potential terminological relationships. There are significant challenges in developing a generic terminological relationship discovery approach. First, a set of discriminative features that can distinguish between noisy and meaningful potential relationship paths has to be found. To develop a problem-independent approach, the discriminating features must be intrinsic to the UMLS and not based on specific instantiations of UMLS concepts (or corresponding terminologies) in external resources such as patient records or biomedical literature. This approach differs significantly from an existing body of work 5,12 that uses the UMLS or terminology resources in conjunction with external data sources. Second, an exponential growth in the number of relationship paths occurs when traversing the indirect relationships in the UMLS metathesaurus. The UMLS metathesaurus is a densely interconnected graph, 6 which implies that transitive traversal with depths of 2 or 3 steps can generate thousands of potential candidates. For example, hemoglobin in the UMLS has 868 direct relationships to other concepts, which in turn have 450,692 relationships to a different set of concepts. Third, evaluation of the feasibility of the proposed approach for generic relationship discovery requires the development of a problem-independent gold-standard.
We approach the problem by modeling semantic features based on our prior work on using the UMLS relationship types and attributes to classify cross-terminology paths for terminology translation and information retrieval. 16 In the study reported in this paper, we investigate whether these semantic features can enable relationship discovery by identifying new potential terminological relationship paths. We then choose various network theoretical measures that may, if present, provide structure clues for potential terminological relationships. We hypothesize that there is a set of semantic and structural features in the UMLS between two given concepts that can indicate the presence of a potential relationship, where the semantic and structure features are defined as:
1 Semantic features: The abstraction of an indirect relationship path using UMLS relationship types/attributes and Semantic Types.
2 Structural features: A set of network theoretical properties such as topological overlap, preferential attachment, graph partitioning and number of relationship paths across a pair of concepts.
To evaluate the proposed approach, we first perform a relationship “diff”, that is, we identify newly added relationships between versions of the UMLS metathesaurus released in different years. Then, we analyze the proposed semantic and structural features of the “diff” relationships in comparison to the (respective) older version of the UMLS to evaluate the prediction accuracy.
Background
Network Analysis
The field of social network analysis 7 provides methods for studying the social relationships among a set of actors (people, groups, or organizations). These methods have been successfully used to study networks found in different domains 8 such as biological networks, the Internet, and biomedical terminologies. 9 In our previous work, 6 we showed that the UMLS metathesaurus is a scale-free network. Scale-free networks are characterized by two properties:
• Power-law distribution: the probability p (k) that a given node in the network is connected to k other nodes is proportional to k−c, where c is generally between 2 and 3 for scale-free networks. The power law implies that a few “hub” nodes are connected to a large number of nodes and that most other nodes in the network have only a small number of connections.
• Preferential growth: A new node when added to the network is more likely to be connected to some hub node. Hence, over time the hub nodes accumulate a higher number of connections.
Another important notion is that of “weak ties”, developed in social sciences, 10 and often used to characterize the strength of social relationships. The topological overlap measures this strength for two given nodes based on the commonality of their neighbors in a network. A high topological overlap indicates that the nodes are connected to the same group of other nodes. For example, for two given nodes vi and vj, the topological overlap (tij) is measured by:
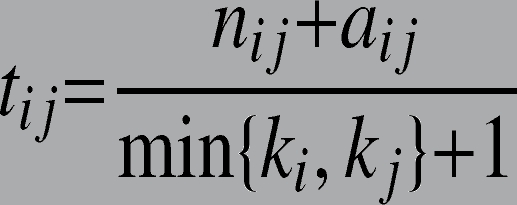 |
where, ki and kj are the degrees (the number of neighbors) of nodes vi and vj, respectively, and nij is the number of common neighbors of vi and vj. aij (adjacency) is 1 if both nodes are connected or 0 otherwise. This measure has been used widely in social networks to characterize gene and protein interaction networks. 11
The UMLS Metathesaurus
The metathesaurus component of the UMLS 1 merges terms from different source terminologies with the same meaning into concepts with unique identifiers (CUI). In the 2007AC version, there were 6,134,676 term tokens (from source terminologies) and 1,516,299 concepts. All the concepts in the UMLS are associated with one or more of the 135 Semantic Types in the UMLS Semantic Network. The relationships between the terms asserted in source terminologies are represented in the UMLS metathesaurus both at their original level of granularity and, if applicable, as relationships between UMLS concepts. The UMLS metathesaurus categorizes relationships in 11 broad categories such as broader, narrower, parent, other, and so on. Some of the relationships also have an attribute associated with them such as “caused-by”, “associated_with” and so on, which defines the nature of the relationship more specifically.
Related Work
Various methods have been developed for using the existing knowledge in the UMLS to discover novel relationships between biomedical concepts. Hristovski, et al 5 developed an approach based on association rule mining to discover novel relationships using the co-occurence data and the Semantic Types in the UMLS. Alhers 12 showed a template-based approach that uses the hierarchical relationships in the UMLS along with biomedical literature to discover novel drug mechanisms. Zhang, et al 13 used frequent patterns of indirect relationship paths to identify correspondence between relationships across ontologies. Bodenreider 14 investigated direct and indirect hierarchical relationships in the UMLS to explore redundancies. Generic approaches for relationship mining have also been developed outside the UMLS. 15
In our previous work, we proposed a semantic abstraction approach 16 to learn semantic patterns in the UMLS for mining relevant cross-terminology links. We also presented approaches that use the global (scale-free) 6 and local (clustering coefficient) 17 network-based properties of the UMLS metathesaurus to facilitate terminology translation.
Methods
We first identified a set of semantic and structural features intrinsic to the UMLS. The rationale for choosing a feature was based on a hypothesis that was tested against relationship “diff” datasets across different UMLS versions. We used the Rich Release format (RRF) of the UMLS metathesaurus that allowed us to capture the source specific semantics and the frequency of relationship paths. In this section, we describe the methodology for creating the study datasets, identifying the relevant UMLS features, developing the relationship prediction algorithm, and validating the approach.
Basic Study Datasets
Relationship Diff (RDiff): Potentially Related
A dataset, RDiff, containing the “diff” of relationships between different versions of the UMLS was prepared by finding all pairs of concepts with a direct relationship asserted in the relationships table (MRREL) of a newer UMLS version such that the respective concepts are also present in the MRREL of older UMLS version but did not have any direct relationship in the older version. Datasets were prepared by calculating RDiff for:
UMLS versions, 2002 and 2004AA
UMLS versions 2004 and 2006AA, and
UMLS versions 2005AA and 2007AB.
We selected all the concept pairs from each RDiff dataset to generate 2-Step indirect paths based on the older versions of the UMLS. Note that although these concept pairs did not have any direct relationships in the older versions, they could potentially have an indirect 2-step relationship path between them in the older version.
Strict-2-Step (S2S): Unrelated
A random sample of 10,000 concept pairs was obtained from each of the older versions of the UMLS such that these concepts pairs did not have a direct relationship asserted in the MRREL but did have a 2-Step indirect relationship path between them. We label these as Strict-2-Step (S2S) concept pairs and assume that they are “unrelated” since they lack a direct relationship between them. We identified the 2-step indirect paths for the 10,000 concept pairs identified from each earlier version of the UMLS under study (2002, 2004, 2005AA).
Direct-1-Step (D1S): Strongly Related
A set of 10,000 concept pairs were randomly sampled from the older versions of the UMLS such that these concept pairs had a direct relationship asserted in the MRREL and were labeled as “strongly related” concept pairs.
We approached the problem of characterizing potential relationships in the UMLS by analyzing a set of semantic and structural properties as described below:
Semantic Abstraction
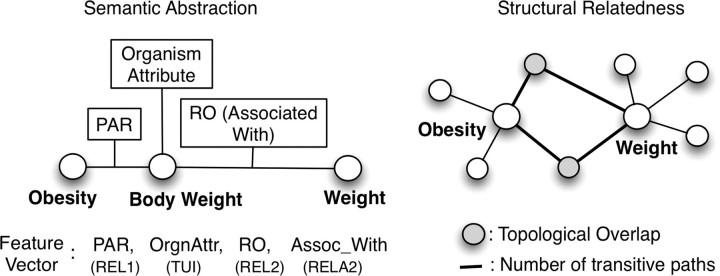
Based on our previous approach, 16 we generated all possible 2-step indirect relationship paths for a given source concept (or between given source and target concepts). Then, we identified the intermediate relationship types (REL), relationship attributes (RELA), and Semantic Types (TUI) of intermediate concepts to create a feature vector. Consider for example, the relationship path shown in ▶ between Obesity and Weight where the feature vector consists only of intermediate concept Semantic Type (Organism Attribute for concept of Body Weight) and relationship types PAR (i.e., has parent), RO (i.e., other relationship) with a relationship attribute (RELA) of “associated with”. The goal was to use this feature vector with a machine learning classifier (such as Naive Bayes or Decision Tree) to determine whether indirect paths with potentially meaningful relationships are distinguishable from other noisy or irrelevant paths. In this study, we used the Decision Tree classifier (C4.5) as it additionally provides the decision tree of features from the training data, which can be used to understand the joint dependencies.
Figure 1.
Characterizing the potential relationship between Obesity and Weight using the semantic abstraction and structured relatedness in the UMLS.
Topological Overlap Analysis
Several existing knowledge discovery methods are essentially based on Swanson's approach 18 of analyzing 2-level deep indirect relationships. The topological overlap quantifies the 2-step neighborhood for a given pair of concepts by measuring the overlap among their neighbors. For example, concepts Obesity and Weight have a topological overlap of 0.05 (with 3 overlapping neighbors). We hypothesized that strongly related concepts will show a high topological overlap, noisy or unrelated concept pairs will have very low topological overlap whereas concept pairs with a potential relationship will show an intermediate or medium topological overlap. Secondly, we also measured the number of indirect relationship paths, again with similar hypothesis that potentially related concepts will have on an average some intermediate number of relationship paths when compared to strongly related and unrelated concept pairs (Obesity and Weight have 3 indirect relationship paths across them).
Scale-free Analysis
To identify the hub concepts in the UMLS metathesaurus, we sorted the concepts in descending order based on the degree. 6 For a given concept, the degree is defined as the number of unique concepts with which the given concept has relationships in the MRREL (▶). A cut-off parameter, k, was used to label the top concepts with degree greater than k as hub concepts. Formally,
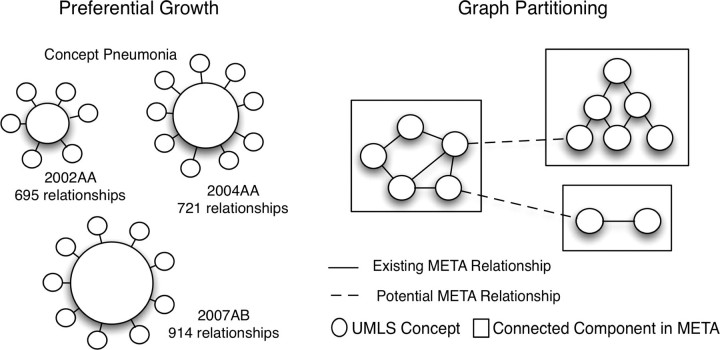
Figure 2.
Diagram showing preferential growth of relationships for concept Pneumonia over different versions of the UMLS and the notion of graph partitioning to analyze the potential relationship across different components in the CleanedMETA.
where degree(C1) = |{C2 UMLS Concepts: C1 and C2 have a relationship in MRREL}| and k is an arbitrary cut off parameter.
We hypothesized that a potential relationship is more likely to contain a source concept or a target concept that is a hub concept node. This is based on the observation that the UMLS metathesaurus is a scale-free network (hence follows the preferential attachment property), which implies that new nodes are more likely to be connected to a hub concept.
Graph Partitioning Analysis
We removed the noninformative and noisy relationships in the UMLS metathesaurus such as sibling (SB), allowed qualifier (AQ), and qualified by (QB) to create different version of metathesaurus labeled as CleanMETA. The CleanMETA contains several “islands” of connected concepts, for example, a component (set of connected concepts) with 36 concepts, nerve-fibers, Neurons, axotomy, Entire axon, Nerve Tissue, and so on. Due to semantic locality, 19 these components indicate a meaningful domain based partitioning of concepts such as neuro-anatomy component, cellular structures component, and ocular disorder component. A relationship across the components or subdomains can be viewed as an important source of new knowledge. We analyzed the potential relationships based on whether they tend to be created within a given component or across the components (▶).
Combined Predictive Model
Finally, we combined the aforementioned semantic and structural features into a predictive model to enable identification of a potential relationship between a given pair of concepts. Towards this goal, we first trained models for all the features using the training datasets (RDiff, S2S and D1S) as follows:
• Semantic Abstraction Model (SAM): the training dataset RDiff+ S2S was used to extract semantic abstraction features and learn a Decision Tree classifier model.
• Topological Overlap Model (TOM): the average range of topological overlap for concepts in the respective training datasets was calculated.
• Preferential Growth Model (PGM): the average likelihood of source or target concept as hub across concepts in the training datasets was calculated.
• Graph Partitioning Model (GPM): the average likelihood of concepts in training datasets having a relationship within the same or across different components was calculated.
Given these models, the proposed relationship prediction algorithm (see below) takes as input a UMLS concept and returns a ranked list of concepts that can have a potential relationship to the input concept. The algorithm starts by identifying all possible 2-step indirect paths to a set of target concepts (limited to specific target terminology). The target concepts are analyzed based on the structural models (developed using the training datasets) to evaluate the likelihood of having a potential relationship, for example, for input concept Obesity and target concept Muscle, the topological overlap is 0.05 and assuming our average TOM is 0.15 (± 0.03), then the concept Muscle is eliminated since it is lower (less than 0.15–0.03 = 0.12) than the topological overlap in the training dataset. Next the Semantic Abstraction features are calculated over the filtered target concepts and SAM is applied to classify the target concepts into class RDiff/Potentially related or S2S/Unrelated. The classified concepts are ranked based on the confidence score obtained from the Decision Tree classifier.
Relationship prediction algorithm:
Input: C = UMLS Concept, semantic and structural models (SAM, TOM, PGM, GPM)
Output: Ranked list of target concepts with potential relationship to C
1 (indirect path calculation): Identify the 2-step indirect paths from C to all other target concepts in the UMLS metathesaurus. Exclude noisy hubs in the traversal.6
2 (structural model based filtering): Based on the TOM, PGM, and GPM eliminate the target concepts with respective model parameters.
3 (semantic model based classification): For the remaining target concepts, calculate Semantic Abstraction features and use SAM with Decision Tree classifier to classify the concepts.
4 (ranking): Rank the list of target concepts based on confidence score from Decision Tree classifier.
To evaluate the proposed UMLS properties with training datasets, we performed the following experimental steps:
1 Using the semantic abstraction approach, a set of feature vectors was obtained from the 2-step indirect relationship paths in the RDiff dataset and S2S dataset. The corresponding classification labels were added to these feature vectors (“Potential_Rel” and “Unrelated”). A binary classifier was created using the C4.5 Decision Tree algorithm in the Weka implementation.20 A 10-fold cross-validation experiment was performed to calculate the classification accuracy and area under the curve (AUC).
2 The structural features were calculated using the sample of 10,000 concept pairs in S2S and D1S datasets and all RDiff relationships over corresponding older versions of the entire UMLS metathesaurus.
3 The algorithm was evaluated over the 2005–07 datasets, which were divided into 80% training and 20% testing sets. The training set was used to train SAM, TOM, PGM and GPM. For scale-free analysis, the hub cut off parameter k was set to subset the top 10% of concepts sorted by degree of relatedness in each respective UMLS version. The algorithm was evaluated over the test source concepts to identify potentially related concepts. Rank precision was calculated for top 5 and top 10 positions.
Results
In the RDiff data generation, we found 2,057,724 concept pairs in RDiff for the 2002 and 2004AA UMLS versions, 2,076,017 concept pairs in RDiff for the 2004 and 2006AA UMLS versions, and 208,929 concept pairs in RDiff for the 2005AA and 2007AB UMLS versions. Over the years under study, the source terminology contributing the highest number of new relationships was SNOMED CT (20–40%), which was added to the metathesaurus in 2004, followed by MeSH and LOINC.
Semantic abstraction strongly discriminates between potential relationships and unrelated indirect paths as indicated by the cross-validation results shown in ▶. High classification accuracy was observed across the different datasets. The analysis of the C4.5 Decision tree allowed identification of the underlying semantic patterns responsible for the result, for example, the path (across RDiff concepts) Abdomen, RO, Abdominal Pain, RQ (related and possibly synonymous), mapped-to, Gastric Pain was correctly classified as a potential relationship.
Table 1.
Table 1 Cross-validation Results for RDiff and S2S Connection Datasets Using a C4.5 Decision Tree Classifier and Sample Semantic Patterns
| Training Dataset | Classification Accuracy (Kappa) | Sample Semantic Patterns Generated Using C4. 5 Decision Tree Classifier |
|---|---|---|
| 2002–2004 | 90% (0.79) | REL1 = AQ, TUI = functional-concept (potential-rel) |
| REL1 = PAR, REL2 = PAR (unrelated) | ||
| 2004–2006 | 91% (0.79) | REL1 = RO, RELA2 = mapped-from (potential rel) |
| REL1 = PAR, REL2 = CHD (unrelated) | ||
| 2005–2007 | 93% (0.81) | REL1 = RN, RELA2 = inverse-isa (potential rel) |
| REL1 = PAR, RELA2 = inverse-isa(unrelated) |
RO = Relationship Other; CHD = Child; PAR = Parent; SIB = Sibling; RN = Narrower Relationship.
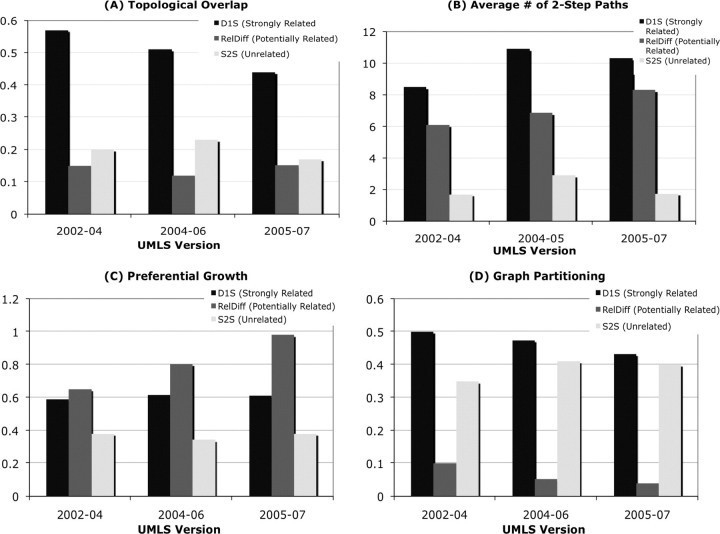
Topological overlap does not distinguish between the potential relationship and unrelated indirect paths as shown in ▶a. However, there were significant differences in the topological overlap of strong relationships and potential relationships, for example, acne vulgaris and eruption second (in RDiff) had overlap of 0.2 whereas D1S concept pairs Blood insulin and Blood calcitonin had overlap score of 0.89. The results indicate that newer relationships are more likely to be created outside the immediate neighborhood of concept pairs.
Figure 3.
A). The topological overlap does not distinguish between potentially related and unrelated concept pairs across the UMLS versions. B). Average number of 2-step indirect relationship for potentially related concepts is higher than unrelated concept pairs. C). The probability of a source or a target concept being a hub is significantly higher for potentially related concepts which corroborates with preferential growth theory for scale free networks. D). The probability of a relationship across concepts in the same component (obtained after graph partitioning) is significantly lower for potential relationships indicating that new relationships are created outside the immediate neighborhood of a given concept.
Average number of paths is a powerful discriminator of potential relationships and unrelated indirect paths as shown in ▶b. The average number of 2-step relationships for potential relationships was significantly higher than unrelated indirect paths. For example, S2S concept Occupational tattoo had 2 paths to Nutritional disorders whereas Abdomen had a potential relationship hernia of abdominal cavity (in RDiff) via 8 indirect paths. The potentially related concepts have a higher number of indirect paths despite of the previous finding that they do not exist in the same neighborhood.
Preferential growth is a strong discriminator of potential relationships, which are more likely to be created with a hub concept as shown in ▶c. This finding supports the preferential growth theory for the UMLS metathesaurus, which is a scale-free network. Graph partitioning shows a significant difference in the probability of potentially related concept pairs belonging to same components when compared against both D1S and S2S datasets (▶d). This result corroborates the topological overlap result that new relationships are more likely to be created outside the immediate neighborhood of the concept and across different subdomains.
The relationship prediction evaluation using the Relationship Prediction Algorithm resulted in 43% target potential concepts ranked in the top 5 positions and 56% in the top 10 positions for the all gold standard concepts reachable in 2-steps. For example, given the input concept, Obesity, the algorithm correctly predicts potential concepts in top 10 results such as Disease, Weight, Metabolic Syndrome and so on. Considering all the 2-step relationship paths for a given input concept, the algorithm's top 10 precision drops to 20%, given the large number of reachable target concepts. ▶ shows the evaluation based on top 10 results for the input concept Mitotic Apparatus considering all relationships paths. This implies that semantic and structural features learned using the training dataset could be used to rank and discover potential relationships in the UMLS.
Table 2.
Table 2 The Results of Potentially Related Concepts Predicted by Relationship Prediction Algorithm for Input Concept as Mitotic Apparatus Using UMLS 2005AA Version and Evaluated against the Newly Added Relationships in the UMLS 2007AB Version
| Sr No. | Input Source Concept | Ranked List of Target Concepts from 2005AA | Has a New Relationship in 2007AB |
|---|---|---|---|
| 1 | mitotic apparatus | mitotic spindle | NO |
| 2 | mitotic apparatus | cytoplasmic matrix | yes |
| 3 | mitotic apparatus | cellular structures | NO |
| 4 | mitotic apparatus | microtubules | NO |
| 5 | mitotic apparatus | cytoplasmic filaments, tubules, centrioles and associated structures | yes |
| 6 | mitotic apparatus | obsolete cellular components | NO |
| 7 | mitotic apparatus | Nonmembraneous cytoplasmic organelle | NO |
| 8 | mitotic apparatus | Pole of spindle | yes |
| 9 | mitotic apparatus | cytoplasmic filaments | yes |
| 10 | mitotic apparatus | cell physiology | NO |
Discussion
We present a problem-independent approach that uses the existing semantic and structural knowledge in the UMLS to discover potential terminological relationships. Nevertheless, the analysis presented in this paper is based on a “future” gold standard, i.e., we only try to predict whether a relationship will become explicit in the metathesaurus, which does not necessarily imply discovery of new knowledge. The proposed approach is capable of discovering non-trivial relationships as indicated by the distribution of the top 10 correctly predicted relationship types, where 24% of relationships were of type RO (relationship other).
Results Analysis
At a broader level, the proposed semantic and structural features represent intuitive hypotheses about the process of creation of new terminological relationships. There are only a limited set of combinations of indirect relationships and Semantic Types that can lead to a valid direct relationship, while most combinations produce invalid relationships. For, example, an indirect relationship from a source concept with a “has parent” (PAR) relationship to intermediate concept with a “has child” (CHD) relationship to a target concept is not an interesting potential relationship since it implies a sibling (SIB) relationship. Hence, once we learn the valid combinations from the training sample, the Semantic Abstraction Model acts as a powerful feature to filter out uninteresting indirect relationships. This is similar to the work presented in Burgun, et al 21 in which such relationship combinations were explicitly analyzed to study co-occurring concepts. In the structural features, the property of higher average number of 2-step paths for potential relationships indicates the semantic locality 19 principle in the UMLS. The low topological overlap for potential relationships corroborates the preferential growth property that real world networks progressively shrink in size towards creating a small-world network. The graph partitioning results signify that new relationships are more likely to be created within a given domain (adhering to the semantic locality principle), thereby contributing small cross-domain knowledge. We combine these properties into a predictive model to discover potential terminological relationships.
Semantic abstraction is a promising approach for identifying potential terminological relationships. In our previous work, 16 we used the semantic abstraction approach to perform information retrieval and terminology translation. When we replaced the 80% training set used in this study with a training dataset from our previous information retrieval study, we found that the ranking precision dropped down to 29% (top 5) and 40% (top 10). This indicates that there exist unique semantic patterns pertaining to terminological relationship discovery within the UMLS.
Our hypothesis that the potentially related concepts will have higher topological overlap than the unrelated concepts was not supported by the experimental results. One possible explanation is due to the definition of “unrelated” concepts, which are concepts related by strict-2-step relationships. The topological overlap measures the overlap of the neighbors for the given 2 nodes and by our definition the S2S relationships will always have one common neighbor. We tested a dataset of strict-3-step relationships (S3S) where the source and target concepts can only be reached by a 2-step path, which resulted in further decrease of topological overlap. One interesting observation is that topological overlap of S2S and RDiff was found to be nearly identical across different years, corroborating Swanson's observation that there is a higher probability of potential relationships over 2-step deep indirect paths.
Most existing research efforts using the UMLS for relationship discovery start from scratch, requiring significant expertise and resources. Our approach provides a generic solution to bootstrap a set of potential relationships for further exploration that can be coupled with problem-specific knowledge from external sources such as biomedical literature or clinical data. The long-term goal of our research is to provide an automated set of tools to allow users to easily perform relationship discovery and other knowledge intensive terminology tasks over the UMLS.
Applications
One of the potential applications of the proposed approach is terminology development. A knowledge engineer can supply an initial set of terms to the relationship prediction algorithm and get a list of potentially related terms. For example, given an input concept such as Dermatitis, the potentially related concepts are Skin, Infectious Skin Diseases, Lichenoid drug eruption, iododerma, and so on. Specific semantic relationships such as anatomical-location, isa, caused-by, etc can then be assigned manually. The target concepts in the algorithm can also be limited to a specific terminology such as SNOMED CT.
Limitations
One important limitation in applying the proposed relationship prediction algorithm is the requirement of an indirect path between the given pair of concepts. The limitation is due to the semantic features that use the abstraction of the path to perform the classification. Hence, our algorithm fails if there are no indirect connections between potentially related concepts. Furthermore, our findings based on average number of 2-step paths and graph partitioning results suggest that new relationships are more likely to be created outside the immediate neighbors of the concept, which implies that indirect relationship paths may not exist. Note that our evaluation for recall was only based on new relationships added over the years to the UMLS. An overall recall and completeness evaluation is a practically infeasible task, which would require a domain expert to analyze all other (∼1.5 million −1) UMLS concepts for existence of a potential relationship, or to wait an infinite number of years for all relationships to eventually be added explicitly. The evaluation and results presented in this paper do not reflect the actual usage or usefulness of the relationships identified for real world applications. We assume that the new relationships added in source vocabularies indicate a potential need or use in the respective domains. However, further research is needed to evaluate the cost-benefit of using proposed automated method towards suggesting useful new terminological relationships.
Conclusions
The semantic and structural properties in the UMLS were analyzed to discover potential terminological relationships. These potential relationships have distinct semantic patterns amenable to automated classification and have higher-than-average numbers of indirect paths. The potential relationships are more likely to be created outside the immediate neighborhood of a given concept. Given a pair of indirectly related concepts, the proposed approach provides a powerful mechanism to predict the existence of a direct relationship using the existing semantic and structural knowledge in the UMLS.
Acknowledgments
The authors thank the anonymous reviewers of AMIA 2008 Annual Symposium for their insightful comments and the National Library of Medicine for their grant support (R21LM009638) that funded this research. Dr. Cimino is supported by funds from the Intramural Research Program at the NIH Clinical Center and the National Library of Medicine.
Footnotes
Throughout this paper, controlled terms will be presented in italics.
References
- 1.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system Methods Inf Med 1993;32(4):281-291Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng Q, Cimino JJ. Automated knowledge extraction from the UMLS Proc AMIA Symp 1998:568-572. [PMC free article] [PubMed]
- 3.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network Nat Biotechnol 2006;24(1):55-62Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor MN, Sarkar IN, Bodenreider O, Lussier YA, Genestrace. phenomic knowledge discovery via structured terminology Pac Symp Biocomput 2005:103-114. [PMC free article] [PubMed]
- 5.Hristovski D, Stare J, Peterlin B, Dzeroski S. Supporting discovery in medicine by association rule mining in MEDLINE and UMLS Medinfo 2001;10(2):1344-1348. [PubMed] [Google Scholar]
- 6.Patel CO, Cimino JJ. Scale-Free A Network View of the UMLS to Learn Terminology. Translations. In Proc. Medinfo. 2007:689-693. [PubMed]
- 7.Wasserman S, Faust K. Social Networks Analysis: Methods and applicationsUK: Cambridge University Press; 1994.
- 8.Barabasi A, Albert R. Emergence of scaling in random networks Science 1999;286(5439):509-512. [DOI] [PubMed] [Google Scholar]
- 9.Bales ME, Lussier YA, Johnson SB. Topological analysis of large-scale biomedical terminology structures J Am Med Inform Assoc. 2007;14(6):788-797Nov–Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granovetter M. The strength of weak ties Am J Sociol 1973;78(6):1360-1380. [Google Scholar]
- 11.Li A, Horvath S. Network Neighborhood Analysis with the Multi-Node Topological Overlap Measure BioInformatics 2007;23(2):222-231. [DOI] [PubMed] [Google Scholar]
- 12.Ahlers C, Hristovski D, Kilicoglu H, Rindflesch T. Using the literature-based discovery paradigm to investigate drug mechanisms Proc AMIA Symp 2007:6-10. [PMC free article] [PubMed]
- 13.Zhang S, Bodenreider O. Comparing associative relationships among equivalent concepts across ontologies Stud Health Technol Inform 2004;107(1):459-466. [PMC free article] [PubMed] [Google Scholar]
- 14.Bodenreider O. Strength in numbers: Exploring redundancy in hierarchical relations across biomedical terminologies AMIA Annu Symp Proc 2003:101-105. [PMC free article] [PubMed]
- 15.Getoor L. Link mining: A new data mining challenge. SIGKDD explor. Newsletter 2003;5(1):84-89. [Google Scholar]
- 16.Patel CO, Cimino JJ. Links in the UMLSMining c-t Proc AMIA Annu Symp PROC 2006:624-628. [PMC free article] [PubMed]
- 17.Patel CO, Cimino JJ. Decompositional Terminology Translation Using Network Analysis Proc AMIA Annu Symp 2007:588-592. [PMC free article] [PubMed]
- 18.Swanson DR. Medical literature as a potential source of new knowledge Bull Med Libr Association 1990;78(1):29-37. [PMC free article] [PubMed] [Google Scholar]
- 19.Bodenreider O, Nelson SJ, Hole WT, Chang HF. Beyond synonymy: Exploiting the UMLS semantics in mapping vocabularies Proc AMIA Symp 1998:815-819. [PMC free article] [PubMed]
- 20.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and TechniquesSan Franciso: Morgan Kaufmann; 2005.
- 21.Burgun A, Bodenreider O. Methods for exploring the semantics of the relationships between co-occurring UMLS concepts Stud Health Technol Inform 2001;84(1):171-175. [PMC free article] [PubMed] [Google Scholar]