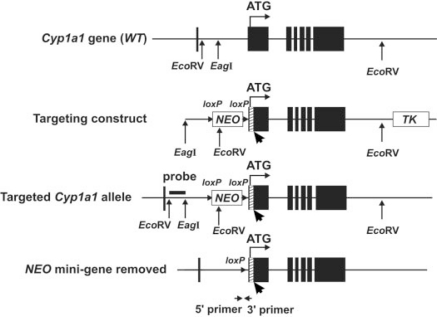
In the above article [ Dong H, Dalton TP, Miller ML, Chen Y, Uno S, Shi Z, Shertzer HG, Bansal S, Avadhani NG, and Nebert DW (2009) Mol Pharmacol75:555–567], Fig. 1A is incorrect as originally drawn; presented here is the corrected version. The EcoRV and EagI sites that were erroneously shown as upstream of exon 1 are actually within intron 1. The EcoRV site is 44 base pairs (bp) downstream of exon 1, the distance between the EcoRV and EagI sites in intron 1 is 883 bp, and the EagI site is 1453 bp from the beginning of exon 2, in the 5′ direction. The distance from the EagI site in intron 1 to the EcoRV site beyond exon 7 is 7580 bp. In this corrected version, the lengths of the probe (792 bp; third line) and the NEO minicassette (1249 bp; second and third lines) are also drawn more proportionately to their actual sizes. The 34-bp loxP sites (arrowheads) and the exon-2 insertions (bold arrows pointing to hatched rectangles representing the organelle-specific variations) in the last three lines, and the primers (arrows) in the last line, are exaggerated in size so that they can be more easily seen.
Fig. 1.
Generation of three Cyp1a1 knock-in mouse lines. A, scheme showing the WT Cyp1a1 gene (top line), targeting construct carrying the organelle-specific alterations (second line), targeted Cyp1a1 alleles after removal of the TK gene (third line), and the final targeted alleles (bottom line) after the NEO gene had been removed via Cre-mediated recombination to create the Cyp1a1(mc), Cyp1a1(mtt) and Cyp1a1(mtp) alleles. NEO (G418-resistant minicassette) and TK (thymidine kinase minicassette) represent genes used as selectable markers. ▪ denotes the seven exons; exon 2 (where ATG translation start-site is located) spans 851 bp. The ATG start-site for translation (denoted as vertical line and horizontal arrow) is located 15 nucleotides into exon 2. Exon 2 insertions (bold arrows pointing to hatched rectangles, not drawn completely to scale) represent the organelle-specific variations. EcoRV and EagI sites and the inserted loxP sites (orientation denoted by small arrowheads) are shown. (legend continues)
The authors regret this error and apologize for any confusion or inconvenience it may have caused.